Genomic Characterization of SNPs for Genetic Differentiation and Selection in Populations from the American Oil Palm [Elaeis oleifera (Kunth) Cortés] Germplasm Bank from Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genotyping by Sequencing and SNP Selection
2.3. Genetic Analysis
2.4. Alignment of SNPs Sequences to Elaeis guineensis and E. oleifera Genomes
2.5. Genomic Characterization and Functional Annotation
3. Results
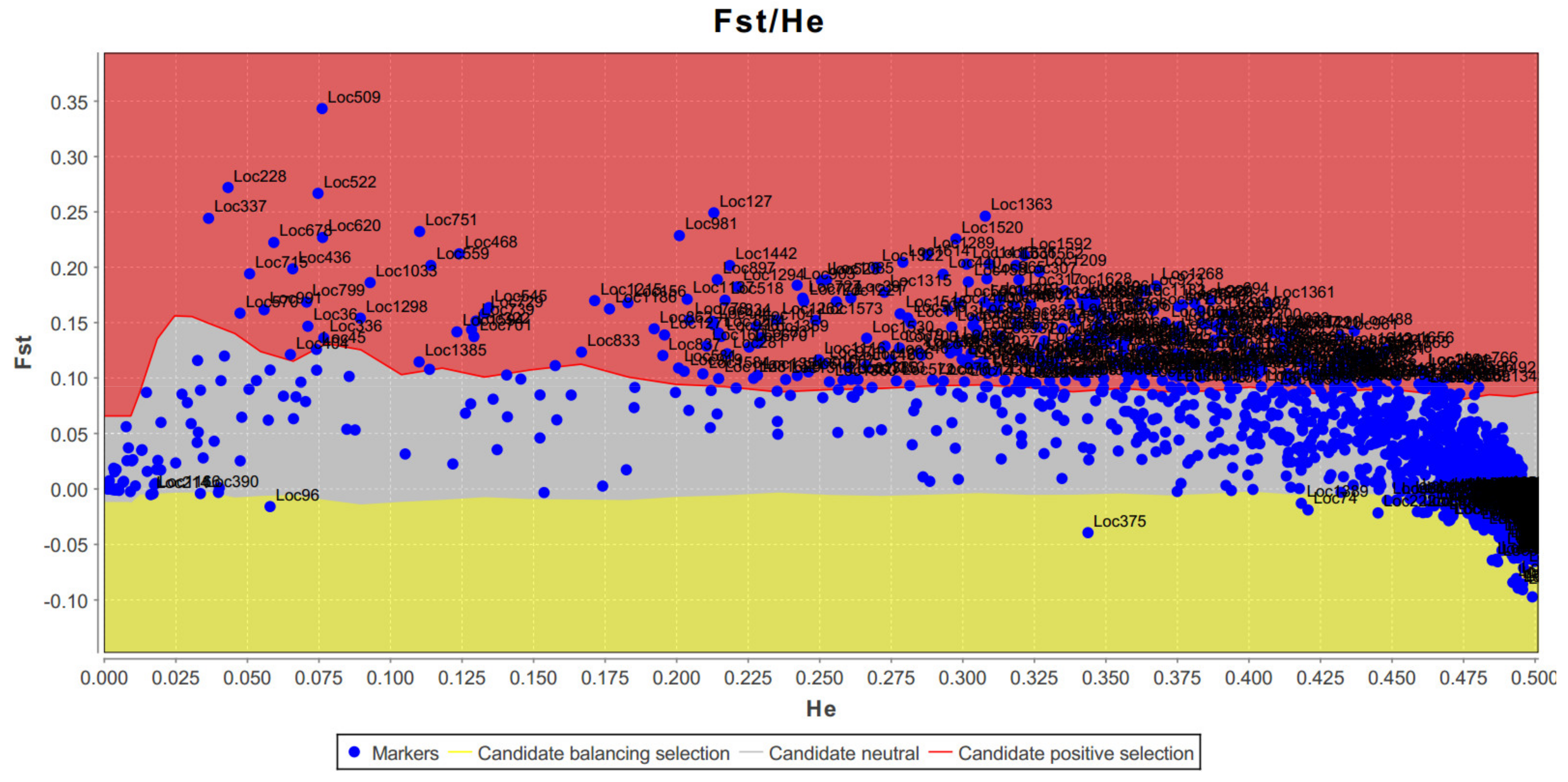
3.1. Genetics Analysis—Genotypic Differentiation in E. oleifera
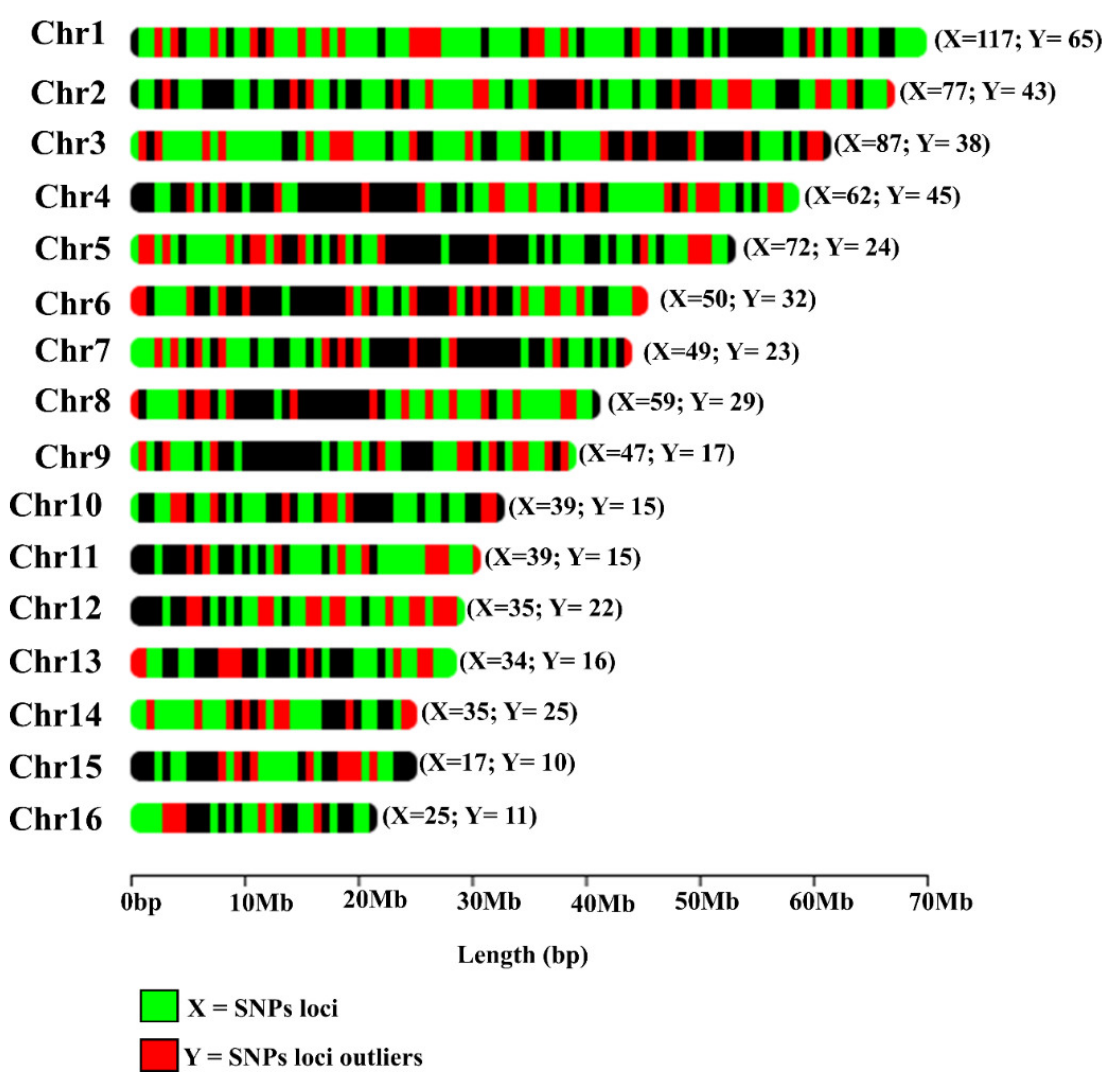
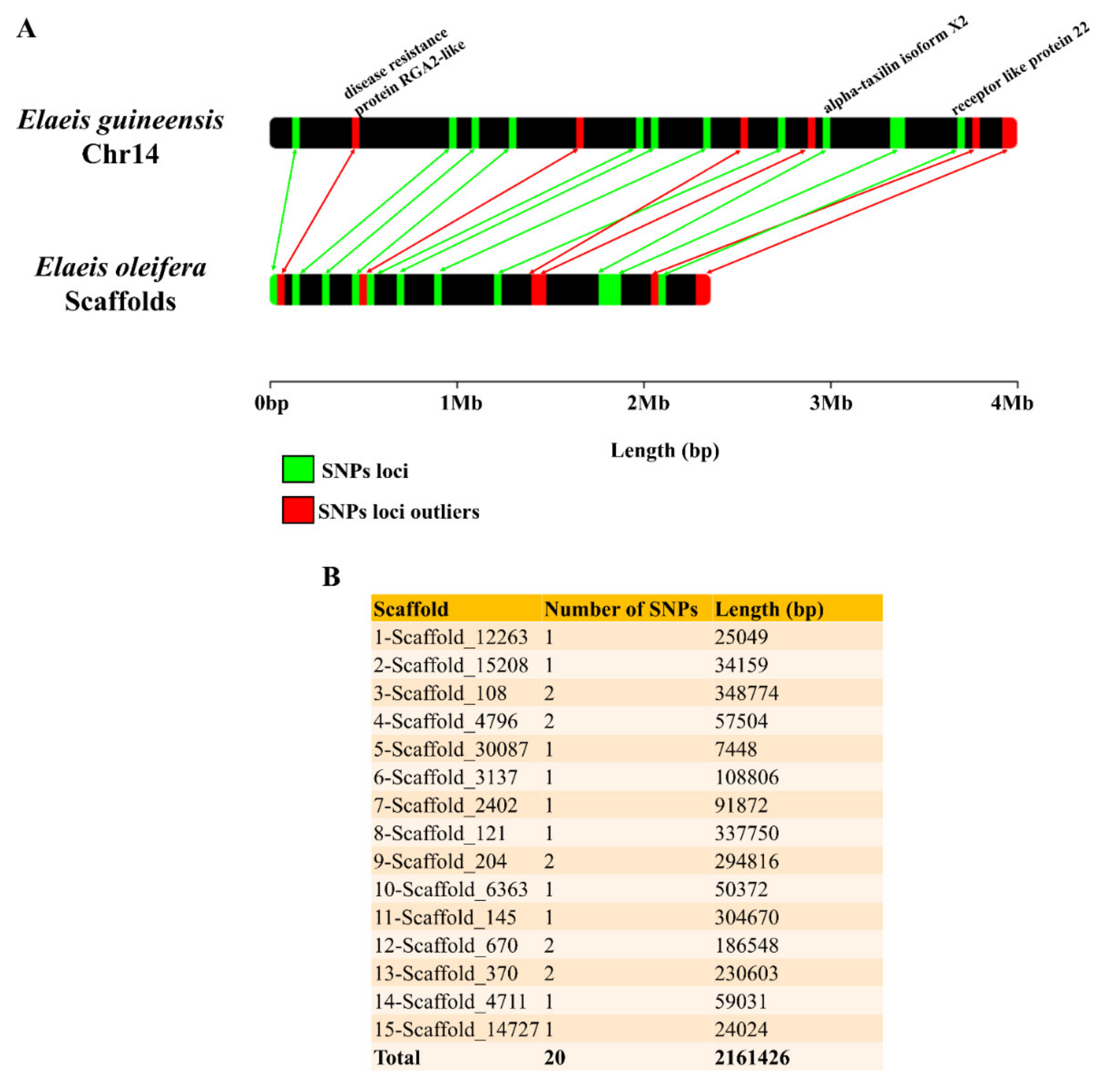
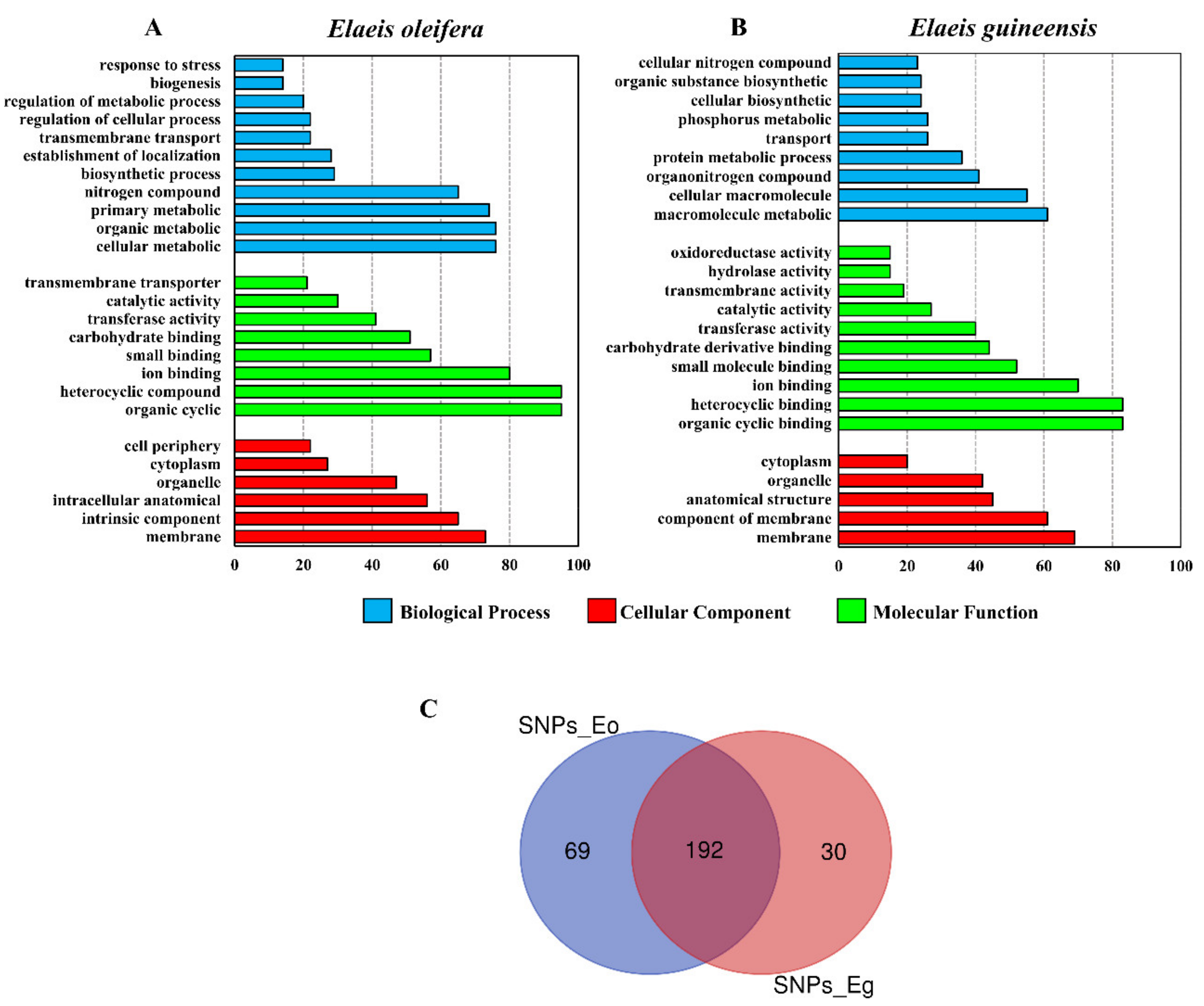
3.2. Genomic Characterization of SNPs in E. oleifera and E. guineensis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.J. The future of oil palm as a major global crop: Opportunities and challenges. J. Oil Palm Res. 2014, 26, 1–24. [Google Scholar]
- Barcelos, E.; Amblard, P.; Berthaud, J.; Seguin, M. Genetic diversity and relationship in American and African oil palm as revealed by RFLP and AFLP molecular markers. Pesqui. Agropecu. Bras. 2002, 37, 1105–1114. [Google Scholar] [CrossRef]
- Dransfield, J.; Uhl, N.W.; Asmussen, C.B.; Baker, W.J.; Harley, M.M.; Lewis, C.E. A new phylogenetic classification of the palm family, Arecaceae. Kew Bull. 2005, 60, 559–569. [Google Scholar]
- Cunha, R.D.; Lopes, R.; Rocha, R.; Lima, W.; Teixeira, P.; Barcelos, E.; Rodrigues, M. Domesticação e melhoramento de caiaué. In Domesticação e Melhoramento: Espécies Amazônicas; Editora da Universidade de Viçosa: Viçosa, Brazil, 2009; pp. 275–296. [Google Scholar]
