Is Coloburiscidae (Ephemeroptera) Monophyletic? A Comparison of Datasets
Abstract
:1. Introduction
1.1. Review of Taxonomy
1.2. Review of Relationships
2. Materials and Methods
2.1. Taxonomic Sampling
2.2. 5-Gene Sanger Dataset and Analysis
2.3. Phylogenomic Dataset and Analysis
2.4. Combined Dataset and Analysis
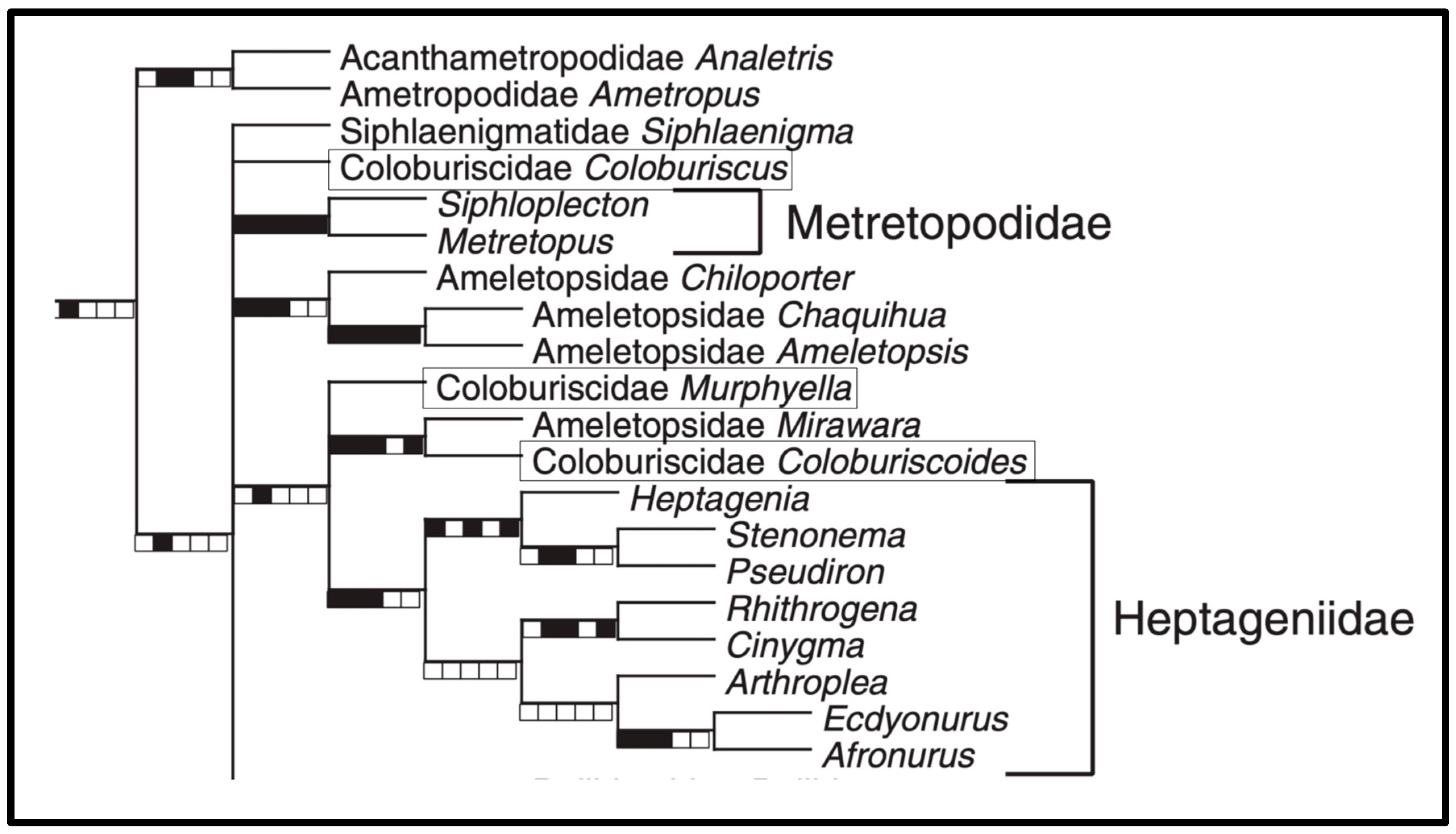
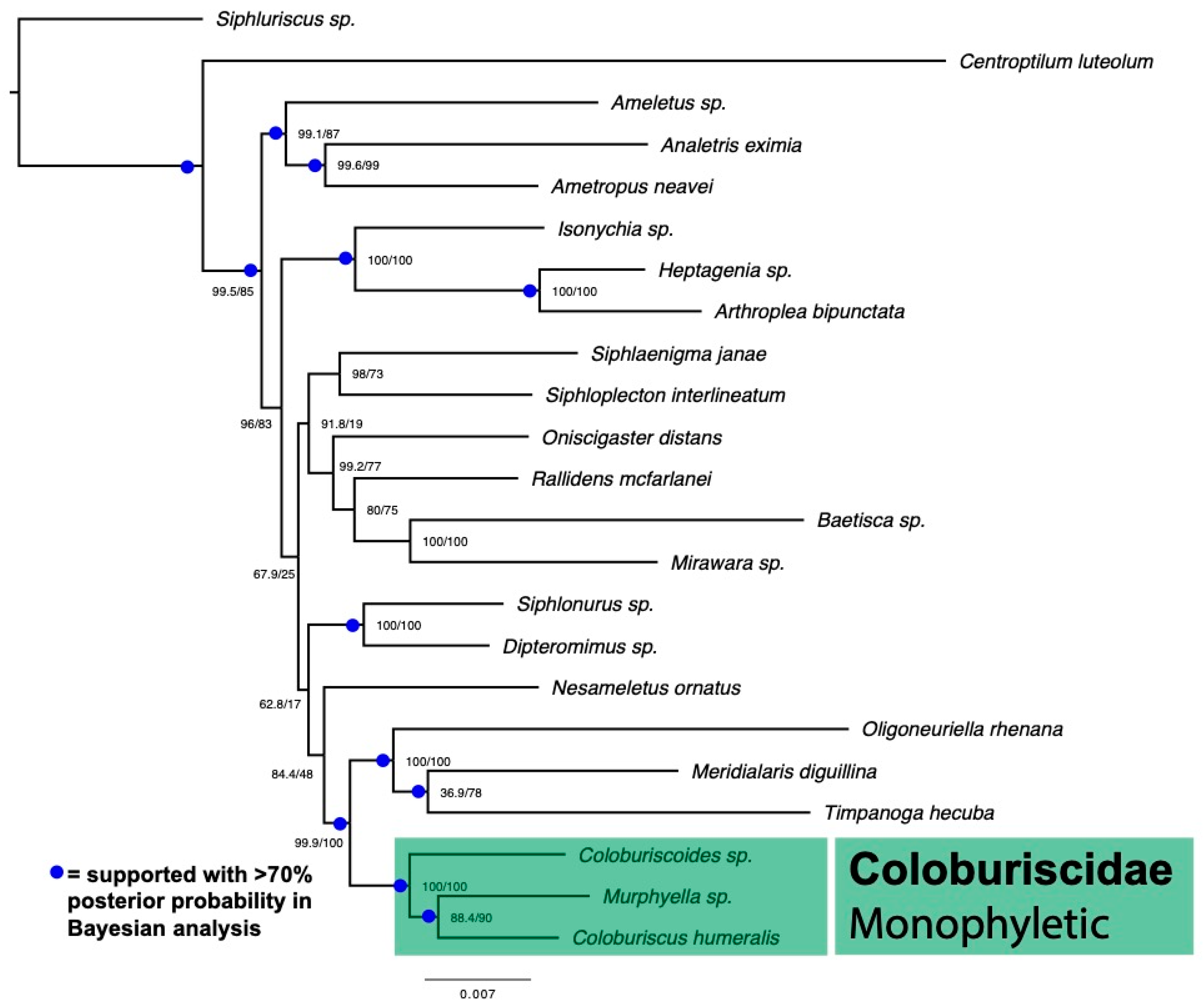
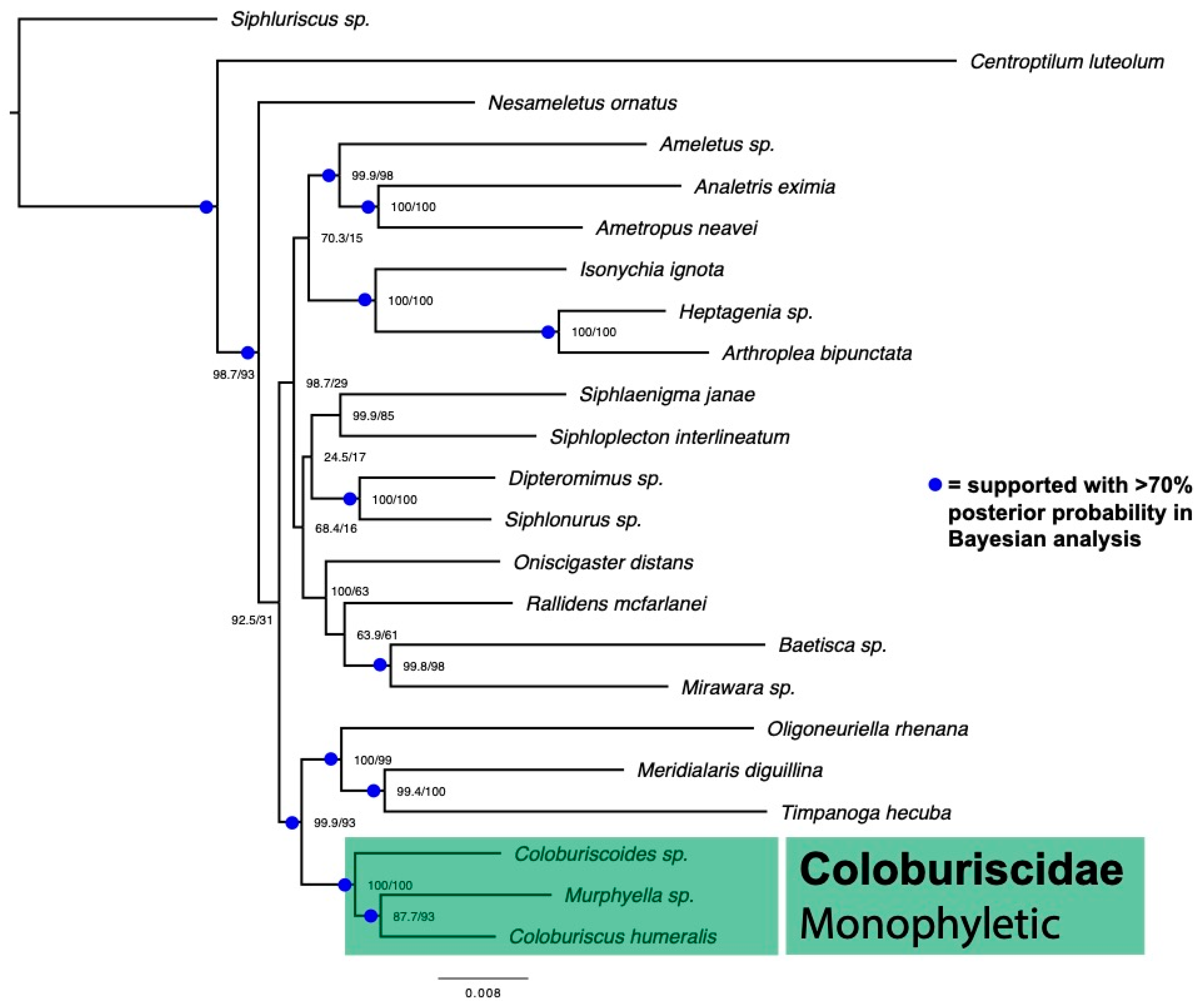
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kukalová-Peck, J. Ephemeroid wing venation based upon new gigantic Carboniferous mayflies and basic morphology, phylogeny, and metamorphosis of pterygote insects (Insecta, Ephemerida). Can. J. Zool. 1985, 63, 933–955. [Google Scholar] [CrossRef] [Green Version]
- Bauernfeind, E.; Soldan, T. The Mayflies of Europe (Ephemeroptera); Brill NV: Ollerup, Denmark, 2012; pp. 513–515. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.B.; Bartlett, S.; Sartori, M.; Breinholt, J.W.; Ogden, T.H. Anchored phylogenomics of burrowing mayflies (Ephemeroptera) and the evolution of tusks. Syst. Entomol. 2018, 43, 692–701. [Google Scholar] [CrossRef]
- Barber-James, H.M.; Gattolliat, J.L.; Sartori, M.; Hubbard, M.D. Global Diversity of Mayflies (Ephemeroptera, In-secta) in Freshwater. In Freshwater Animal Diversity Assessment; Springer: Berlin/Heidelberg, Germany, 2017; pp. 339–350. [Google Scholar]
- Ogden, T.; Gattolliat, J.L.; Sartori, M.; Staniczek, A.; Soldán, T.; Whiting, M. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): Combined analysis of morphological and molecular data. Syst. Entomol. 2009, 34, 616–634. [Google Scholar] [CrossRef]
