Molecular Phylogeny of Selected Kenyan Eucalyptus Species Inferred from MatK, rbcL and TrnL-F Genes and Their Suitability for Power Transmission Poles
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Determination of Phenotypic Traits
2.2. DNA Extraction, PCR, and Sequencing
2.3. Phylogenetic Analysis
3. Results
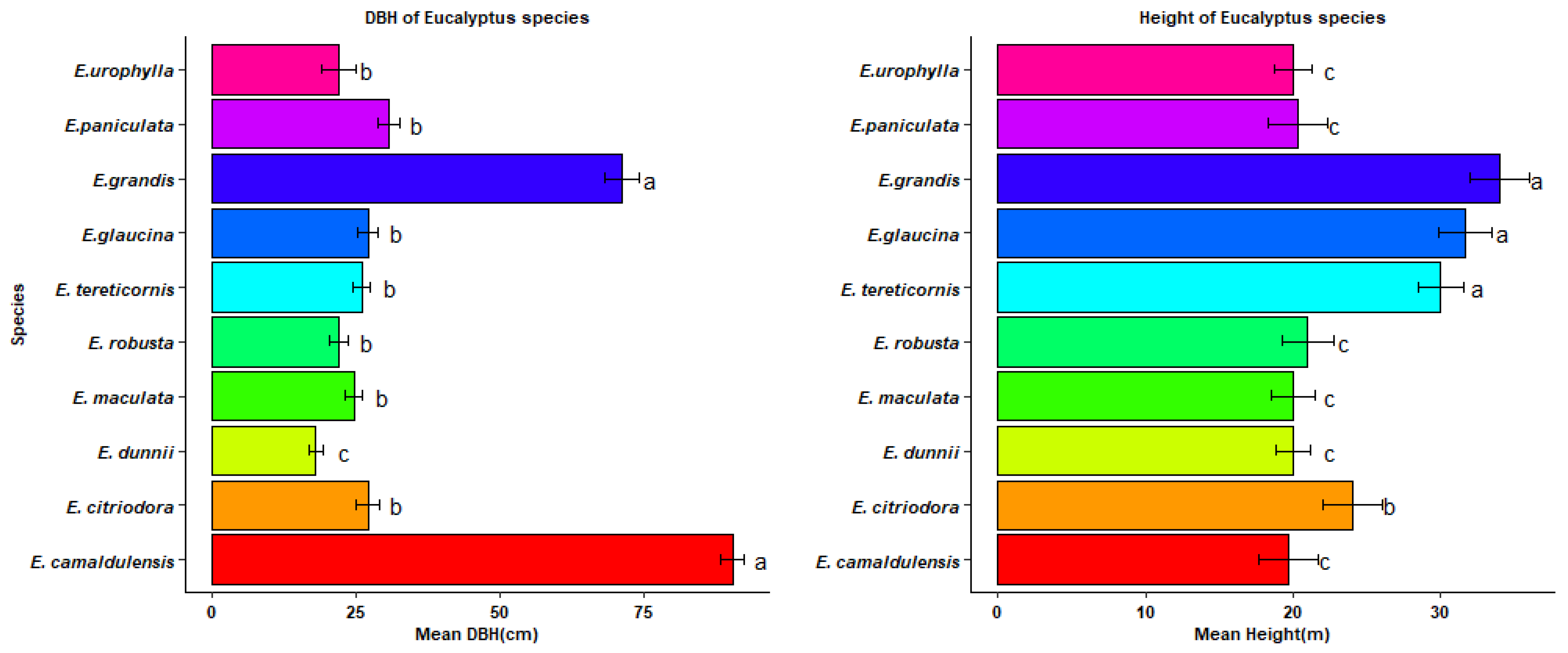
3.1. Phenotypic Traits of Eucalyptus Species
