Abstract
This survey reports on the DNA identification and occurrence of Culex torrentium and Cx. pipiens s.s. in Belgium. These native disease-vector mosquito species are morphologically difficult to separate, and the biotypes of Cx. pipiens s.s. are morphologically indistinguishable. Culex torrentium and Cx. pipiens s.s. were identified using the COI and ACE2 loci. We recorded 1248 Cx. pipiens s.s. and 401 Cx. torrentium specimens from 24 locations in Belgium (collected between 2017 and 2019). Culex pipiens biotypes pipiens and molestus, and their hybrids, were differentiated using fragment-size analysis of the CQ11 locus (956 pipiens and 227 molestus biotype specimens, 29 hybrids). Hybrids were observed at 13 out of 16 sympatric sites. These results confirm that both species are widespread in Belgium, but while Cx. torrentium revealed many COI haplotypes, Cx. pipiens s.s. showed only one abundant haplotype. This latter observation may either reflect a recent population-wide demographic or range expansion, or a recent bottleneck, possibly linked to a Wolbachia infection. Finally, new evidence is provided for the asymmetric but limited introgression of the molestus biotype into the pipiens biotype.
1. Introduction
Culex pipiens s.l. is a complex of three species, viz. Cx. australicus Dobrotworsky & Drummond, 1953; Cx. pipiens s.s. Linnaeus, 1758; and Cx. quinquefasciatus Say, 1823. The latter species is common in (sub)tropical regions with no known established populations in Europe [1], but has been introduced in the Netherlands with airplane traffic [2]. In contrast, Culex australicus is endemic to Australia. In Europe, hybrids between Cx. quinquefasciatus and Cx. pipiens s.s. were characterized in southern coastal regions and the Mediterranean region by applying DNA methods [3]. Despite morphological similarities with Cx. pipiens s.s. [4], Culex torrentium Martini, 1925 is no longer considered as belonging to the Cx. pipiens species complex [5,6]. Nevertheless, both species occur throughout Europe; Cx. torrentium is more common in northern Europe and at high elevations further south, whereas Cx. pipiens s.s. is more common in the south, but the exact species distribution limits are still unclear [7]. The two species occur in sympatry and are native in Belgium, where Cx. pipiens s.s. appears to be more abundant and widespread [8,9,10,11,12,13]. Within Cx. pipiens s.s., two biotypes are recognized, viz. Cx. pipiens biotype pipiens Linnaeus, 1758, and Cx. pipiens biotype molestus Forskål, 1775 [14].
The identification of Cx. torrentium and Cx. pipiens s.s. is difficult as the two species differ by a few subtle morphological characteristics only [4,15]. The biotypes of Cx. pipiens s.s. are morphologically indistinguishable [14], but they show four key behavioural differences [16,17,18,19]. Females of Cx. pipiens biotype pipiens need a bloodmeal to produce their first batch of viable eggs, prefer feeding on birds, breed in open spaces, and overwinter in a state of diapause. In contrast, females of Cx. pipiens biotype molestus can produce a first batch of viable eggs without a bloodmeal, prefer feeding on mammals, can breed in confined mating spaces, and do not overwinter in a state of diapause. In temperate regions of Europe, including Belgium, both biotypes co-occur in open aboveground spaces, but Cx. pipiens biotype molestus has a preference for confined spaces such as cellars, cesspits, human-made basements, or subways, where these mosquitoes mate and remain active throughout the year [20,21]. Hybrids between biotypes have been reported [13,19,22], displaying a combination of the behavioural traits of both biotypes [20,23]. However, since hybrids are less frequent than expected under random mating, the biotypes may show some degree of reproductive isolation [24,25]. Across the Mediterranean basin, populations in open spaces are genetically more homogenous, with individuals displaying mixed biotype ancestry and a mix of the four key behavioural traits [20]. Thus, the genetic differentiation between biotypes decreases gradually from north to south across the western Palearctic. This may be linked to less severe winters, allowing the non-diapausing molestus biotype to survive in open-space environments and admix [20].
In Europe, Culex pipiens s.s. is the principal vector for West Nile Virus (WNV), and several other arboviruses [26]. Culex pipiens s.s. biotype hybrids with an opportunistic feeding behaviour seem to transmit WNV between birds and humans more easily than non-hybrid Cx. pipiens biotypes [23,27,28,29]. Culex torrentium, in turn, is an important vector for Sindbis virus (SINV) in Sweden [30]. However, Cx. torrentium also has a high potential to transmit WNV [31,32]. In view of the recent outbreaks of WNV infections in Germany and the Netherlands [33], it is important to closely monitor competent Culex vectors.
The distinction between Cx. torrentium, Cx. pipiens s.s., and Cx. pipiens biotypes, has not been investigated systematically. Hence, the distribution and identity of these taxa is still poorly known in most European countries [34]. However, this information is essential to establish reliable risk projection and control programmes, particularly for the early detection of WNV vectors and their potential spread in Europe [35,36]. Therefore, the present paper reports on the DNA-based identification, occurrence, and diversity of Cx. torrentium and Cx. pipiens s.s., as well as Cx. pipiens biotypes molestus and pipiens and their hybrids, in Belgium.
2. Materials and Methods
2.1. Sampling
Adult and larval mosquitoes were collected in 2017 (August–November), 2018, and 2019 (both April–November) in the framework of the MEMO project (Monitoring of Exotic MOsquito species in Belgium [11]). Thirty-one potential points of entry (PoEs) for exotic mosquitoes in Belgium were surveyed using different sampling and trapping methods [11]. The PoEs included ports and airports, used tire and lucky bamboo import companies, parking lots along highways, wholesale markets, a flower auction, an allotment garden, an industrial area, and cemeteries along the border with Germany. Specimens were morphologically identified as Cx. pipiens s.l./Cx. torrentium using the keys of [4,37]. A random subset of 1689 Cx. pipiens s.l./Cx. torrentium specimens were selected for DNA-based identification (Table S1), using the sample_frac function of the dplyr package in R v4.03 [38].