- Jacobus, L.M.; Macadam, C.R.; Sartori, M. Mayflies (Ephemeroptera) and Their Contributions to Ecosystem Services. Insects 2019, 10, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blakey, R.C.; Fielding, C.R.; Frank, T.D.; Isbell, J.L. Gondwana paleogeography from assembly to breakup—A 500 my odyssey. Geol. Soc. Am. Spec. Pap. 2008, 441, 1–28. [Google Scholar] [CrossRef]
- Derkarabetian, S.; Baker, C.M.; Giribet, G. Complex patterns of Gondwanan biogeography revealed in a dispersal-limited arachnid. J. Biogeogr. 2021, 48, 1336–1352. [Google Scholar] [CrossRef]
- Harley, S.; Fitzsimons, I.; Zhao, Y. Antarctica and supercontinent evolution: Historical perspectives, recent advances and unresolved issues. Geol. Soc. Lond. Spec. Publ. 2013, 383, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Edmunds, G.F.; Allen, R.K.; Peters, W.L. An Annotated Key to the Nymphs of the Families and Subfamilies of May-Flies (Ephemeroptera); University of Utah Biology: Salt Lake City, UT, USA, 1963; Volume 13, pp. 1–49. [Google Scholar]
- Riek, E.F. The classification of the Ephemeroptera. In Proceedings of the First International Conference on Ephemeroptera, Ephemer, Tallahassee, 17–20 August 1970; pp. 160–178. [Google Scholar]
- Landa, V. A contribution to the evolution of the order Ephemeroptera based on comparative anatomy. In Proceedings of the First International Conference on Ephemeroptera, Ephemer, Tallahassee, 17–20 August 1970; pp. 155–159. [Google Scholar]
- McCafferty, W.P. The Cladistics, Classification, and Evolution of the Heptagenioidea (Ephemeroptera). Ephemer. Plecoptera 1991, 87–102. [Google Scholar]
- McCafferty, W.P. Toward a Phylogenetic Classification of the Ephemeroptera (Insecta): A Commentary on Systematics. Ann. Entomol. Soc. Am. 1991, 84, 343–360. [Google Scholar] [CrossRef]