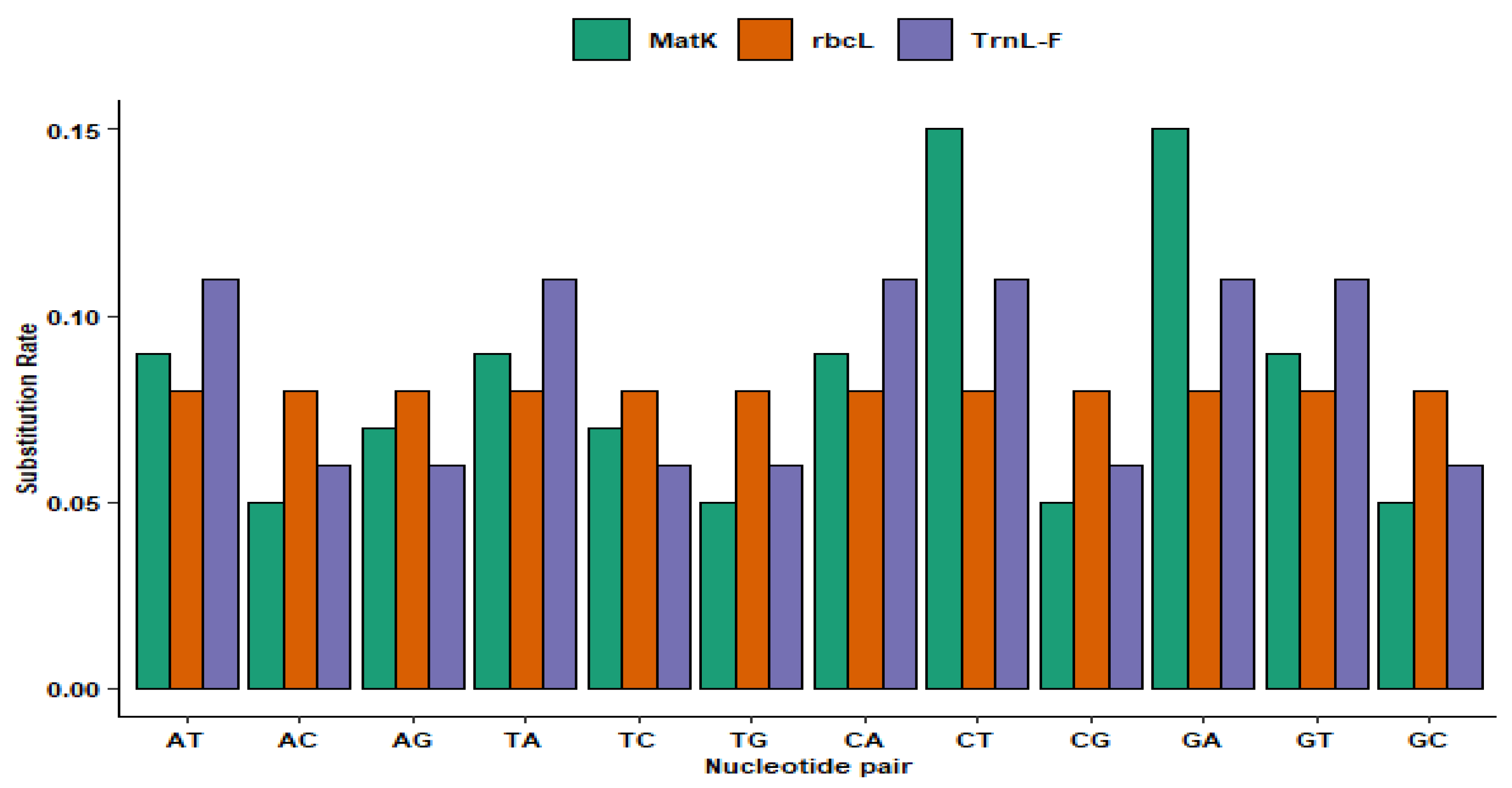
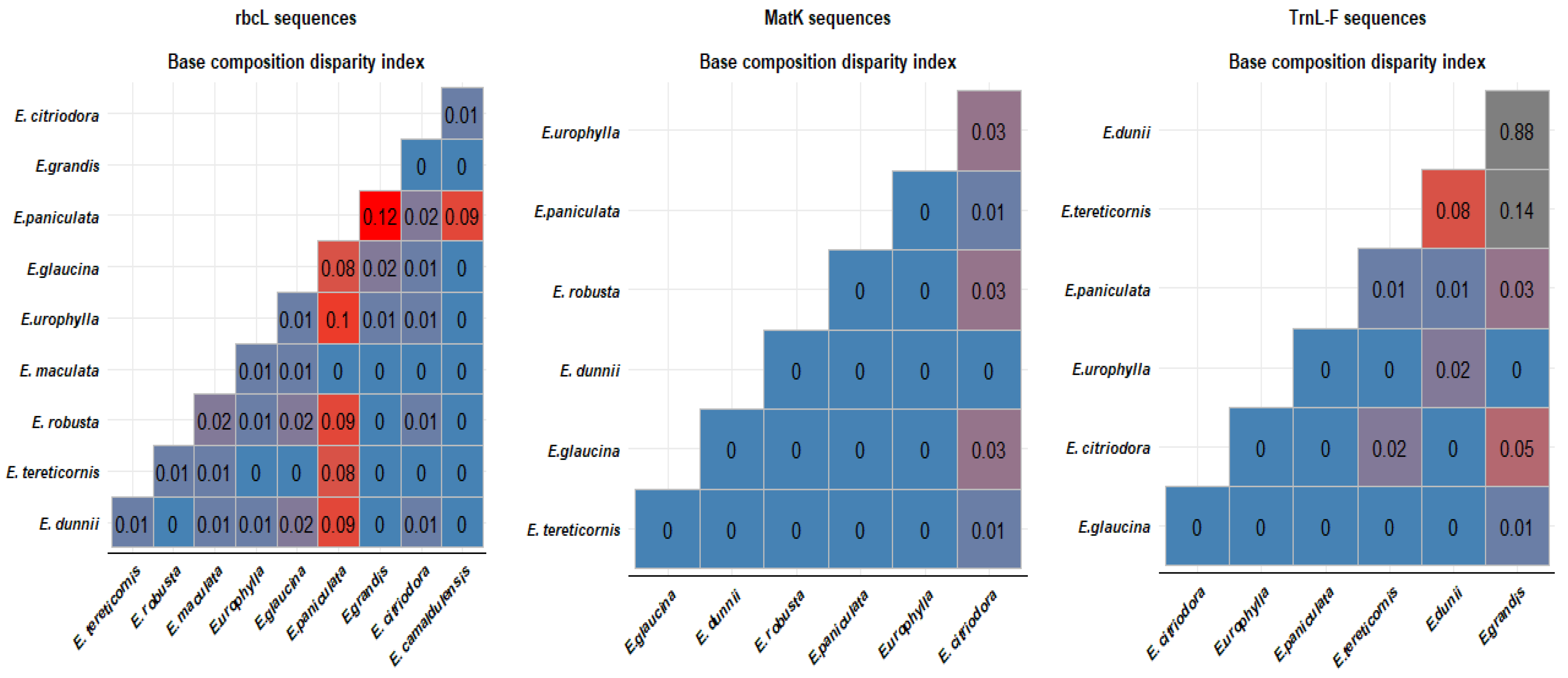
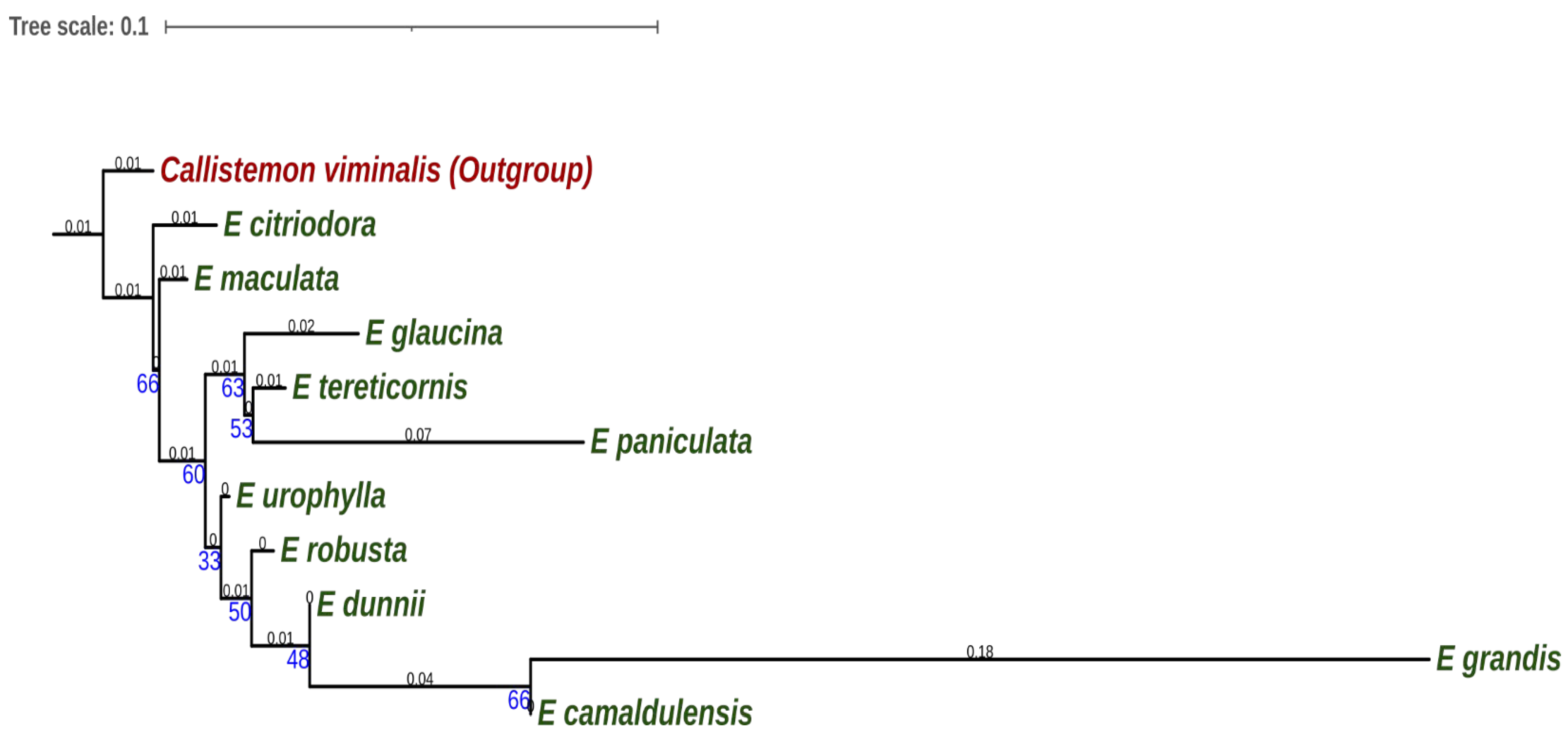
3.2. Molecular Phylogeny of Eucalyptus Species
4. Discussion
4.1. Phenotypic Traits of Eucalyptus Species
4.2. Phylogeny of Eucalyptus Species
5. Conclusions and Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kluthe, B.G. Eucalyptus in Kenya; Impacts on Environment and Society; University of Arkansas: Fayetteville, AR, USA, 2016. [Google Scholar]
- Birhanu, S.; Kumsa, F. Review on expansion of Eucalyptus, its economic value and related environmental issues in Ethiopia. Int. J. Res. Environ. Sci. 2018, 4, 41–46. [Google Scholar]
- Brooker, M.I.H. A new classification of the genus Eucalyptus L’Her. (Myrtaceae). Aust. Syst. Bot. 2000, 13, 79–148. [Google Scholar] [CrossRef]
- Langat, D.; Muchiri, M.N. Power Transmission Poles and Firewood in Kenya Financial Analysis of GROWING Eucalyptus grandis for Production of Medium Size Power Transmission Poles and Firewood in Kenya. 2015. Available online: https://www.researchgate.net/publication/328567812_Financial_analysis_of_growing_Eucalyptus_grandis_for_production_of_medium_size_power_transmission_poles_and_firewood_in_Kenya (accessed on 5 April 2022).