- de Farias, M.P.; de Capdeville, G.; Falcão, R.; de Moraes, P.B.; Leão, A.P.; Camillo, J.; da Cunha, R.N.V.; Alves, A.A.; Júnior, M.T.S. Microscopic characterization of American oil palm (Elaeis oleifera (Kunth) Cortés) floral development. Flora 2018, 243, 88–100. [Google Scholar] [CrossRef]
- Meléndez, M.R.; Ponce, W.P. Pollination in the oil palms Elaeis guineensis, E. oleifera and their hybrids (OxG), in tropical America. Pesqui. Agropecu. Trop. 2016, 46, 102–110. [Google Scholar] [CrossRef]
- Bittencourt, C.B.; de Castro Lins, P.; de Jesus Boari, A.; Quirino, B.F.; Teixeira, W.G.; Junior, M.T.S. Oil Palm Fatal Yellowing (FY), a Disease with an Elusive Causal Agent; IntechOpen: London, UK, 2021. [Google Scholar]
- Okoye, M.; Bakoumé, C.; Uguru, M.; Singh, R.; Okwuagwu, C. Genetic relationships between elite oil palms from Nigeria and selected breeding and germplasm materials from Malaysia via Simple Sequence Repeat (SSR) Markers. J. Agric. Sci. 2016, 8, 159. [Google Scholar] [CrossRef]
- Cardona, C.C.C.; Coronado, Y.M.; Conronado, A.C.M.; Ochoa, I. Genetic diversity in oil palm (Elaeis guineensis Jacq) using RAM (Random Amplified Microsatellites). Bragantia 2018, 77, 546–556. [Google Scholar] [CrossRef]
- Budiman, L.F.; Apriyanto, A.; Pancoro, A.; Sudarsono, S. Genetic diversity analysis of Tenera × Tenera and Tenera × Pisifera Crosses and D self of oil palm (Elaeis guineensis) parental populations originating from Cameroon. Biodivers. J. Biol. Divers. 2019, 20, 937–949. [Google Scholar] [CrossRef]
- Moretzsohn, M.d.C.; Ferreira, M.; Amaral, Z.; Coelho, P.J.d.A.; Grattapaglia, D.; Ferreira, M.E. Genetic diversity of Brazilian oil palm (Elaeis oleifera HBK) germplasm collected in the Amazon Forest. Euphytica 2002, 124, 35–45. [Google Scholar] [CrossRef]