2.2. DNA Extraction and COI Amplification
DNA was extracted from legs (adults) or abdomen (larvae) using the NucleoSpin® Tissue DNA extraction kit (Macherey-Nagel), following the manufacturer’s protocol, but with an elution volume of 70 µL. Remaining parts of the specimens and dried DNA extracts are stored at the Royal Belgian Institute of Natural Sciences (Collection Identifier: IG34179). The universal primers LCO1490 and HCO2198 [39] were used to amplify the mitochondrial cytochrome c oxidase subunit I (COI) barcode region (658 bp) [40]. If this was unsuccessful, the C1N-2191 and C1J-1718 primer combination [41] was used to amplify a 472 bp fragment of the COI barcode region. All PCR mixtures, cycling conditions, purification, and sequencing details are as described by [42] (Table S2). Raw sequences were trimmed, corrected, translated into amino acids, and assembled using Geneious® v.10.0.4 (Biomatters Ltd., Auckland, New Zealand). A consensus sequence was generated for each specimen.
2.3. Fragment-Size Analyses
To distinguish between Cx. pipiens s.s. and Cx. torrentium, a fragment of the Acetylcholinesterase-2 locus (ACE2) was amplified in a 10 µL PCR reaction volume as described by [43] (Table S2). This method also allows for detection of the eventual presence of introduced exotic Cx. quinquefasciatus. Using the forward primer B1246s and the reverse primers ACEpip, ACEquin, and ACEtorr, species-specific fragment sizes were produced, viz. 610 bp for Cx. pipiens s.s., 416 bp for Cx. torrentium, and 274 bp for Cx. quinquefasciatus [43]. PCR products were checked on a 2.5% agarose gel (45 min; 90 V).
To identify the two Cx. pipiens s.s. biotypes and their hybrids, the CQ11 microsatellite locus was amplified using the forward primer CQ11F2 and the reverse primers pipCQ11R and molCQ11R, following [44] (Table S2). PCR products were checked on a 2.5% agarose gel (45 min; 90V), with a band at 200 bp for Cx. pipiens biotype pipiens, and at 250 bp for Cx. pipiens biotype molestus. Hybrids showed both bands. Such hybrids were subsequently re-extracted and re-amplified for the CQ11 locus to exclude possible DNA contamination and confirm their status by visualisation of the two bands.
2.4. COI Data Analyses
The species identification engine of BOLD was used (www.boldsystems.org, accessed on 24 February 2020) with the species-level barcode records option to find the closest matching reference sequence. A Neighbour-Joining (NJ) tree was constructed to examine the clustering support of each Culex species occurring in Belgium [12,45] (Geneious® v10.0.4. (Biomatters Ltd., Auckland, New Zealand), Tamura-Nei distance model, 1000 bootstrap replicates). To do so, all publicly available COI sequences (http://www.boldsystems.org/index.php/databases, 16 March 2020) for these Culex species were aligned, using ClustalW in Geneious® v10.0.4 (Biomatters Ltd., Auckland, New Zealand), with the newly generated COI sequences in this study. COI sequences of four species of the genus Coquillettidia Dyar, 1905 were included as outgroup (GenBank accession numbers: GQ165785, GQ165801, GQ165802, and GQ165803). The alignment was checked for stop codons and trimmed to retain 658 bp. Sequences of less than 300 bp and conspecific identical sequences were discarded.
Pairwise differences in COI nucleotide frequencies between species, biotypes, and biotype hybrids were evaluated using Wright’s F-statistics in Arlequin v3.5 (1000 random permutations for significance, with subsequent standard Bonferroni correction) [46]. Haplotype frequencies, mean number of pairwise nucleotide differences (k) and nucleotide diversity (Pi) were also estimated with Arlequin v3.5, excluding sequences with ambiguous sites.
2.5. Habitat Characterization: Land-Cover Classes
The percentage of Corine Land-Cover (CLC) classes (© European Union, Copernicus Land Monitoring Service 2021, European Environment Agency (EEA)) was calculated in a 2.5 km buffer zone around each sampling location. The latest raster file (CLC 2018) was used, and calculation was performed in Q-GIS and R v4.03. The levels were grouped into five main CLC classes, i.e., artificial or urban areas, agricultural areas, forest and seminatural areas, wetlands, and water bodies.
3. Results
In total, 34,401 specimens from 27 out of 31 PoEs were morphologically identified as Cx. pipiens s.l./Cx. torrentium of which 1,689 specimens from 24 sites were selected for DNA-based verification. Of these, 573 were adults and 1113 were larvae. Adults were collected using a Mosquito Magnet trap (N = 242; 42.2%), Frommer Updraft Gravid trap (N = 59; 10.3%) and BG-Sentinel trap (N = 272; 47.5%). The four PoEs where these species were not collected were only surveyed using oviposition traps. Based on the BOLD similarity percentages, the COI NJ-tree (Figure S1), and the ACE2 fragment sizes (Figure S2), 401 specimens were identified as Cx. torrentium (Nadult = 40; Nlarva = 361—Table S1), and 1248 as Cx. pipiens s.s. Thirty-seven specimens did not provide ACE2 results and were therefore considered as Cx. pipiens s.l./Cx. torrentium. Three sequences were of too low quality for identification. The ACE2 fragment-size analysis provided no evidence of Cx. quinquefasciatus. In the NJ-tree, Cx. torrentium forms a cluster with 74.9% bootstrap support inside the Cx. pipiens s.s./Cx. torrentium group (Figure S1). The Cx. torrentium cluster includes all generated and downloaded (BOLD) COI sequences.