- Oballa, P.O.; Muchiri, M.N.; Kigomo, B.N. Facts on Growing and Use of Eucalyptus; Kenya Forestry Research Institute: Nairobi, Kenya, 2010. [Google Scholar]
- Girijashankar, V. Genetic transformation of Eucalyptus. Physiol. Mol. Biol. Plants 2011, 17, 9–23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sivananthawerl, T.; Mitlöhner, R. Eucalyptus grandis and other important Eucalyptus species: A case study from Sri Lanka. In Silviculture in the Tropics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 463–472. [Google Scholar]
- Luna, R.K. Eucalypts in agroforestry. In Eucalypts in India; ENVIS Centre on Forestry: Dehradun, India, 2009; p. 209. [Google Scholar]
- Turnbull, J.W.; Booth, T.H. Eucalypts in cultivation: An overview. In Eucalyptus Genus Eucalyptus; Taylor & Francis: London, UK, 2002; pp. 52–74. [Google Scholar]
- Muthike, G.; Ali, G. Concrete vs. Wooden Poles: Effects of the Shift to Concrete Poles on Tree Growers; Kenya Forestry Research Institute: Nairobi, Kenya, 2021. [Google Scholar]
- Thumma, B.R.; Nolan, M.F.; Evans, R.; Moran, G.F. Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 2005, 171, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Marco de Lima, B.; Cappa, E.P.; Silva-Junior, O.B.; Garcia, C.; Mansfield, S.D.; Grattapaglia, D. Quantitative genetic parameters for growth and wood properties in Eucalyptus “urograndis” hybrid using near-infrared phenotyping and genome-wide SNP-based relationships. PLoS ONE 2019, 14, e0218747. [Google Scholar] [CrossRef] [PubMed]
- Alper, K.; Tekin, K.; Karagöz, S.; Ragauskas, A.J. Sustainable energy and fuels from biomass: A review focusing on hydrothermal biomass processing. Sustain. Energy Fuels 2020, 4, 4390–4414. [Google Scholar] [CrossRef]
- Sandak, J.; Sandak, A.; Meder, R. Assessing trees, wood and derived products with near infrared spectroscopy: Hints and tips. J. Near Infrared Spectrosc. 2016, 24, 485–505. [Google Scholar] [CrossRef]
- Schimleck, L.R.; Raymond, C.A.; Beadle, C.L.; Downes, G.M.; Kube, P.D.; French, J. Applications of NIR spectroscopy to forest research. Appita J. 2000, 53, 458–464. [Google Scholar]
- Hamilton, M.G.; Raymond, C.A.; Harwood, C.E.; Potts, B.M. Genetic variation in Eucalyptus nitens pulpwood and wood shrinkage traits. Tree Genet. Genomes 2009, 5, 307–316. [Google Scholar] [CrossRef]
- Musila, F.M.; Lukhoba, C.W.; Nguta, J.M.; Dossaji, S.F. Phylogeny of Ten Kenyan Plectranthus Species in the Coleus Clade Inferred from Leaf Micromorphology, Rbcl and MatK Genes. J. Bot. 2017, 2017, 4369029. [Google Scholar] [CrossRef]
- Cheboiwo, J.K. Modelling Future Production. Processing and Trade in Transmission Poles in Kenya and East Africa; Kenya Forestry Research Institute: Nairobi, Kenya, 2014. [Google Scholar]
- Cheboiwo, J.K. Unexploited possibilities: The Status of Forest Resources and Trade Opportunities in Wood Products in East and Central Africa; Kenya Forestry Research Institute: Nairobi, Kenya, 2009. [Google Scholar]
