Abstract
Ants of the «Formica picea—F. candida» complex are widespread across Eurasia. However, it is still a matter of debate if it constitutes one or two species. In this study, we collected a sample of specimens from different parts of Eurasia, sequenced the mitochondrial cox1 and cytb genes, as well as three nuclear loci—wg, Top1, and ITS2—and analyzed the available published data. We found this complex to contain a new, yet undescribed, taxon that has a large distribution in Siberia and East Asia. Thus, the «Formica picea—F. candida» complex consists of at least three taxa with distinct distributions.
1. Introduction
The taxonomic position of many widespread and abundant ant species remains uncertain [1,2]. Among them are the species of the «black bog ant» complex, Formica picea Nylander, 1846 and F. candida Smith, 1878. Members of this complex are found from West Europe to Kamchatka, as well as from the valleys of northern rivers to Tibet (Figure 1).
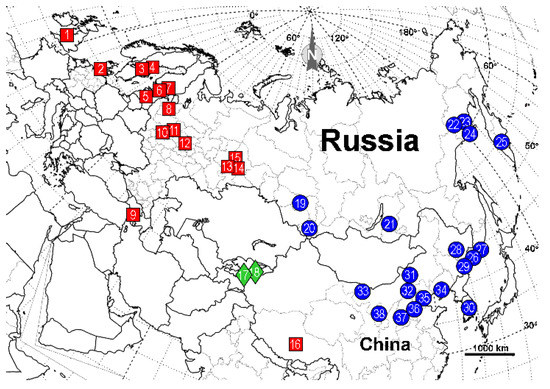
Figure 1.
Specimens of the «F. picea—F. candida» complex used in this study. Red squares, F. picea; green diamonds, F. candida; blue circles, Formica sp. Specimen numbers refer to Table 1.
Morphological variations within the complex were studied by Dlussky [11] based on extensive collections. Dlussky [11] concluded that there is no ground to split it into several species. Later on, Bolton [12] suggested that the name F. candida has the priority over F. picea; this was picked up by some—but not all—of the scientists. Seifert [13] suggested that F. picea and F. candida are two separate species, based on the features of pubescence and some morphometric parameters. According to Seifert [13], the distribution of F. picea includes Europe, the Caucasus, and the West Siberian Lowlands, and that of F. candida is in Central Asia, the Altai Mountains, and East Siberia. The habitats of F. picea include the bogs and European mountains above the forest belt, while F. candida lives in diverse habitats, including river floodplains and dry steppes of Asia.
However, Zakharov et al. [14] refuted Seifert’s [13] conclusion, stating that the variation of pubescence within the European populations of F. picea is higher than the proposed differences between the species. They also stated that there is a smooth gradient of morphological variation from Europe to the Far East. Based on this, the authors returned to treating F. picea and F. candida as synonyms.
Molecular genetic analysis is increasingly used to resolve problematic questions in ant taxonomy, due to its high resolution and its potential to detect cryptic taxa. [15] There were several molecular genetic attempts to resolve the relationships in the «F. picea—F. candida». Goropashnaya [3] performed a phylogenetic analysis based on the mitochondrial cytb gene. She demonstrated that the complex is split into two clades: one restricted to Europe and West Siberia and the other, to Central Asia. Later Goropashnaya et al. referred to these clades as a pair of species [5]. Antonov and Bukin [16] studied a sample of cytb sequences of specimens from several regions of the Palearctic identified as F. candida and F. picea. They concluded that genetic distances within F. candida are higher than those between the two species. Important datasets were also obtained by Chen et al. [7] and Schär et al. [4], based on the mitochondrial cox1 gene.
Therefore, the status of the «F. picea—F. candida» species complex remains uncertain. The problem with molecular data is that very similar sequences can be deposited under different species names depending on the authors’ viewpoint on this matter. Moreover, some authors sequenced the cytb gene, and the others sequenced cox1, so the resulting samples cannot be compared directly.