Based on the CQ11 fragment-size analysis 956 specimens were assigned to Cx. pipiens biotype pipiens (Nadult = 315 (33%); Nlarva = 641 (67%)) and 227 specimens to Cx. pipiens biotype molestus (Nadult = 187 (82%); Nlarva = 40 (18%)) (Figure S3, Table S1). More adults of the molestus biotype were collected than larvae, and the pipiens biotype. Additionally, 29 specimens were identified as hybrids between both biotypes (Nadult = 8 (28%); Nlarva = 21 (72%)), while the biotypes of 36 Cx. pipiens s.s. sequences were not determined due to missing CQ11 results. These sequences, together with those identified as Cx. pipiens s.l./Cx. torrentium (N = 37), were excluded from further analyses. The abundance of each taxon at each sampling site is shown in Figure 1. In most sites where both biotypes co-occur (N = 16), crossbreeding was identified, with hybrids detected at 13 sampling locations (Figure 1, Table S3). Sites where biotypes co-occurred included environments dominated by urban (Kallo, Charleroi, Zeebrugge and Zaventem), agricultural (Villers-Le-Bouillet, Vrasene, Frameries, Aische-en-Refail, Büllingen and Natoye), and forest and seminatural (Eupen, Dilsen-Stokkem, Houyet and Maasmechelen) areas (Figure 2). At these sites, larval stages of both biotypes were collected on the same days in the same types of larval habitats, viz. used tires, drainage holes, plastic containers, and cemetery flower vases, on multiple occasions. Once they were also found together in a large artificial pond. Culex pipiens s.s. and Cx. torrentium were collected on the same days in the following same types of larval habitats: used tires, drainage holes, cemetery flower vases, plastic sheets, and metal and plastic containers. The new COI sequences were deposited in GenBank (accession numbers: Cx. torrentium—OM749168-OM749568; Cx. pipiens biotype pipiens—OM748139-OM749094; Cx. pipiens biotype molestus—OM747912-OM748138; Cx. pipiens biotype pipiens X Cx. pipiens biotype molestus—OM747883-OM747911; Cx. pipiens s.s.: OM749132-OM749167; Cx. pipiens s.l./Cx. torrentium—OM749095-OM749131).
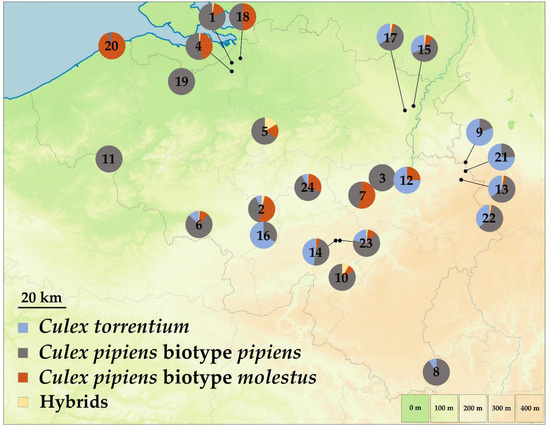
Figure 1.
Distribution of Culex specimens identified using DNA-based techniques and collected during the MEMO survey 2017–2019 [11]. Hybrids = Cx. pipiens biotype pipiens X Cx. pipiens biotype molestus. The numbers indicated on the pie charts are the collection site numbers used in Table S3.
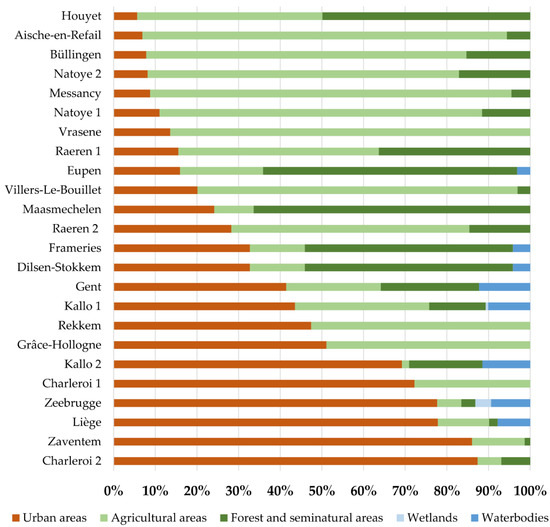
Figure 2.
The percentage of main Corine Land-Cover classes in a 2.5 km buffer zone around the sampling locations (group levels based on five classes).
The pairwise FST values, i.e., a measure of the average genetic differentiation, between Cx. pipiens biotype pipiens and Cx. pipiens biotype molestus, were significantly different from zero, which was also the case between the latter and the hybrids (Table 1). The hybrids and Cx. pipiens biotype pipiens showed the smallest average pairwise nucleotide differences and the lowest average nucleotide diversities (Table 2), despite Cx. pipiens biotype pipiens being the most widespread taxon in this survey (Figure 1; Table S4). Culex torrentium showed higher average pairwise nucleotide differences and nucleotide diversities than Cx. pipiens biotype pipiens (Table 2).

Table 1.
Pairwise FST estimates between biotypes and biotype hybrids of Culex pipiens s.s. based on COI sequences, calculated using Arlequin v3.5. Hybrids = Cx. pipiens biotype molestus X Cx. pipiens biotype pipiens. Significant values after standard Bonferroni correction marked by an asterisk (p < 0.0005).

Table 2.
COI sequence diversity of Culex pipiens s.s. biotypes and Cx. torrentium, calculated using Arlequin v3.5. Hybrids = Cx. pipiens biotype molestus x Cx. pipiens biotype pipiens, NCOI = number of generated COI sequences, NHap = number of haplotypes, NUnsharedHap = number of haplotypes specific to the taxon, S = number of polymorphic sites, k = average pairwise nucleotide differences, Pi = nucleotide diversity.
The most common COI haplotypes in Cx. pipiens s.s. were H1 (698 out of 1248 sequences, including 19 hybrids, 91 Cx. pipiens biotype molestus, and 588 Cx. pipiens biotype pipiens) and H2 (84 out of 1248 sequences, including 82 Cx. pipiens biotype molestus and 2 Cx. pipiens biotype pipiens) (Figure 3). The most common COI haplotype in Cx. torrentium was H3 (125 out of 401 sequences) (Figure 3). The haplotype of 509 sequences could not be identified because of ambiguous sites or short fragment lengths (NCx. torrentium = 122; NCx. pipiens biotype pipiens = 330; NCx. pipiens biotype molestus = 49; Nhybrids = 8).