- Kagombe, J.K.; Kiprop, J.; Langat, D.; Cheboiwo, J.K.; Wekesa, L.; Ongugo, P.O.; Mbuvi, M.T.; Leley, N. Socio-Economic Impact of Forest Harvesting Moratorium in Kenya; KEFRI: Nairobi, Kenya, 2020. [Google Scholar]
- Muthike, G.M.; Githiomi, J. Review of the Wood Industry in Kenya; Technology Development, Challenges and Opportunities. Int. J. Res. Stud. Agric. Sci. 2017, 8, 44–52. [Google Scholar]
- Cheboiwo, J.K. The Status of the Poles Sector; Production, Processing and Trade in Transmission Posts in East Africa; Kenya Forestry Research Institute: Nairobi, Kenya, 2014. [Google Scholar]
- Steane, D.A. Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae). DNA Res. 2005, 12, 215–220. [Google Scholar] [CrossRef]
- Barthet, M.M.; Hilu, K.W.; Barthet, M.M.; Hilu, K.W. Expression of matK: Functional and Evolutionary Implications Linked references are available on JSTOR for this article: Expression of matK: Functional and evolutionary implications. Am. J. Bot. 2019, 94, 1402–1412. [Google Scholar] [CrossRef]
- Suzuki, Y.; Makino, A. Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol. 2012, 160, 533–540. [Google Scholar] [CrossRef]
- Payn, K.G.; Dvorak, W.S.; Janse, B.J.H.; Myburg, A.A. Microsatellite diversity and genetic structure of the commercially important tropical tree species Eucalyptus urophylla, endemic to seven islands in eastern Indonesia. Tree Genet. Genomes 2008, 4, 519–530. [Google Scholar] [CrossRef]
- McCallum, M.L. Vertebrate biodiversity losses point to a sixth mass extinction. Biodivers. Conserv. 2015, 24, 2497–2519. [Google Scholar] [CrossRef]
- Assis, T.; Warburton, P.; Harwood, C. Artificially induced protogyny: An advance in the controlled pollination of Eucalyptus. Aust. For. 2005, 68, 27–33. [Google Scholar] [CrossRef]
- Andersen, H.-E.; Reutebuch, S.E.; McGaughey, R.J. A rigorous assessment of tree height measurements obtained using airborne lidar and conventional field methods. Can. J. Remote Sens. 2006, 32, 355–366. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Heckenhauer, J.; Barfuss, M.H.J.; Samuel, R. Universal multiplexable matK primers for DNA barcoding of angiosperms. Appl. Plant Sci. 2016, 4, 1500137. [Google Scholar] [CrossRef]
- Chiang, T.-Y.; Schaal, B.A.; Peng, C.-I. Universal primers for amplification and sequencing a noncoding spacer between the atpB and rbcL genes of chloroplast DNA. Bot. Bull. Acad. Sin. 1998, 39, 249–250. [Google Scholar]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L. ). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Michałek, M.; Ventura, E. Phylogenetic complexity of the Kimura 3-parameter model. Adv. Math. 2019, 343, 640–680. [Google Scholar] [CrossRef]
- Erickson, K. The jukes-cantor model of molecular evolution. Primus 2010, 20, 438–445. [Google Scholar] [CrossRef]
- Tamura, K. Model selection in the estimation of the number of nucleotide substitutions. Mol. Biol. Evol. 1994, 11, 154–157. [Google Scholar]
