Abstract
The need for herbal medicinal plants is steadily increasing. Hence, the accurate identification of plant material has become vital for safe usage, avoiding adulteration, and medicinal plant trading. DNA barcoding has shown to be a valuable molecular identification tool for medicinal plants, ensuring the safety and efficacy of plant materials of therapeutic significance. Using morphological characters in genera with closely related species, species delimitation is often difficult. Here, we evaluated the capability of the nuclear barcode ITS2 and plastid DNA barcodes rbcL and matK to identify 20 medicinally important plant species of Caryophyllales. In our analysis, we applied an integrative approach for species discrimination using pairwise distance-based unsupervised operational taxonomic unit “OTU picking” methods, viz., ABGD (Automated Barcode Gap Analysis) and ASAP (Assemble Species by Automatic Partitioning). Along with the unsupervised OTU picking methods, Supervised Machine Learning methods (SML) were also implemented to recognize divergent taxa. Our results indicated that ITS2 was more successful in distinguishing between examined species, implying that it could be used to detect the contamination and adulteration of these medicinally important plants. Moreover, this study suggests that the combination of more than one method could assist in the resolution of morphologically similar or closely related taxa.
1. Introduction
Large numbers of people in developing countries rely on wild plant species for their medicinal needs. Over thousands of plant species are used in traditional medicine in different parts of the world. During ancient and modern culture, the healing properties of certain plants have been identified, and these plants currently play a significant role in the treatment of various diseases [1]. Due to their medicinal properties, there is a continuous and perpetual interest in researching and utilizing these valuable natural resources, as demonstrated by a plethora of literature (e.g., [2,3,4,5,6,7,8,9,10]). Different plant species have been used in ethnomedicine in the Arabian Peninsula since ancient times [5,10]. Sakkir [11] provided an overview of the medicinal plants in the United Arab Emirates (UAE) flora and indicated that roughly 18% of the total plant species identified have medicinal values. These authors attributed such a low representation of medicinal plants in UAE flora to be most likely due to the unknown medicinal properties of the remaining taxa or lack of documentation of their traditional uses. Cybulska et al. [12] reviewed the available information on the medicinal uses of the halophytes in the UAE flora and highlighted the presence of valuable medicinal plants. These medicinally important plants display a specific tolerance to environmental stresses such as high temperature, drought, and salinity. It is expected that these medicinal halophytes might represent a valuable source of phytochemicals in salt marshes, where harsh conditions induce the production of both enzymes and phytochemicals in response to Reactive Oxygen Species (ROS) [12].
A recent survey on the ethnomedicinal knowledge of commonly used medicinal plants in a part of the UAE highlighted the importance of traditional medicinal plants and the need for knowledge documentation [13]. Further, due to various threats to medicinal plants, such as habitat loss and alteration, and overgrazing, there is a need for proper identification and conservation. Phondani et al. [8] documented the ethnobotanical uses of 58 medicinally important plants of the Arabian deserts. They emphasized the need to document ethnobotanical knowledge for sustainability and scientific validation to conserve these valuable medicinal plants native to the Arabian deserts. The above proves the importance of using many UAE plants in folk medicine.
There is a continued increase in the demand for herbal medicinal plants. Therefore, there are some accidental or intentional contaminations and adulteration with non-medicinal plants or other undesirable plant tissues [14,15]. Such contamination could reduce the effectiveness of the active ingredients, which might lead to detrimental health consequences [16,17]. The authentication of plant material has become necessary for safe use, avoiding adulteration, and trade in medicinal plants. Therefore, there is a need for fast authentication methods to authenticate dried herbal medicinal plants from the other components [18]. The detection of adulteration requires accurate, fast molecular techniques for plant identification, especially if it is difficult to discriminate between closely related plants morphologically [19]. In addition, the proper identification and documentation of medicinal plants in the region could add to their conservation and sustainable utilization.