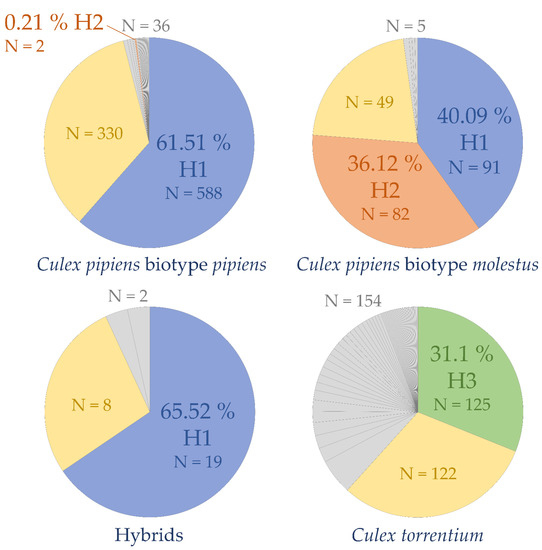
Figure 3.
COI haplotype frequencies calculated with Arlequin v.3.5. [46]. Hybrids = Culex pipiens biotype pipiens X Cx. pipiens biotype molestus, blue = Cx. pipiens s.s. haplotype H1, orange = Cx. pipiens s.s. haplotype H2, green = Cx. torrentium haplotype H3, yellow = sequences to which no haplotypes could be assigned, grey = haplotypes with a frequency < 6%.
4. Discussion
4.1. Species Occurrence in Belgium
In line with previous studies [8,9,13], Cx. pipiens s.s. appears to be more common in Belgium than Cx. torrentium. Likewise, Culex pipiens biotype pipiens is more common and widespread in Belgium than Cx. pipiens biotype molestus, as the latter comprises only 13% of the specimens (Table S4). However, industrial areas were overrepresented in this survey for the early detection of exotic Aedes species [11]; thus, the sampling may have been biased. As such, the molestus biotype, with its preference for hypogean habitats [21] and highly eutrophic waters in confined mating spaces [20,21], may be underrepresented, as these environments were not surveyed during the MEMO project [11]. Thus, more targeted surveys are needed to determine the actual prevalence and distribution of Cx. pipiens biotype molestus in Belgium. Nevertheless, this study confirms the co-occurrence of both biotypes in urban, agricultural, and forest and seminatural habitats [13,47,48,49,50] (Figure 2). Despite the sympatric occurrence of both biotypes, only few hybrid specimens were found (1.7%; Tables S3 and S4), i.e., less than in Germany (4.2%) [19], Portugal (8–10%) [47], and Italy (14.4%) [49]. These low hybridisation rates suggest at least partial reproductive/ecological isolation between biotypes [20,47], with some rare haplotypes identified as specific to one biotype (Table 2, see next section). The sympatric co-occurrence of both biotypes and their hybrids with their opportunistic feeding behaviour [47,51] hints at the potential danger of viral transmissions from birds to humans (i.e., to act as bridge for disease vectors). However, the low frequency of hybrids likely limits their potential epidemiological role in WNV outbreaks.
While Cx. pipiens s.s. and Cx. torrentium are sympatric in some areas, the latter species was not collected in the north of Belgium (Figure 1). Again, this may be a sampling artefact, since the species was collected all over Belgium during the nationwide MODIRISK mosquito survey (2007–2010) [9,48]. Both species were equally observed in different habitats and have adapted to a life in human neighbourhoods [52], with Cx. torrentium and Cx. pipiens s.s. larvae often found in small artificial and nutrient-rich bodies of water [7,52]. Thus, both species are widespread in Belgium, but their exact distribution limits in Europe remain to be determined.
4.2. COI Haplotype Composition and Genetic Variability
Belgian Cx. torrentium showed a higher COI variability than Cx. pipiens s.s., which is in line with [52,53]. However, we found no evidence of further sub-structuring or taxonomic differentiation within Cx. torrentium, while previous studies reported some morphological variability within the species [53,54]. The limited intraspecific variation within Cx. pipiens s.s. is consistent with [54,55]. Hence, COI haplotype H1 had a prevalence of 61.51% in Cx. pipiens biotype pipiens and of 40.09% in Cx. pipiens biotye molestus. Similar prevalences were reported by [52] (H1 = haplotype 1), [3] (H1 = haplotype A/C), and [56] (H1 = haplotype H). As such, haplotype H1 is widespread and most frequent in northern temperate climates (Germany, Japan, North America, and Russia) [3,52,56]. Situations in which populations show limited genetic variation and consist of a highly frequent haplotype, jointly with a few rare haplotypes, can be explained by either a recent population-wide demographic or range expansion, or a recent bottleneck, possibly in combination with a Wolbachia infection [52]. Such a Wolbachia infection can severely reduce mitochondrial diversity [52,57,58]. This might, in part, explain the limited COI diversity in Cx. pipiens s.s., which shows Wolbachia infection rates of >90% [52,59,60,61], whereas COI diversity might have been retained in Cx. torrentium, within which Wolbachia infections appear to be very rare [59,62].
In Belgium, haplotype H2 was almost exclusively found in Cx. pipiens biotype molestus. This is somewhat unexpected, as this haplotype is rarely found in temperate climates, but associated with (sub)tropical climates ([3] H2 = haplotype E/E1; [56] H2 = haplotype C). Elsewhere, haplotype H2 is prevalent in Cx. quinquefasciatus (42%) and its hybrids with Cx. pipiens s.s. (32%) [3]. Hence, COI haplotypes in Cx. pipiens s.l. are not species-specific [56].