- Araya, E.; Alvarado, A.; Escobar, R. Use of DNA markers for fingerprinting compact clones and determining the genetic relationship between Elaeis oleifera germplasm origins. In Proceedings of the International Society for Oil Palm Breeders (ISOPB), Kuala Lumpur, Malaysia, 9–12 November 2009; pp. 4–5. [Google Scholar]
- Arias, D.; Montoya, C.; Romero, H. Molecular characterization of oil palm Elaeis guineensis Jacq. materials from Cameroon. Plant Genet. Resour. 2013, 11, 140–148. [Google Scholar] [CrossRef]
- Ithnin, M.; Teh, C.-K.; Ratnam, W. Genetic diversity of Elaeis oleifera (HBK) Cortes populations using cross species SSRs: Implication’s for germplasm utilization and conservation. BMC Genet. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Natawijaya, A.; Ardie, S.W.; Syukur, M.; Maskromo, I.; Hartana, A.; Sudarsono, S. Genetic structure and diversity between and within African and American oil palm species based on microsatellite markers. Biodivers. J. Biol. Divers. 2019, 20, 3365. [Google Scholar] [CrossRef]
- Pereira, V.M.; Filho, J.A.F.; Leão, A.P.; Vargas, L.H.G.; de Farias, M.P.; Rios, S.d.A.; da Cunha, R.N.V.; Formighieri, E.F.; Alves, A.A.; Souza, M.T. American oil palm from Brazil: Genetic diversity, population structure, and core collection. Crop Sci. 2020, 60, 3212–3227. [Google Scholar] [CrossRef]
- Rios, S.d.A.; da Cunha, R.; Lopes, R.; da Silva, E. Recursos Genéticos de Palma de Óleo (Elaeis guineensis Jacq.) e Caiuaé (Elaeis oleifera (HBK) Cortes); Embrapa Amazônia Ocidental-Documentos (INFOTECA-E): Itacoatiara, Brazil, 2012. [Google Scholar]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations: Vol. 2. The Theory of Gene Frequencies; The University of Chicago Press: Chicago, IL, USA, 1969. [Google Scholar]
- Hastings, W.K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 1970, 57, 97–109. [Google Scholar] [CrossRef]