The classical taxonomic techniques based on morphology and anatomy complement molecular techniques for accurately identifying morphologically similar closely related plant taxa. Currently, there are initiatives for generating DNA barcode libraries of vascular plant flora. Completing such libraries and making them available will help fast, accurate identification, which would lead to the better conservation and utilization of native plants, particularly those used in herbal medicine [20,21,22]. In this context, a tool such as DNA barcoding could help resolve these issues and lead to the rapid and accurate identification of medicinal species. Moreover, DNA barcoding could be helpful in the identification of medicinal plants in trade, as most herbal material is traded as dried leaves, roots, and bark or in powdered form, thus contributing to their safe use and avoiding adulteration [23].
DNA barcoding has become a useful complementary tool in diverse disciplines of biological sciences. The application of plant DNA barcodes, especially in floristic investigations, ecology, evolution, and conservation fields, has gained momentum over the last decade [24,25]. Several studies have highlighted the potential applications of DNA barcoding in the accurate identification of taxa, discovery of cryptic species, and as an crucial component in phylogenetics investigations [24,26,27,28]. However, this technique also has some potential limitations, especially in plants where the selected barcode region might lack enough information to provide DNA level species-specific differences and the concurrent observation of such differences at the secondary metabolite level, similar to that observed by Celiński et al. [29].
In medicinal plant research, DNA barcoding is emerging as a valuable molecular identification technique that has greatly ensured the safety and effectiveness of plant materials of medicinal value [30]. The reviews by Techen et al. [31] and Nazar et al. [32] have discussed the selection of the genomic regions as possible barcodes for medicinal plants, including new achievements in the field of DNA barcoding. Those reviews provide a comprehensive overview of DNA-based methods, technologies, and a combination of three or more genomic regions that were investigated for medicinal plant identification by various researchers worldwide.
Over the years, researchers have suggested different coding and noncoding genes in the nuclear and plastid genomes as potential barcodes for plants [33,34,35,36,37]. The types of DNA barcode markers used for plant identification range from a single chloroplast region to a combination of different regions (see [30,31,38]). Significant progress has been made in the identification of medicinal plants using DNA barcoding (e.g., [25,31,39,40,41,42,43,44,45,46,47]. For medicinal plant identification, some researchers have used a combination of markers between matK, rbcL, trnH-psbA, and ITS2 sequences. For example, Schori et al. [48] analyzed the rbcL, matK, and psbA-trnH loci of fourteen species of medicinal plants and found that depending on the plant to be identified, one region was preferred over the other, as a single barcode region is not enough to ensure the species identification.
Moreover, along with the selection of efficient DNA barcode markers for species identification, it is necessary to utilize competent methods for effective species discrimination. The results produced by one or more methods sometimes differ, which could require the implementation of more than one method that must be applied and compared jointly [49,50]. The most conventional DNA barcode analysis method is the pairwise distance-based unsupervised Operational Taxonomic Unit (OTU) picking method, where Automated Barcode Gap Discovery (ABGD) is the most widely used tool, followed by the recently developed Assemble Species by Automatic Partitioning (ASAP) [50,51]. Comparatively, some studies have shown a higher rate of species discrimination using the supervised learning approach [21,52,53,54,55]. Thus, in this study, we used a comparative approach of implementing both unsupervised and supervised methods for species delimitation.
The UAE has not received much attention to digitally record flora in the form of DNA barcodes [22], as there are only three studies cataloging flora of the UAE to date. Moreover, there are scarce studies on the DNA barcodes of medicinal plants [56], and existing studies have amplified three barcode loci for the coding genes matK, rbcL, and rpoC1 in 10 flowering plants from the UAE. Maloukh et al. [57] focused on authenticating the morphological identification of 51 plant species using rbcL and matK regions. Further, Mosa et al. [15] provided evidence that DNA barcoding was efficient in the detection of adulteration in plant-based herbal products in the UAE. Based on the results obtained, these authors also suggested rbcL as a promising barcode locus for resolving their studied species.