Currently, the biotypes pipiens and molestus of Cx. pipiens s.s. are regarded as different monophyletic evolutionary units undergoing incipient ecological speciation, so that they may be distinct phylogenetic entities [22,27,47,63,64,65]. This was supported by the significant FST estimates found in the present study. The different mating behaviours of both biotypes was considered as an initial factor of a sympatric speciation process [47]. The limited level of hybridisation is not bidirectional, with a mainly male-mediated introgression from molestus to the pipiens biotype [47], which explains the prevalence of H1 (typical of pipiens) and absence of H2 (typical of molestus) in hybrids. This asymmetric introgression may reflect a mating strategy wherein stenogamous molestus males mate with both molestus and pipiens females in above-ground habitats, while pipiens males mate (via specialised swarming behaviour) in open spaces and, therefore, have a higher disposition to mate with pipiens females [47]. An experimental study revealed at least one reproductive isolating mechanism, with females actively avoiding copulation with males of the other biotype, and pipiens females being unsuccessful in receiving molestus males’ sperm [66].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14060486/s1, Figure S1: Neighbour-Joining tree based on the COI sequences available on BOLD/GenBank for Culex species present in Belgium [12,45] including the sequences generated in this study, and identified based on the ACE2 fragment-size analysis. Bootstrap values are indicated at the branches. Sequences were collapsed in species clusters. N = total number of unique sequences included, following Geneious® v10.0.4; Figure S2: Example of ACE2 fragment-size analysis on a 2.5% agarose gel. PCR multiplex including the primers B1246s (Forward), ACEpip (Reverse), ACEtorr (Reverse), and ACEquin (Reverse). Cycling conditions are provided in Table S2. P = Cx. pipiens s.s. (610 bp); T = Cx. torrentium (416 bp); Figure S3: Example of CQ11 fragment-size analysis on a 2.5% agarose gel. PCR multiplex including the primers CQ11F2 (Forward), molCQ11R (Reverse) and pipCQ11R (Reverse). Cycling conditions are provided in Table S2. PP = Culex pipiens biotype pipiens (200 bp); PM = Culex pipiens biotype molestus (250 bp); H = Cx. pipiens biotype pipiens X Cx. pipiens biotype molestus (200 bp and 250 bp); Table S1: Detailed list of DNA-based identified specimens, including their life stage at collection and the trapping method; Table S2: Summary of PCR cycling conditions for the amplification of the COI, ACE2, and CQ11 loci; Table S3: Map codes, municipalities, and coordinates of sampling localities with taxon occurrence; Table S4: Overview of the COI sequencing success per year. N = number of specimens. Reference [67] are cited in the Supplementary Materials.
Author Contributions
Conceptualization: N.S., I.D., K.D.W., W.D., R.M. and W.V.B.; Methodology: N.S., K.D.W., I.D., A.V. (Adwine Vanslembrouck), J.D.W., A.S., I.V. and W.V.B.; Formal Analysis and Software: N.S., A.V. (Ann Vanderheyden), K.M. and S.G.; Data Curation: W.D., N.S. and I.D.; Writing—Original Draft Preparation: A.V. (Ann Vanderheyden) and N.S.; Writing—Review and Editing: I.D., K.D.W., W.D., K.M., S.G., A.V. (Adwine Vanslembrouck), J.D.W., A.S., I.V., M.D.M., T.B., R.M. and W.V.B.; Visualization: A.V. (Ann Vanderheyden) and N.S.; Supervision: I.D., W.V.B., R.M., T.B. and M.D.M.; Project Administration and Funding Acquisition: I.D., W.V.B., R.M., T.B. and M.D.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work is part of the MEMO project (tender: CES-2016-02), funded by the Flemish, Walloon and Brussels regional governments and the Federal Public Service (FPS) Public Health, Food Chain Safety and Environment in the context of the National Environment and Health Action Plan (NEHAP) (Belgium). The Barcoding Facility for Organisms and Tissues of Policy Concern (BopCo-http://bopco.myspecies.info/, accessed 15 May 2022) is financed by the Belgian Science Policy Office (Belspo). The Outbreak Research Team of the Institute of Tropical Medicine is funded by the Department of Economy, Science and Innovation of the Flemish government.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accessed 15 May 2022, reference number: OM747883-OM749568.
Acknowledgments
We would like to thank all cooperating companies for giving access to their private property during mosquito sampling surveys. We would like to thank the laboratory and technical staff at the Institute of Tropical Medicine Antwerp (ITM), at the Royal Belgian Institute of Natural Sciences (RBINS), and at the Royal Museum for Central Africa (RMCA).
Conflicts of Interest
The authors declare no conflict of interest.
References
- ECDC. Culex pipiens Factsheet. 2020. Available online: https://www.ecdc.europa.eu/en/all-topics-z/disease-vectors/facts/mosquito-factsheets/culex-pipiens-factsheet-experts (accessed on 16 February 2022).