- Levene, H. Robust tests for equality of variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling; Olkin, I., Ghurye, S.G., Hoeffding, W., Madow, W.G., Mann, H.B., Eds.; Stanford University Press: Redwood City, CA, USA, 1960. [Google Scholar]
- Antao, T.; Lopes, A.; Lopes, R.J.; Beja-Pereira, A.; Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a F ST-outlier method. BMC Bioinform. 2008, 9, 1–5. [Google Scholar] [CrossRef]
- Singh, R.; Ong-Abdullah, M.; Low, E.-T.L.; Manaf, M.A.A.; Rosli, R.; Nookiah, R.; Ooi, L.C.-L.; Ooi, S.E.; Chan, K.-L.; Halim, M.A. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 2013, 500, 335–339. [Google Scholar] [CrossRef]
- Filho, J.A.F.; de Brito, L.S.; Leão, A.P.; Alves, A.A.; Formighieri, E.F.; Souza, M.T. In silico approach for characterization and comparison of repeats in the genomes of oil and date palms. Bioinform. Biol. Insights 2017, 11, 1177932217702388. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Slatkin, M. Rare alleles as indicators of gene flow. Evolution 1985, 39, 53–65. [Google Scholar] [CrossRef]
- Barton, N.; Slatkin, M. A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 1986, 56, 409–415. [Google Scholar] [CrossRef]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef]
- Kim, C.; Guo, H.; Kong, W.; Chandnani, R.; Shuang, L.-S.; Paterson, A.H. Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci. 2016, 242, 14–22. [Google Scholar] [CrossRef]
- Steele, K.A.; Quinton-Tulloch, M.J.; Amgai, R.B.; Dhakal, R.; Khatiwada, S.P.; Vyas, D.; Heine, M.; Witcombe, J.R. Accelerating public sector rice breeding with high-density KASP markers derived from whole genome sequencing of indica rice. Mol. Breed. 2018, 38, 1–13. [Google Scholar] [CrossRef]