- Kluge, N. The Phylogenetic System of Ephemeroptera; Kluwer Academic: Dordrecht, The Netherlands, 2004. [Google Scholar] [CrossRef]
- Ogden, T.H.; Whiting, M.F. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol. Phylogenetics Evol. 2005, 37, 625–643. [Google Scholar] [CrossRef]
- Ogden, T.; Whiting, M.F. The problem with “the Paleoptera Problem:” sense and sensitivity. Cladistics 2003, 19, 432–442. [Google Scholar] [CrossRef]
- Ogden, T.H.; Breinholt, J.W.; Bybee, S.M.; Miller, D.B.; Sartori, M.; Shiozawa, D.; Whiting, M.F. Mayfly phylo-genomics: Initial evaluation of anchored hybrid enrichment data for the order Ephemeroptera. Zoosymposia 2019, 16, 167–181. [Google Scholar] [CrossRef]
- Genecodes. Sequencher; Version 5.2.4; Genecodes Co.: Ann Arbor, MI, USA, 1999. [Google Scholar]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Breinholt, J.W.; Earl, C.; Lemmon, A.R.; Lemmon, E.M.; Xiao, L.; Kawahara, A.Y. Resolving Relationships among the Megadiverse Butterflies and Moths with a Novel Pipeline for Anchored Phylogenomics. Syst. Biol. 2017, 67, 78–93. [Google Scholar] [CrossRef] [Green Version]
- Veevers, J. Gondwanaland from 650–500 Ma assembly through 320 Ma merger in Pangea to 185–100 Ma breakup: Supercontinental tectonics via stratigraphy and radiometric dating. Earth-Science Rev. 2004, 68, 1–132. [Google Scholar] [CrossRef]
- Yoshida, M.; Santosh, M. Voyage of the Indian subcontinent since Pangea breakup and driving force of supercontinent cycles: Insights on dynamics from numerical modeling. Geosci. Front. 2018, 9, 1279–1292. [Google Scholar] [CrossRef]
- Barker, N.P.; Weston, P.; Rutschmann, F.; Sauquet, H. Molecular dating of the ‘Gondwanan’ plant family Proteaceae is only partially congruent with the timing of the break-up of Gondwana. J. Biogeogr. 2007, 34, 2012–2027. [Google Scholar] [CrossRef]