- Scholte, E.; Ibáñez-Justicia, A.; Stroo, A.; Zeeuw, J.D.; Hartog, W.D.; Reusken, C. Mosquito collections on incoming intercontinental flights at Schiphol International airport, the Netherlands, 2010–2011. J. Eur. Mosq. Control Assoc. 2014, 32, 17–21. [Google Scholar]
- Shaikevich, E.V.; Vinogradova, E.B.; Bouattour, A.; Gouveia de Almeida, A.P. Genetic diversity of Culex pipiens mosquitoes in distinct populations from Europe: Contribution of Cx. quinquefasciatus in Mediterranean populations. Parasites Vectors 2016, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Petric, D.; Zgomba, M.; Boase, C.; Madon, M.; Dahl, C.; Kaiser, A. Mosquitoes and Their Control; Springer: Heidelberg, Germany; New York, NY, USA, 2010. [Google Scholar]
- Miller, B.R.; Crabtree, M.B.; Savage, H.M. Phylogeny of fourteen Culex mosquito species, including the Culex pipiens complex, inferred from the internal transcribed spacers of ribosomal DNA. Insect Mol. Biol. 1996, 5, 93–107. [Google Scholar] [CrossRef]
- Harbach, R.E. Culex pipiens: Species versus species complex taxonomic history and perspective. J. Am. Mosq. Control Assoc. 2012, 28, 10–23. [Google Scholar] [CrossRef]
- Hesson, J.C.; Rettich, F.; Merdić, E.; Vignjević, G.; Östman, Ö.; Schäfer, M.; Schaffner, F.; Foussadier, R.; Besnard, G.; Medlock, J.; et al. The arbovirus vector Culex torrentium is more prevalent than Culex pipiens in northern and central Europe. Med. Vet. Entomol. 2014, 28, 179–186. [Google Scholar] [CrossRef]
- Dekoninck, W.; Hendrickx, F.; Versteirt, V.; Coosemans, M.; De Clercq, E.M.; Hendrickx, G.; Hance, T.; Grootaert, P. Changes in species richness and spatial distribution of mosquitoes (Diptera: Culicidae) inferred from museum specimen records and a recent inventory: A case study from Belgium suggests recent expanded distribution of arbovirus and malaria vectors. J. Med. Entomol. 2013, 50, 237–243. [Google Scholar] [CrossRef]
- Versteirt, V.; Boyer, S.; Damiens, D.; De Clercq, E.; Dekoninck, W.; Ducheyne, E.; Grootaert, P.; Garros, C.; Hance, T.; Hendrickx, G. Nationwide inventory of mosquito biodiversity (Diptera: Culicidae) in Belgium, Europe. Bull. Èntomol. Res. 2013, 103, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Deblauwe, I.; Sohier, C.; Schaffner, F.; Rakotoarivony, L.M.; Coosemans, M. Implementation of surveillance of invasive mosquitoes in Belgium according to the ECDC guidelines. Parasites Vectors 2014, 7, 201. [Google Scholar] [CrossRef]
- Deblauwe, I.; De Wolf, K.; Smitz, N.; Vanslembrouck, A.; Schneider, A.; De Witte, J.; Verlé, I.; Dekoninck, W.; De Meyer, M.; Backeljau, T.; et al. Monitoring of Exotic Mosquitoes in Belgium (MEMO): Final Report Phase 7 Part 1: MEMO Results; Institute of Tropical Medicine: Antwerp, Belgium, 2020. [Google Scholar]
- Boukraa, S.; Dekoninck, W.; Versteirt, V.; Schaffner, F.; Coosemans, M.; Haubruge, E.; Francis, F. Updated checklist of the mosquitoes (Diptera: Culicidae) of Belgium. J. Vector Ecol. 2015, 40, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Boukraa, S.; de La Grandiere, M.A.; Bawin, T.; Raharimalala, F.N.; Zimmer, J.Y.; Haubruge, E. Diversity and ecology survey of mosquitoes potential vectors in Belgian equestrian farms: A threat prevention of mosquito-borne equine arboviruses. Prev. Vet. Med. 2016, 124, 58–68. [Google Scholar] [CrossRef]
- Harbach, R.E.; Harrison, B.A.; Gad, A.M. Culex (Culex) molestus Forskål (Diptera: Culicidae): Neotype designation, description, variation, and taxonomic status. Proc. Entomol. Soc. Wash. 1984, 86, 521–542. [Google Scholar]
- Razygraev, A.V.; Sulesco, T.M. The Use of the Bayes Factor for identification of Culex pipiens and C. torrentium (Diptera: Culicidae) based on morphometric wing characters. Èntomol. Rev. 2020, 100, 220–227. [Google Scholar] [CrossRef]
- Byrne, K.; Nichols, R.A. Culex pipiens in London Underground tunnels: Differentiation between surface and subterranean populations. Heredity 1999, 82, 7–15. [Google Scholar] [CrossRef]
- Lundström, J. Mosquito-borne viruses in western Europe: A review. J. Vector Ecol. 1999, 24, 1–39. [Google Scholar] [PubMed]
- Vinogradova, E.B. Culex Pipiens Pipiens Mosquitoes: Taxonomy, Distribution, Ecology, Physiology, Genetics, Applied Importance and control; Pensoft: Sofia, Bulgaria, 2000. [Google Scholar]
- Rudolf, M.; Czajka, C.; Börstler, J.; Melaun, C.; Jöst, H.; von Thien, H.; Badusche, M.; Becker, N.; Schmidt-Chanasit, J.; Krüger, A.; et al. First nationwide surveillance of Culex pipiens Complex and Culex torrentium mosquitoes demonstrated the presence of Culex pipiens biotype pipiens/molestus hybrids in Germany. PLoS ONE 2013, 8, e71832. [Google Scholar] [CrossRef]
- Haba, Y.; McBride, L. Origin and status of Culex pipiens mosquito ecotypes. Curr. Biol. 2022, 32, R237–R246. [Google Scholar] [CrossRef]
- Becker, N.; Jöst, A.; Weitzel, T. The Culex pipiens complex in Europe. J. Am. Mosq. Control Assoc. 2012, 28, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, A.A.; Masri, R.A.; Khrabrova, N.V.; Sibataev, A.K.; Fritz, M.L.; Sharakhova, M.V. Genomic differentiation and intercontinental population structure of mosquito vectors Culex pipiens pipiens and Culex pipiens molestus. Sci. Rep. 2020, 10, 7504. [Google Scholar] [CrossRef] [PubMed]
- Fritz, M.L.; Walker, E.D.; Miller, J.R.; Severson, D.W.; Dworkin, I. Divergent host preferences of above- and below-ground Culex pipiens mosquitoes and their hybrid offspring. Med. Vet. Entomol. 2015, 29, 115–123. [Google Scholar] [CrossRef]