- Heim, C.B.; Gillman, J.D. Genotyping-by-sequencing-based investigation of the genetic architecture responsible for a∼ sevenfold increase in soybean seed stearic acid. G3 Genes Genomes Genet. 2017, 7, 299–308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gouesnard, B.; Negro, S.; Laffray, A.; Glaubitz, J.; Melchinger, A.; Revilla, P.; Moreno-Gonzalez, J.; Madur, D.; Combes, V.; Tollon-Cordet, C. Genotyping-by-sequencing highlights original diversity patterns within a European collection of 1191 maize flint lines, as compared to the maize USDA genebank. Theor. Appl. Genet. 2017, 130, 2165–2189. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Wang, W.; Gong, S.; Zuo, J.; Li, S.; Xu, S. High density linkage map construction and mapping of yield trait QTLs in maize (Zea mays) using the genotyping-by-sequencing (GBS) technology. Front. Plant Sci. 2017, 8, 706. [Google Scholar] [CrossRef] [PubMed]
- Eltaher, S.; Sallam, A.; Belamkar, V.; Emara, H.A.; Nower, A.A.; Salem, K.F.; Poland, J.; Baenziger, P.S. Genetic diversity and population structure of F3: 6 Nebraska winter wheat genotypes using genotyping-by-sequencing. Front. Genet. 2018, 9, 76. [Google Scholar] [CrossRef]
- Hussain, W.; Baenziger, P.S.; Belamkar, V.; Guttieri, M.J.; Venegas, J.P.; Easterly, A.; Sallam, A.; Poland, J. Genotyping-by-sequencing derived high-density linkage map and its application to QTL mapping of flag leaf traits in bread wheat. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Slatkin, M.; Barton, N.H. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 1989, 43, 1349–1368. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Ridley, M. Evolução; Artmed Editora: São Paulo, Brazil, 2009. [Google Scholar]