We collected a sample of the «F. picea—F. candida» complex specimens from the Northern Palearctic. Moreover, we studied the diversity, geographic and habitat distribution, and ecology of its population from the north of the Russian Far East [17]. The most important specimens were identified by Dlussky and Radchenko, who are specialists in the genus Formica; both consider this complex a single species (Radchenko: [18], personal communication). Here, we attempt to resolve the issue on the «F. picea—F. candida» complex using molecular analysis, based on the mitochondrial cox1 and cytb genes, as well as three nuclear loci, with the integration of the available GenBank sequences obtained by other authors.
2. Materials and Methods
We collected a set of Formica specimens from Eurasia (Figure 1, Table 1). DNA was isolated from single ethanol-fixed worker ants using the commercial silica columns (BioSilica, Novosibirsk, Russia), as described in [4]. A fragment of the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene was amplified using the universal primers LCO1490m (5′-TACTC-AACAA-ATCAC-AAAGA-TATTG-G-3′; modified from [10]) and HCO2198 (5′-TAAAC-TTCAG-GGTGA-CCAAA-AAATC-A-3′; [19]). A part of the cytochrome b (cytb) was amplified using the primers Fcbl-F (5′-ACCCT-CACCT-GTAAA-TATTT-CTT-3′) and Fcbl-R (GGAAT-AGATC-GTAAA-ATTGC-AT-3′) designed in this study. PCR was performed using the Biomaster HS-Taq PCR Mix (Biolabmix, Novosibirsk, Russia).

Table 1.
Specimens used in this study.
Three nuclear loci were amplified using the primers designed in this study: a fragment of the wingless (wg) gene, with FWg-F (5′-CGTGG-TCGGC-GATAA-TCTA-3′) and FWg-R (5′-CACCA-CCACC-TCCTG-AGTCT-3′); topoisomerase 1 (Top1), with FTPO-F (5′-GAACC-ATTGC-CACCC-ATAGT-3′) and FTPO-R (5′-AGCGC-CAGCT-TGTCA- ATAAA-3′); the ribosomal internal transcribed spacer (ITS2), with FITS2-Fw (5′-TCATT-AACGT-TCCGG-AGGTC-3′) and FITS2-Rv (5′-TAAAA-TCGTT-GGCCC-TTACG-3′).
The obtained DNA fragments were visualized by agarose gel electrophoresis; unincorporated primers and nucleotide phosphates were removed using the shrimp alkaline phosphatase/E. coli exonuclease I mix (New England Biolabs, Ipswich, MA, USA). Sanger sequencing was performed on a 3130xl DNA Analyzer (Applied Biosystems, Framingham, MA, USA) in the SB RAS Genomics Core Facility (ICBFM SB RAS, Novosibirsk, Russia) using both forward and reverse primers. The obtained sequences were deposited in GenBank under accession numbers ON228270-ON228284, ON220872-ON220887, and ON862896-ON862909. Sequences obtained by other authors were also used in this study (see Table 1 for GenBank accessions). This dataset includes the cytb sequence for the F. candida neotype (AY786154).
Phylogenetic trees were constructed using the Maximum Likelihood (ML) algorithm and Bayesian Inference, as described in [20]. ML trees were built in RAxML v. 8.2.12 (A. Stamatikis, Hedelberg, Germany) [21], using the GTR + I + G model, as suggested by MrModeltest v.2.0 (J. Nylander, Uppsala, Sweden) [22]. Bayesian analysis was made in MrBayes v. 3.4 (F. Ronquist et al., Stockholm, Sweden) [23] as two simultaneous independent analyses from different random starting trees. There were 20 million generations performed; 25% of the generations were discarded as burn-in.
3. Results
We sequenced fragments of the mitochondrial cox1 and cytb loci for our sample (Table 1) and constructed phylogenetic trees. The final alignment for cox1 contained 593 bp; for cytb, 693 bp. On the cytb tree (Figure 2) the «F. picea—F. candida» complex was split into two big clades. Nucleotide distances between them were as large as those among other Formica species. One of the clades included the specimens from Kyrgyzstan, Europe, the Urals, the Caucasus, and Tibet. The sequences from Kyrgyzstan that included the F. candida neotype fell into a well-supported subclade. The rest of the specimens (shown on Figure 2 as F. picea) formed a group with no statistical support. This group included F. picea (including a specimen from southern Finland, the terra typica of F. picea), as well as those submitted to GenBank as F. candida. The second clade (shown on Figure 2 as Formica sp.) contained the specimens from West and East Siberia, the Russian Far East, China, and Korea. For cytb, the average p-distances between F. candida and F. picea were 1.34%; between F. picea and Formica sp., 3.25%; between F. candida and Formica sp., 3.35%. The mean p-distance within F. picea was 0.81%; within F. candida, 0.06%; within Formica sp., 1.05%.