- Osório, H.C.; Zé-Zé, L.; Amaro, F.; Nunes, A.; Alves, M.J. Sympatric occurrence of Culex pipiens (Diptera, Culicidae) biotypes pipiens, molestus and their hybrids in Portugal, Western Europe: Feeding patterns and habitat determinants. Med. Vet. Entomol. 2014, 28, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Sousa, C.A.; Vicente, J.L.; Pinho, L.; Calderón, I.; Arez, E.; Almeida, A.P.; Donnelly, M.J.; Pinto, J. Feeding patterns of molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in a region of high hybridization. Parasites Vectors 2013, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Brugman, V.A.; Hernández-Triana, L.M.; Medlock, J.M.; Fooks, A.R.; Carpenter, S.; Johnson, N. The role of Culex pipiens L. (Diptera: Culicidae) in virus transmission in Europe. Int. J. Environ. Res. Public Health 2018, 15, 389. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, D.M.; Keyghobadi, N.; Malcolm, C.A.; Mehmet, C.; Schaffner, F.; Mogi, M.; Fleischer, R.C.; Wilkerson, R.C. Emerging Vectors in the Culex pipiens Complex. Science 2004, 303, 1535–1538. [Google Scholar] [CrossRef]
- Gomes, B.; Kioulos, E.; Papa, A.; Almeida, A.P.G.; Vontas, J.; Pinto, J. Distribution and hybridization of Culex pipiens forms in Greece during the West Nile virus outbreak of 2010. Infect. Genet. Evol. 2013, 16, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Ciota, A.T.; Chin, P.A.; Kramer, L.D. The effect of hybridization of Culex pipiens complex mosquitoes on transmission of West Nile virus. Parasites Vectors 2013, 6, 305. [Google Scholar] [CrossRef]
- Hesson, J.C.; Verner-Carlsson, J.; Larsson, A.; Ahmed, R.; Lundkvist, Å.; Lundström, J.O. Culex torrentium mosquito role as major enzootic vector defined by rate of Sindbis virus infection, Sweden, 2009. Emerg. Infect. Dis. 2015, 21, 875. [Google Scholar] [CrossRef] [PubMed]
- Leggewie, M.; Badusche, M.; Rudolf, M.; Jansen, S.; Börstler, J.; Krumkamp, R.; Huber, K.; Krüger, A.; Schmidt-Chanasit, J.; Tannich, E.; et al. Culex pipiens and Culex torrentium populations from Central Europe are susceptible to West Nile virus infection. One Health 2016, 2, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Heitmann, A.; Lühken, R.; Leggewie, M.; Helms, M.; Badusche, M.; Rossini, G.; Schmidt-Chanasit, J.; Tannich, E. Culex torrentium: A potent vector for the transmission of west nile virus in central Europe. Viruses 2019, 11, 492. [Google Scholar] [CrossRef]
- ECDC. Surveillance and Disease Data for West Nile Virus Infections. 2020. Available online: https://www.ecdc.europa.eu/en/west-nile-fever/surveillance-and-disease-data (accessed on 21 February 2022).
- Weitzel, T.; Braun, K.; Collado, A.; Jöst, A.; Becker, N. Distribution and frequency of Culex pipiens and Culex torrentium (Culicidae) in Europe and diagnostic allozyme markers. Eur. Mosq. Bull. 2011, 29, 22–37. [Google Scholar]
- Calistri, P.; Giovannini, A.; Hubalek, Z.; Ionescu, A.; Monaco, F.; Savini, G.; Lelli, R. Epidemiology of West Nile in Europe and in the Mediterranean Basin. Open Virol. J. 2010, 4, 29. [Google Scholar] [CrossRef]
- García-Carrasco, J.M.; Muñoz, A.R.; Olivero, J.; Segura, M.; Real, R. Predicting the spatio-temporal spread of West Nile virus in Europe. PLoS Negl. Trop. Dis. 2021, 15, e0009022. [Google Scholar] [CrossRef] [PubMed]
- Gunay, F.; Picard, M.; Robert, V. MosKeyTool, An Interactive Identification Key for Mosquitoes of Euro-Mediterranean. 2018. Available online: http://medilabsecure.com/moskeytool (accessed on 5 August 2020).
- Wickham, H.; François, R.; Henry, L.; Müller, K. A Grammar of Data Manipulation dplyr 1.0.8. 2020. Available online: https://dplyr.tidyverse.org/index.html (accessed on 1 April 2022).
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hebert, P.; Cywinska, A.; Ball, S.; DeWaard, J. Biological identifications through DNA barcodes. Proc. R. Soc. London Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenback, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Èntomol. Soc. Am. 1994, 87, 653–701. [Google Scholar] [CrossRef]
- Ibáñez-Justicia, A.; Smitz, N.; den Hartog, W.; van de Vossenberg, B.; De Wolf, K.; Deblauwe, I.; Van Bortel, W.; Jacobs, F.; Vaux, A.G.C.; Medlock, J.M.; et al. Detection of exotic mosquito species (Diptera: Culicidae) at international airports in Europe. Int. J. Environ. Res. Public Health 2020, 17, 3450. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Fonseca, D.M. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae). Am. J. Trop. Med. Hyg. 2004, 70, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Bahnck, C.M.; Fonseca, D.M. Rapid assay to identify the two genetic forms of Culex (Culex) pipiens L. (Diptera: Culicidae) and hybrid populations. Am. J. Trop. Med. Hyg. 2006, 75, 251–255. [Google Scholar] [CrossRef]
- De Wolf, K.; Vanderheyden, A.; Deblauwe, I.; Smitz, N.; Gombeer, S.; Vanslembrouck, A.; Meganck, K.; Dekoninck, W.; DE Meyer, M.; Backeljau, T.; et al. First record of the West Nile virus bridge vector Culex modestus Ficalbi (Diptera: Culicidae) in Belgium, validated by DNA barcoding. Zootaxa 2021, 4920, 131–139. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Gomes, B.; Sousa, C.A.; Novo, M.T.; Freitas, F.B.; Alves, R.; Côrte-Real, A.R.; Salgueiro, P.; Donnelly, M.J.; Almeida, A.P.; Pinto, J. Asymmetric introgression between sympatric molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in the Comporta region, Portugal. BMC Evol. Biol. 2009, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Versteirt, V.; De Clercq, E.; Dekoninck, W.; Damiens, D.; Ayrinhac, A.; Jacobs, F.; Bortel, W.V. MODIRISK: Mosquito Vectors of Disease: Spatial Biodiversity, Drivers of Change, and Risk. Antwerp, Belgium. 2011. Available online: https://www.belspo.be/belspo/SSD/science/Reports/MODIRISK_Phase 1 Summary.pdf (accessed on 31 January 2017).