- Arias, D.; González, M.; Prada, F.; Restrepo, E.; Romero, H. Morpho-agronomic and molecular characterisation of oil palm Elaeis guineensis Jacq. material from Angola. Tree Genet. Genomes 2013, 9, 1283–1294. [Google Scholar] [CrossRef]
- Arias, D.; González, M.; Prada, F.; Ayala-Diaz, I.; Montoya, C.; Daza, E.; Romero, H.M. Genetic and phenotypic diversity of natural American oil palm (Elaeis oleifera (HBK) Cortés) accessions. Tree Genet. Genomes 2015, 11, 1–13. [Google Scholar] [CrossRef]
- Billotte, N.; Risterucci, A.-M.; Barcelos, E.; Noyer, J.-L.; Amblard, P.; Baurens, F.-C. Development, characterisation, and across-taxa utility of oil palm (Elaeis guineensis Jacq.) microsatellite markers. Genome 2001, 44, 413–425. [Google Scholar] [CrossRef]
- Zaki, N.M.; Ismail, I.; Rosli, R.; Chin, T.N.; Singh, R. Development and characterization of Elaeis oleifera microsatellite markers. Sains Malays. 2010, 39, 909–912. [Google Scholar]
- Zaki, N.M.; Singh, R.; Rosli, R.; Ismail, I. Elaeis oleifera genomic-SSR markers: Exploitation in oil palm germplasm diversity and cross-amplification in Arecaceae. Int. J. Mol. Sci. 2012, 13, 4069–4088. [Google Scholar] [CrossRef]
- Bakoumé, C.; Wickneswari, R.; Siju, S.; Rajanaidu, N.; Kushairi, A.; Billotte, N. Genetic diversity of the world’s largest oil palm (Elaeis guineensis Jacq.) field genebank accessions using microsatellite markers. Genet. Resour. Crop Evol. 2015, 62, 349–360. [Google Scholar] [CrossRef]
- Niu, S.; Song, Q.; Koiwa, H.; Qiao, D.; Zhao, D.; Chen, Z.; Liu, X.; Wen, X. Genetic diversity, linkage disequilibrium, and population structure analysis of the tea plant (Camellia sinensis) from an origin center, Guizhou plateau, using genome-wide SNPs developed by genotyping-by-sequencing. BMC Plant Biol. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Thurow, L.B.; Gasic, K.; Raseira, M.d.C.B.; Bonow, S.; Castro, C.M. Genome-wide SNP discovery through genotyping by sequencing, population structure, and linkage disequilibrium in Brazilian peach breeding germplasm. Tree Genet. Genomes 2020, 16, 1–14. [Google Scholar] [CrossRef]
- Delfini, J.; Moda-Cirino, V.; dos Santos Neto, J.; Ruas, P.M.; Sant’Ana, G.C.; Gepts, P.; Gonçalves, L.S.A. Population structure, genetic diversity and genomic selection signatures among a Brazilian common bean germplasm. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
| Population | Geographic Region | Locality | Number of Subsamples | Number of Plants |
|---|---|---|---|---|
| 1 | Manaus | Caldeirão | 7 | 18 |
| 2 | Careiro | 25 | 68 | |
| 3 | Manacapuru | 1 | 3 | |
| 4 | Iranduba | 2 | 6 | |
| Subtotal | 35 | 95 | ||
| 5 | Rio Amazonas | Amatari | 11 | 31 |
| 6 | Autazes | 11 | 28 | |
| 7 | Maués | 11 | 32 | |
| Subtotal | 33 | 91 | ||
| 8 | Rio Solimões | Anori | 3 | 9 |
| 9 | B. Constant | 1 | 3 | |
| 10 | Coari | 19 | 54 | |
| 11 | Tefé | 5 | 14 | |
| 12 | Tonantins | 4 | 12 | |
| Subtotal | 32 | 92 | ||
| 13 | Rio Negro | Acajatuba | 10 | 29 |
| 14 | Barcelos | 2 | 2 | |
| 15 | Moura | 11 | 32 | |
| Subtotal | 23 | 63 | ||
| 16 | Caracaraí | BR174 | 12 | 35 |
| 17 | Vila Moderna | 6 | 18 | |
| Subtotal | 18 | 53 | ||
| 18 | Rio Madeira | Manicoré | 58 | 140 |
| 19 | Novo Aripuanã | 7 | 19 | |
| Subtotal | 65 | 159 | ||
| TOTAL | 206 | 553 | ||
| Population | 1-Qintra | 1-Qinter | Fis |
|---|---|---|---|
| 1 | 0.617 | 0.353 | −0.745 |
| 2 | 0.629 | 0.378 | −0.665 |
| 3 | 0.629 | 0.382 | −0.645 |
| 4 | 0.629 | 0.380 | −0.655 |
| 5 | 0.634 | 0.384 | −0.650 |
| 6 | 0.611 | 0.377 | −0.619 |
| 7 | 0.594 | 0.361 | −0.644 |
| 8 | 0.623 | 0.376 | −0.656 |