- Toussaint, E.F.A.; Bloom, D.; Short, A.E.Z. Cretaceous West Gondwana vicariance shaped giant water scavenger beetle biogeography. J. Biogeogr. 2017, 44, 1952–1965. [Google Scholar] [CrossRef]
- Gamble, T.; Bauer, A.M.; Greenbaum, E.; Jackman, T. Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J. Biogeogr. 2007, 35, 88–104. [Google Scholar] [CrossRef]
- Noben, S.; Kessler, M.; Quandt, D.; Weigand, A.; Wicke, S.; Krug, M.; Lehnert, M. Biogeography of the Gondwanan tree fern family Dicksoniaceae-A tale of vicariance, dispersal and extinction. J. Biogeogr. 2017, 44, 2648–2659. [Google Scholar] [CrossRef]
- Waters, J.M.; Craw, D. Goodbye Gondwana? New Zealand Biogeography, Geology, and the Problem of Circularity. Syst. Biol. 2006, 55, 351–356. [Google Scholar] [CrossRef] [Green Version]
- Knapp, M.; Stöckler, K.; Havell, D.; Delsuc, F.; Sebastiani, F.; Lockhart, P.J. Relaxed Molecular Clock Provides Evidence for Long-Distance Dispersal of Nothofagus (Southern Beech). PLOS Biol. 2005, 3, e14. [Google Scholar] [CrossRef]

| Genus | Specific Name | Location |
|---|---|---|
| Coloburiscoides | giganteus Tillyard (1933) | Australia |
| Coloburiscoides | haleuticus Eaton (1871) | Australia |
| Coloburiscoides | munionga Tillyard (1933) | Australia |
| Coloburiscus | humeralis Walker (1853) | New Zealand |
| Coloburiscus | tonnoiri Lestage (1935) | New Zealand |
| Coloburiscus | remota Walker (1853) | New Zealand |
| Murphyella | needhami Lestage (1930) | Chile |
| Family | Genus | Specific Name | Number of Exons Captured |
|---|---|---|---|
| Baetiscidae | Baetisca | sp. | 421 |
| Acanthametropodidae | Analetris | exima | 422 |