- Di Luca, M.; Toma, L.; Boccolini, D.; Severini, F.; La Rosa, G.; Minelli, G.; Bongiorno, G.; Montarsi, F.; Arnoldi, D.; Capelli, G.; et al. Ecological Distribution and CQ11 Genetic Structure of Culex pipiens Complex (Diptera: Culicidae) in Italy. PLoS ONE 2016, 11, e0146476. [Google Scholar] [CrossRef]
- Zittra, C.; Flechl, E.; Kothmayer, M.; Vitecek, S.; Rossiter, H.; Zechmeister, T.; Fuehrer, H.-P. Ecological characterization and molecular differentiation of Culex pipiens complex taxa and Culex torrentium in eastern Austria. Parasites Vectors 2016, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Hamer, G.L.; Molaei, G.; Walker, E.D.; Goldberg, T.L.; Kitron, U.D.; Andreadis, T.G. Genetic variation associated with mammalian feeding in Culex pipiens from a West Nile virus epidemic region in Chicago, Illinois. Vector-Borne Zoonotic Dis. 2009, 9, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Werblow, A.; Klimpel, S.; Bolius, S.; Dorresteijn, A.W.C.; Sauer, J.; Melaun, C. Population structure and distribution patterns of the sibling mosquito species Culex pipiens and Culex torrentium (Diptera: Culicidae) reveal different evolutionary paths. PLoS ONE 2014, 9, e102158. [Google Scholar] [CrossRef] [PubMed]
- Werblow, A.; Bolius, S.; Dorresteijn, A.W.C.; Melaun, C.; Klimpel, S. Diversity of Culex torrentium Martini, 1925—A potential vector of arboviruses and filaria in Europe. Parasitol. Res. 2013, 112, 2495–2501. [Google Scholar] [CrossRef]
- Fedorova, M.V.; Shaikevich, E.V. Morphological and molecular-genetic distinctions between adult mosquitoes Culex torrentium Martini and C. pipiens Linnaeus (Diptera, Culicidae) from Moscow Province. Èntomol. Rev. 2007, 87, 127–135. [Google Scholar] [CrossRef]
- Guillemaud, T.; Pasteur, N.; Rousset, F. Contrasting levels of variability between cytoplasmic genomes and incompatibility types in the mosquito Culex pipiens. Proc. R. Soc. London Ser. B Biol. Sci. 1997, 264, 245–251. [Google Scholar] [CrossRef]
- Koosha, M.; Oshaghi, M.A.; Sedaghat, M.M.; Vatandoost, H.; Azari-Hamidian, S.; Abai, M.R.; Hanafi-Bojd, A.A.; Mohtarami, F. Sequence analysis of mtDNA COI barcode region revealed three haplotypes within Culex pipiens assemblage. Exp. Parasitol. 2017, 181, 102–110. [Google Scholar] [CrossRef]
- Cariou, M.; Duret, L.; Charlat, S. The global impact of Wolbachia on mitochondrial diversity and evolution. J. Evol. Biol. 2017, 30, 2204–2210. [Google Scholar] [CrossRef]
- Hurst, G.D.D.; Jiggins, F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proc. R. Soc. B Boil. Sci. 2005, 272, 1525–1534. [Google Scholar] [CrossRef]
- Bergman, A.; Hesson, J.C. Wolbachia prevalence in the vector species Culex pipiens and Culex torrentium in a Sindbis virus-endemic region of Sweden. Parasites Vectors 2021, 14, 428. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Lagnel, J.; Raymond, M.; Bourtzis, K.; Fort, P.; Weill, M. Transposable element polymorphism of Wolbachia in the mosquito Culex pipiens: Evidence of genetic diversity, superinfection and recombination. Mol. Ecol. 2005, 14, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
- Vinogradova, E.B.; Fedorova, M.V.; Shaikevich, E.V. Endosymbiotic bacterium Wolbachia pipientis in synanthropic populations of the mosquito Culex pipiens pipiens L. (Diptera, Culicidae). Dokl. Biol. Sci. 2003, 389, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Leggewie, M.; Krumkamp, R.; Badusche, M.; Heitmann, A.; Jansen, S.; Schmidt-Chanasit, J.; Tannich, E.; Becker, S.C. Culex torrentium mosquitoes from Germany are negative for Wolbachia. Med. Vet. Entomol. 2018, 32, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Shaikevich, E. PCR-RFLP of the COI gene reliably differentiates Cx. pipiens, Cx. pipiens f. molestus and Cx. torrentium of the Pipiens Complex. Eur. Mosq. Bull. 2007, 23, 25–30. [Google Scholar]
- Gomes, B.; Wilding, C.S.; Weetman, D.; Sousa, C.A.; Novo, M.T.; Savage, H.M.; Almeida, A.P.G.; Pinto, J.; Donnelly, M.J. Limited genomic divergence between intraspecific forms of Culex pipiens under different ecological pressures. BMC Evol. Biol. 2015, 15, 197. [Google Scholar] [CrossRef]
- Kothera, L.; Godsey, M.; Mutebi, J.P.; Savage, H.M. A comparison of aboveground and belowground populations of Culex pipiens (Diptera: Culicidae) mosquitoes in Chicago, Illinois, and New York City, New York, using microsatellites. J. Med. Entomol. 2010, 47, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Trocke, S.; Sim, C. Comparative studies of stenogamous behaviour in the mosquito Culex pipiens complex. Med. Vet. Entomol. 2018, 32, 427–435. [Google Scholar] [CrossRef]
- Bourguet, D.; Fonseca, D.; Vourch, G.; Dubois, M.; Chandre, F.; Severini, C.; Raymond, M. The acetylcholinesterase gene Ace: A diagnostic marker for the pipiens and quinquefasciatus forms of the Culex pipiens complex. J. Am. Mosq. Control Assoc. 1998, 14, 390–396. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).