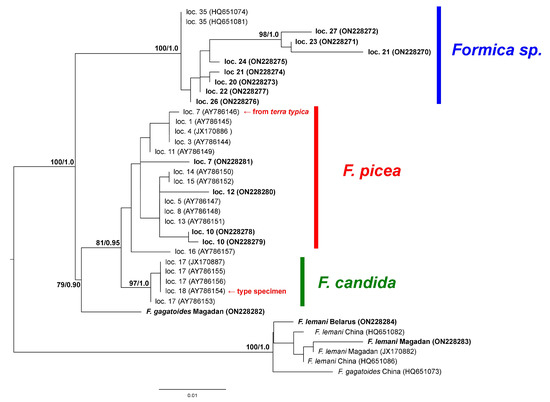
Figure 2.
Phylogenetic tree constructed using cytb sequences. Numbers near branches indicate ML bootstrap support/Bayesian posterior probabilities. GenBank accessions given in parentheses; sequences obtained in this study are shown in bold.
The tree constructed using the cox1 gene (Figure 3) was built on more specimens than the cytb tree. However, there were no data for Kyrgyzstan, where the neotype of F. candida was described, or other regions of Central Asia, and there were few F. picea specimens. On the cox1 tree, the «F. picea—F. candida» complex was also found to contain two clades: one corresponding to F. picea (including terra typica, i.e., the region where the original type specimens were collected) and the other to Formica sp. The average p-distances between F. picea and Formica sp. were 2.52%; within the species, they were 0.57 and 0.38%, respectively. We should also note that the cox1 tree contained as many as three clades, with specimens identified as F. gagatoides.
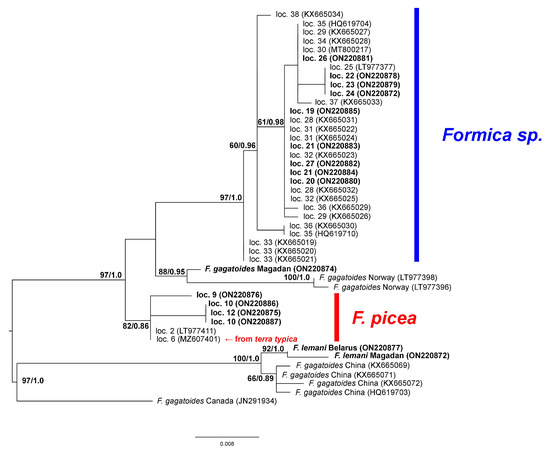
Figure 3.
Phylogenetic tree constructed using cox1 sequences. Numbers near branches indicate ML bootstrap support/Bayesian posterior probabilities. GenBank accessions given in parentheses; sequences obtained in this study are shown in bold.
The final alignment for the wg locus included 312 bp; for Tpo1, 717 bp; for ITS2, 678 bp. Sequences of wg and TPO of F. picea, Formica sp., and F. gagatoides were identical, except for some degenerate positions. In the ITS sequence, F. picea and Formica sp. could be distinguished by one A<>G substitution in position 216 of the alignment, while these two taxa differed from F. gagatoides by two substitutions and one short indel.