- Al-Atiyat, R.M.; Aljumaah, R.S. Genetic distances and phylogenetic trees of different Awassi sheep populations based on DNA sequencing. Genet. Mol. Res. 2014, 13, 6557–6568. [Google Scholar] [CrossRef]
- Page, R.D.M.; Holmes, E.C. Molecular Evolution: A Phylogenetic Approach; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Scott, M.H. The Use of Chemically Treated Wooden Poles for Telephone and Power Transmission Lines in South Africa Poles for Telephone and Power Transmission. J. S. Afr. For. Assoc. 1946, 13, 21–34. [Google Scholar]
- de Nogueira, M.C.; deAlmeida, D.H.; de Araujo, V.A.; Vasconcelos, J.S.; Christoforo, A.L.; de Almeida, T.H.; Lahr, F.A.R. Physical and mechanical properties of Eucalyptus saligna wood for timber structures. Ambiente Constr. 2019, 19, 233–239. [Google Scholar] [CrossRef]
- Wiehle, M.; Prinz, K.; Kehlenbeck, K.; Goenster, S.; Mohamed, S.A.; Finkeldey, R.; Buerkert, A.; Gebauer, J. The African baobab (Adansonia digitata, Malvaceae): Genetic resources in neglected populations of the Nuba Mountains, Sudan. Am. J. Bot. 2014, 101, 1498–1507. [Google Scholar] [CrossRef]
- Jump, A.S.; Marchant, R.; Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 2009, 14, 51–58. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Sreekumar, V.B.; Renuka, C. Assessment of genetic diversity in Calamus thwaitesii BECC. (Arecaceae) using RAPD markers. Biochem. Syst. Ecol. 2006, 34, 397–405. [Google Scholar] [CrossRef]
- De Araújo, E.S.N.N.; Gimenes, M.A.; Lopes, C.R. Phylogenetic relationships among genera Eucalyptus and Corymbia species based on rnDNA internal transcribed spacers sequences. Sci. For. Sci. 2002, 62, 75–85. [Google Scholar]
- Bryant, D.; Hahn, M.W. The Concatenation Question. No Commercial Publisher| Authors Open Access Book; 2020. Available online: https://hahnlab.sitehost.iu.edu/Publications/BryantHahn2020.pdf (accessed on 5 April 2022).
- Udovicic, F.; McFadden, G.I.; Ladiges, P.Y. Phylogeny of Eucalyptus and Angophora based on 5S rDNA spacer sequence data. Mol. Phylogenet. Evol. 1995, 4, 247–256. [Google Scholar] [CrossRef]
- Rozefelds, A.C. Eucalyptus phylogeny and history: A brief summary. Tasforests Hobart. 1996, 8, 15–26. [Google Scholar]
- Butcher, P.A.; McDonald, M.W.; Bell, J.C. Congruence between environmental parameters, morphology and genetic structure in Australia’s most widely distributed eucalypt, Eucalyptus camaldulensis. Tree Genet. Genomes 2009, 5, 189–210. [Google Scholar] [CrossRef]
- Dutkowski, G.W.; Potts, B.M. Genetic variation in the susceptibility of Eucalyptus globulus to drought damage. Tree Genet. Genomes 2012, 8, 757–773. [Google Scholar] [CrossRef]
- Dawson, J.C.; Goldringer, I. Breeding for genetically diverse populations: Variety mixtures and evolutionary populations. In Organic Crop Breeding; Wiley: Hoboken, NJ, USA, 2012; pp. 77–98. [Google Scholar]
- Acosta-Gallegos, J.A.; Kelly, J.D.; Gepts, P. Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci. 2007, 47, S-44. [Google Scholar] [CrossRef]