| 9 | 0.613 | 0.369 | −0.661 |
| 10 | 0.629 | 0.378 | −0.663 |
| 11 | 0.600 | 0.365 | −0.647 |
| 12 | 0.632 | 0.367 | −0.720 |
| 13 | 0.649 | 0.357 | −0.820 |
| 14 | 0.647 | 0.372 | −0.741 |
| 15 | 0.667 | 0.378 | −0.765 |
| 16 | 0.651 | 0.365 | −0.785 |
| 17 | 0.612 | 0.357 | −0.713 |
| 18 | 0.591 | 0.366 | −0.617 |
| 19 | 0.636 | 0.375 | −0.727 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leão, A.P.; Filho, J.A.F.; Pereira, V.M.; Alves, A.A.; Souza Júnior, M.T. Genomic Characterization of SNPs for Genetic Differentiation and Selection in Populations from the American Oil Palm [Elaeis oleifera (Kunth) Cortés] Germplasm Bank from Brazil. Diversity 2022, 14, 270. https://doi.org/10.3390/d14040270
Leão AP, Filho JAF, Pereira VM, Alves AA, Souza Júnior MT. Genomic Characterization of SNPs for Genetic Differentiation and Selection in Populations from the American Oil Palm [Elaeis oleifera (Kunth) Cortés] Germplasm Bank from Brazil. Diversity. 2022; 14(4):270. https://doi.org/10.3390/d14040270
Chicago/Turabian StyleLeão, André Pereira, Jaire Alves Ferreira Filho, Valquiria Martins Pereira, Alexandre Alonso Alves, and Manoel Teixeira Souza Júnior. 2022. "Genomic Characterization of SNPs for Genetic Differentiation and Selection in Populations from the American Oil Palm [Elaeis oleifera (Kunth) Cortés] Germplasm Bank from Brazil" Diversity 14, no. 4: 270. https://doi.org/10.3390/d14040270
APA StyleLeão, A. P., Filho, J. A. F., Pereira, V. M., Alves, A. A., & Souza Júnior, M. T. (2022). Genomic Characterization of SNPs for Genetic Differentiation and Selection in Populations from the American Oil Palm [Elaeis oleifera (Kunth) Cortés] Germplasm Bank from Brazil. Diversity, 14(4), 270. https://doi.org/10.3390/d14040270