| Ameletidae | Ameletus | sp. | 435 |
| Ameletopsidae | Mirawara | sp. | 429 |
| Ametropodidae | Ametropus | naevi | 431 |
| Arthropleidae | Arthroplea | bipunctata | 399 |
| Baetidae | Centroptilum | luteolum | 414 |
| Coloburiscidae | Murphyella | sp. | 438 |
| Coloburiscidae | Coloburiscoides | sp. | 442 |
| Coloburiscidae | Coloburiscus | humeralis | 435 |
| Dipteromimidae | Dipteromimus | sp. | 432 |
| Ephemerellidae | Timpanoga | sp. | 414 |
| Heptageniidae | Heptagenia | sp. | 431 |
| Isonychiidae | Isonychia | sp. | 444 |
| Leptophlebiidae | Meridialaris | diguillina | 413 |
| Metropodidae | Siphloplecton | interlineatum | 432 |
| Nesameletidae | Nesameletus | ornatus | 420 |
| Oligoneuridae | Oligoneuriella | rhenana | 415 |
| Oniscigastridae | Oniscigaster | distans | 441 |
| Rallidentidae | Rallidens | mcfarlanei | 358 |
| Siphlaenigmatidae | Siphlaenigma | janae | 390 |
| Siphloneuridae | Siphlonurus | sp. | 445 |
| Siphluriscidae | Siphluriscus | sp. | 340 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meecham, J.; Ogden, T.H. Is Coloburiscidae (Ephemeroptera) Monophyletic? A Comparison of Datasets. Diversity 2022, 14, 505. https://doi.org/10.3390/d14070505
Meecham J, Ogden TH. Is Coloburiscidae (Ephemeroptera) Monophyletic? A Comparison of Datasets. Diversity. 2022; 14(7):505. https://doi.org/10.3390/d14070505
Chicago/Turabian StyleMeecham, Jarod, and T. Heath Ogden. 2022. "Is Coloburiscidae (Ephemeroptera) Monophyletic? A Comparison of Datasets" Diversity 14, no. 7: 505. https://doi.org/10.3390/d14070505
APA StyleMeecham, J., & Ogden, T. H. (2022). Is Coloburiscidae (Ephemeroptera) Monophyletic? A Comparison of Datasets. Diversity, 14(7), 505. https://doi.org/10.3390/d14070505