4. Discussion
In this study, we analyzed the dataset of the «F. picea—F. candida» complex in order to find out if F. picea and F. candida are synonyms or two distinct species. The results were unexpected. The sample, indeed, contained two clades: one with F. candida and F. picea specimens and the other representing a previously unknown taxon (Figure 2). For mitochondrial sequences, genetic distances between these clades were as high as between the well-established species of the genus Formica. This is in line with earlier findings that recovered multiple cryptic species within the genus Formica [24,25], as well as other well-studied genera, such as Pheidole [26,27], Myrmica [28,29], Lasius and Cardiocondyla [24], etc. For the nuclear sequences, however, the differences were not as high: wg and Tpo1 gene fragments were identical, and only one substitution distinguished F. picea from Formica sp. On the other hand, F. gagatoides was also very close to these taxa, differing from F. picea by two substitutions and one indel in ITS2, with two other genes being identical. It is well known that nuclear DNA has a lower substitution rate compared to the mitochondrial genome, and closely related ant species may have few distinctive positions or none at all. From the data obtained in [4], one can see that there are very few differences in the nuclear sequences of the whole F. picea/F. gagatoides/F. lemani/F. fusca/F. neorufarbis group. This is the usual situation that is found in ants, e.g., in the well-studied Formica rufa group, several species acknowledged by myrmecologists cannot be distinguished by nuclear markers and even show significant interspecific hybridization [30,31]. A similar case can also be observed in the genus Proformica [32]. Thus, we can conclude that the Asian Formica sp. is apparently distinct genetically from F. picea and F. candida, but it is unclear whether it should be regarded as a separate species or as a subspecies.
F. candida was initially described by Smith [33] based on a single specimen. This specimen was collected “On the road across the Pamir, from Sarikol to Panja”, which is probably somewhere near the easternmost part of China, on the border with Tajikistan. According to [13], this specimen was lost, so he fixed a neotype from Kyrgyzstan. This location is quite remote (about 315 km straight line distance in a mountainous area) from the original one. Our sample contained a cytb sequence of the neotype (AY786154). We can state that, so far, the F. candida clade is limited to the available specimens from Kyrgyzstan and was not found elsewhere.The type specimen of F. picea is old and highly damaged [13]. It was collected in the vicinity of Helsingfors (currently Helsinki, Finland) with no precise information on its whereabouts. Both locations available in our sample are several dozen km from Helsinki. Given the fact that no other group of the complex was found in Finland, we can suggest that these accessions represent the «real» F. picea.
Whether F. picea and F. candida are reciprocally monophyletic remains an open question. On our cytb tree F. picea was not supported as a clade (Figure 2), but more data are needed to verify that.
Based on the data on the type specimen origin, we cannot assign the Formica sp. clade to either F. picea or F. candida and have to conclude that it represents a new taxon that is still to be described. From the maps, we can see that all three ant taxa have distinct distributions: Formica sp. is found in Asian Russia, Siberia, and East Asia; F. candida, in Central Asia; F. picea, in Europe, with a single finding in Tibet. As suggested by multiple authors, morphological differences between these taxa are elusive [11,13,18]. There might be some differences in ecological preferences of these species: the European F. picea is found in swamps or in the alpine belt of mountains. The Asian Formica sp. is considered to be eurytopic in Siberia, while in the Russian Far East, it is found mostly in floodplains ([11] and our observations). However, it remains to be proved that these ecological differences coincide with the distribution boundaries of these species.
5. Conclusions
By trying to resolve the relationships within the «F. candida—F. picea» complex, in order to separate these two species, we found a new third member of this complex. It is important to note that genetic distances between F. candida and F. picea are significantly smaller than between them and the newly found taxon. The latter seems to not be the only cryptic taxon in this group: we obtained preliminary evidence that F. gagatoides might also represent as many as three cryptic taxa.
Author Contributions
Conceptualization, Z.A.Z. and D.I.B.; methodology, S.V.S. and T.V.P.; investigation, S.V.S. and T.V.P.; data curation, S.V.S.; writing—original draft preparation, D.I.B.; writing—review and editing, Z.A.Z. and S.V.S.; visualization, D.I.B. and S.V.S.; supervision, D.I.B.; project administration, D.I.B.; funding acquisition, D.I.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the State Budget Projects no. 1021060307698-5 (Faunogenesis and adaptive strategies of poikilothermic animals in extreme environments of the North) and no. FWNR-2022-0022.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The obtained sequences were deposited in GenBank under accession numbers ON228270-ON228284 (cytb) and ON220872-ON220887 (cox1).