- Munthali, C.R.Y.; Chirwa, P.W.; Changadeya, W.J.; Akinnifesi, F.K. Genetic differentiation and diversity of Adansonia digitata L. (baobab) in Malawi using microsatellite markers. Agrofor. Syst. 2013, 87, 117–130. [Google Scholar] [CrossRef][Green Version]
- Nakabonge, G.; Roux, J.; Gryzenhout, M.; Wingfield, M.J. Distribution of Chrysoporthe canker pathogens on Eucalyptus and Syzygium spp. in eastern and southern Africa. Plant Dis. 2006, 90, 734–740. [Google Scholar]

| Gene | Sequence | Reference | |
|---|---|---|---|
| MatK | Forward | 5′-CCTATCCATCTGGAAATCTTA-3′ | [31] |
| Reverse | 3′-GTTCTAGCACAAGAAAGTCG -5′ | ||
| rbcL | Forward | 5′-ATGTCACCACAAACAGAGACTAAAGC-3′ | [32] |
| Reverse | 3′-GTAAAATCAAGTCCACCCG-5′ | ||
| TrnL-F | Forward | 5′-ATCATGTTAATTAATGTCTAGAA-3′ | [33] |
| Reverse | 3′-CGGGATCCTCAAGGGCCACCAAAGCTGT-5′ |
| Eucalyptus Species | Gene | Genbank Accession Number | Gene | Genbank Accession Number | Gene | Genbank Accession Number |
|---|---|---|---|---|---|---|
| E.urophylla | rbcL | OM949975 | MatK | OM949969 | TrnL-F | OM949984 |
| E.paniculata | rbcL | OM949976 | MatK | OM949970 | TrnL-F | OM949985 |
| E. citriodora | rbcL | OM949977 | MatK | OM949971 | TrnL-F | OM949986 |
| E. robusta | rbcL | OM949978 | MatK | OM949972 | TrnL-F | - |
| E. maculata | rbcL | OM949979 | MatK | - | TrnL-F | - |
| E. camaldulensis | rbcL | OM949980 | MatK | - | TrnL-F | - |
| E. dunnii | rbcL | OM949981 | MatK | OM949973 | TrnL-F | OM949987 |
| E.grandis | rbcL | OM949982 | MatK | - | TrnL-F | OM949988 |
| E. tereticornis | rbcL | OM949983 | MatK | OM949974 | TrnL-F | OM949989 |
| E.glaucina | rbcL | OM985042 | MatK | OM985041 | TrnL-F | OM985043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chebet, D.; Musila, F.M.; Kituyi, S.N.; Muthike, G.M.; Kaigongi, M.M. Molecular Phylogeny of Selected Kenyan Eucalyptus Species Inferred from MatK, rbcL and TrnL-F Genes and Their Suitability for Power Transmission Poles. Diversity 2022, 14, 563. https://doi.org/10.3390/d14070563
Chebet D, Musila FM, Kituyi SN, Muthike GM, Kaigongi MM. Molecular Phylogeny of Selected Kenyan Eucalyptus Species Inferred from MatK, rbcL and TrnL-F Genes and Their Suitability for Power Transmission Poles. Diversity. 2022; 14(7):563. https://doi.org/10.3390/d14070563
Chicago/Turabian StyleChebet, Daisy, Fredrick M. Musila, Sarah N. Kituyi, George M. Muthike, and Magrate M. Kaigongi. 2022. "Molecular Phylogeny of Selected Kenyan Eucalyptus Species Inferred from MatK, rbcL and TrnL-F Genes and Their Suitability for Power Transmission Poles" Diversity 14, no. 7: 563. https://doi.org/10.3390/d14070563
APA StyleChebet, D., Musila, F. M., Kituyi, S. N., Muthike, G. M., & Kaigongi, M. M. (2022). Molecular Phylogeny of Selected Kenyan Eucalyptus Species Inferred from MatK, rbcL and TrnL-F Genes and Their Suitability for Power Transmission Poles. Diversity, 14(7), 563. https://doi.org/10.3390/d14070563