Acknowledgments
We are grateful to N.A. Bulakhova, E.B. Fedoseeva, N.G. Gagarinova, T.V. Popkova, Yu. N. Sundukov, S.V. Chesnokova, Z.M. Yusupov and J. Sorvari for their help with specimen collection. Sanger sequencing was performed in SB RAS Genomics Core Facility (ICBFM SB RAS, Novosibirsk, Russia).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Borowiec, M.L.; Moreau, C.S.; Rabeling, C. Ants: Phylogeny and Classification. In Encyclopedia of Social Insects; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–18. [Google Scholar]
- Valenzuela-González, J.E.; Pérez-Toledo, G.R.; García-Martínez, M.A. Adelomyrmex dorae sp. nov. Garcia-Martinez (Hymenoptera: Formicidae): A New Species Supported by Parsimony Analysis of Morphological Characters. Trans. Am. Entomol. Soc. 2017, 143, 713–727. [Google Scholar] [CrossRef]
- Goropashnaya, A. Phylogeographic Structure and Genetic Variation in Formica Ants. Ph.D. Thesis, Uppsala University, Uppsala, Sweden, 20 December 2003. [Google Scholar]
- Schär, S.; Talavera, G.; Espadaler, X.; Rana, J.D.; Andersen, A.; Cover, S.P.; Vila, R. Do Holarctic ant species exist? Trans-Beringian dispersal and homoplasy in the Formicidae. J. Biogeogr. 2018, 45, 1917–1928. [Google Scholar] [CrossRef]
- Goropashnaya, A.V.; Fedorov, V.B.; Seifert, B.; Pamilo, P. Phylogenetic relationships of Palaearctic Formica species (Hymenoptera, Formicidae) based on mitochondrial cytochrome b sequences. PLoS ONE 2012, 7, e41697. [Google Scholar] [CrossRef] [PubMed]
- Roslin, T.; Somervuo, P.; Pentinsaari, M.; Hebert, P.D.N.; Agda, J.; Ahlroth, P.; Anttonen, P.; Aspi, J.; Blagoev, G.; Blanco, S.; et al. A molecular-based identification resource for the arthropods of Finland. Mol. Ecol. Resour. 2022, 22, 803–822. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, S. Phylogenetic relationships based on DNA barcoding among 16 species of the ant genus Formica (Hymenoptera: Formicidae) from China. J. Insect Sci. 2017, 17, 117. [Google Scholar] [CrossRef]
- Park, S.; Noh, P.; Kang, J.-Y. Endosymbionts and phage WO infections in Korean ant species (Hymenoptera: Formicidae). Proc. Natl. Inst. Ecol. Repub. Korea 2020, 1, 52–57. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, S.Y.; Ye, D.; Chen, Y.; Lu, C. Molecular phylogeny of the ant subfamily Formicinae (Hymenoptera, Formicidae) from China Based on mitochondrial genes. Sociobiology 2013, 60, 135–144. [Google Scholar] [CrossRef][Green Version]
- Shekhovtsov, S.V.; Berman, D.I.; Bulakhova, N.A.; Makarova, O.L.; Peltek, S.E. Phylogeography of earthworms from high latitudes of Eurasia. Acta Zool. Acad. Sci. Hung. 2018, 64, 369–382. [Google Scholar] [CrossRef]
- Dlussky, G.M. Ants of the Genus Formica (Hymenoptera: Formicidae g. Formica); Nauka: Moscow, Russia, 1967; p. 236. [Google Scholar]
- Bolton, B. A New General Catalogue of the Ants of the World; Harvard University Press: Cambridge, UK, 1995; p. 502. [Google Scholar]
- Seifert, B. The “Black Bog Ant” Formica picea Nylander, 1846—A species different from Formica candida Smith, 1878 (Hymenoptera: Formicidae). Myrmecolog. Nachr. 2004, 6, 29–38. [Google Scholar]
- Zakharov, A.A.; Dlussky, G.M.; Goryunov, D.N.; Gilyov, A.V.; Zryanin, V.A.; Fedoseeva, E.B.; Gorokhovskaya, E.A.; Radchenko, A.G. Monitoring of the Formica Ants; KMK Scientific Press: Moscow, Russia, 2013; p. 99. [Google Scholar]
- Ward, P.S.; Brady, S.G.; Fisher, B.L.; Schultz, T.R. The evolution of myrmicine ants: Phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst. Entomol. 2015, 40, 61–81. [Google Scholar] [CrossRef]
- Antonov, I.A.; Bukin, Y.S. Molecular phylogenetic analysis of the ant genus Formica L. (Hymenoptera: Formicidae) from Palearctic region. Russ. J. Genet. 2016, 52, 810–820. [Google Scholar] [CrossRef]
- Berman, D.I.; Alfimov, A.V.; Zhigulskaya, Z.A.; Leirikh, A.N. Overwintering and Cold-Hardiness of Ants in the Northeast of Asia; Pensoft Publishers: Sofia-Moscow, Russia, 2010; p. 294. [Google Scholar]
- Radchenko, A.G. Ants (Hymenoptera, Formicidae) of Ukraine; Institute of Zoology of Ukraine NAS: Kyiv, Ukraine, 2016; p. 495. [Google Scholar]
- Folmer, O.; Hoeh, W.R.; Black, M.B.; Vrijenhoek, R.C. Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Shekhovtsov, S.V.; Shipova, A.A.; Poluboyarova, T.V.; Vasiliev, G.V.; Golovanova, E.V.; Geraskina, A.P.; Bulakhova, N.A.; Szederjesi, T.; Peltek, S.E. Species delimitation of the Eisenia nordenskioldi complex (Oligochaeta, Lumbricidae) using transcriptomic data. Front. Genet. 2020, 11, 1508. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest Ver. 2; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Seifert, B. Cryptic species in ants (Hymenoptera: Formicidae) revisited: We need a change in the alpha-taxonomic approach. Myrmecol. News 2009, 12, 149–166. [Google Scholar]
- Bernasconi, C.; Cherix, D.; Seifert, B.; Pamilo, P. Molecular taxonomy of the Formica rufa group (red wood ants) (Hymenoptera: Formicidae): A new cryptic species in the Swiss Alps. Myrmecol. News 2011, 14, 37–47. [Google Scholar]
- Seifert, B. Inconvenient hyperdiversity—The traditional concept of “Pheidole pallidula” includes four cryptic species (Hymenoptera: Formicidae). SOIL Org. 2016, 88, 1–17. [Google Scholar]
- Fournier, D.; Tindo, M.; Kenne, M.; Mbenoun Masse, P.S.; Van Bossche, V.; De Coninck, E.; Aron, S. Genetic structure, nestmate recognition and behaviour of two cryptic species of the invasive big-headed ant Pheidole megacephala. PLoS ONE 2012, 7, e31480. [Google Scholar] [CrossRef]
- Seifert, B.; Yazdi, A.B.; Schultz, R. Myrmica martini sp. n.—A cryptic species of the Myrmica scabrinodis species complex (Hymenoptera: Formicidae) revealed by geometric morphometrics and nest-centroid clustering. Myrmecol. News 2014, 19, 171–183. [Google Scholar]
- Ebsen, J.R.; Boomsma, J.J.; Nash, D.R. Phylogeography and cryptic speciation in the Myrmica scabrinodis Nylander, 1846 species complex (Hymenoptera: Formicidae), and their conservation implications. Insect Conserv. Divers. 2019, 12, 467–480. [Google Scholar] [CrossRef]
- Seifert, B. A taxonomic revision of the Palaearctic members of the Formica rufa group (Hymenoptera: Formicidae)–The famous mound-building red wood ants. Myrmecol. News 2021, 31, 133–179. [Google Scholar] [CrossRef]
- Pamilo, P.; Kulmuni, J. Genetic identification of Formica rufa group species and their putative hybrids in northern Europe. Myrmecol. News 2022, 32. [Google Scholar] [CrossRef]
- Sanllorente, O.; Lorite, P.; Ruano, F.; Palomeque, T.; Tinaut, A. Phylogenetic relationships between the slave-making ants Rossomyrmex and their Proformica hosts in relation to other genera of the ant tribe Formicini (Hymenoptera: Formicidae). J. Zool. Syst. Evol. Res. 2018, 56, 48–60. [Google Scholar] [CrossRef]
- Smith, F. Scientific Results of the Second Yarkand Mission; Based upon the Collections and Notes of the Late Ferdinand Stoliczka, Ph.D.; Superintendent of Government Printing (Government of India): Calcutta, India, 1878; p. 22. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).