Since 2018, the Sharjah Seed Bank and Herbarium have engaged in the process of DNA barcoding the entire UAE flora [20,21,22]. Here, we assessed the capability of plastid DNA barcode markers rbcL and matK, and a nuclear marker, ITS2, for the identification of 20 medicinally important plant species belonging to the order Caryophyllales. The core Caryophyllales represent one of the largest eudicot orders with about 12,000 species and 30 families worldwide [58], and some species are used medicinally [59,60]. Various molecular systematic studies on Caryophyllales are available that have substantially increased our knowledge of their phylogeny [61]. The Caryophyllales is represented in the UAE’s flora by 11 families. These are Aizoaceae, Amaranthaceae, Caryophyllaceae, Frankeniaceae, Gisekiaceae, Molluginaceae, Nyctaginaceae, Plumbaginaceae, Polygonaceae, Portulacaceae, and Tamaricaceae [62,63]. Among the families of Caryophyllales that have difficulties in morphological discrimination, especially during vegetative stages of the life cycles, are Amaranthaceae (e.g., Haloxylon persicum, H. salicornicum, Salicornia persica), Polygonaceae (e.g., Calligonum comosum and C. crinitum), and Tamaricaceae (Tamarix aucheriana and T. nilotica).
The objective of the present study was to barcode the medicinal plant species, compare the discriminatory power of the standard barcode regions, and explore the taxonomic implications in the studied taxa. Establishing a DNA barcoding system could facilitate the conservation of the UAE’s medicinal taxa, help overcome the limitations of morphological characters, and contribute to species identification for their efficient utilization. The study results could help generate DNA barcode libraries of the UAE vascular plant flora, which could be a step toward completing the UAE and global DNA barcoding libraries.
2. Materials and Methods
2.1. Specimen Collection and Data Acquisition
Twenty species from the order Caryophyllales were included in this study. Of these, 13 species (36 samples) were collected from the field between 2019 and 2020, and 7 species were retrieved from the NCBI GenBank. All the studied species are from the United Arab Emirates (Figure 1), and their information is provided in Table S1. Overall, we collected between one and eight specimens per species. Our collection did not involve protected areas or endangered species.
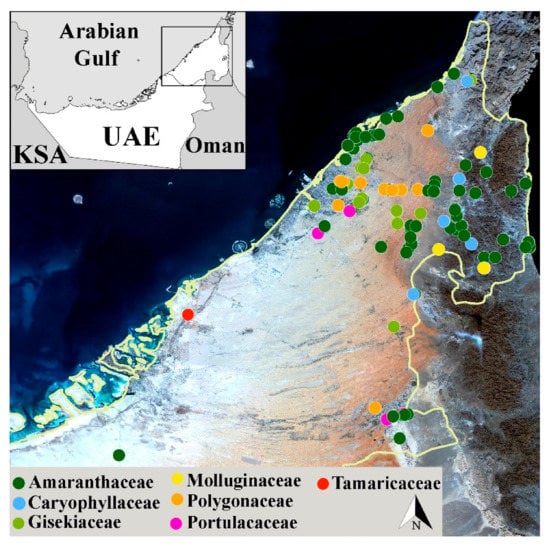
Figure 1.
Map showing collection sites of plant samples for DNA barcoding and seed bank collections.
The collected species included herbaceous plants, shrubs, and trees, which occur in a range of habitats, including sandy, coastal plains, mountain ridges, and wadi beds. Species such as Haloxylon salicornicum, Gisekia pharnacioides, and Aerva javanica had the highest number of accessions, while Tamarix nilotica and Amaranthus hybridus had the lowest number. Morphological identification of all plant species was based on reliable diagnostic characters and available literature on local flora [62,63,64,65], produced by researchers who have worked in the field exploring UAE’s flora for about one decade. The voucher specimens for collected species were curated by the Sharjah Seedbank and Herbarium (SSBH), Environment and Protected Areas Authority (EPAA), for record and references. According to the literature survey, the plant species included in the present study have medicinal values and are used in the treatment of various ailments (Table S2).
2.2. Tissue Sampling and DNA Extraction
Plant tissues were sampled from 36 individuals and dried immediately with silica gel at room temperature for DNA extraction. Unique identifiers were provided to the specimens and the tissue samples. Further, the tissue samples were ground to a fine powder using BeadBlaster 24 microcentrifuge homogenizer. Genomic DNA extraction was then performed using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with minor modifications, where proteinase K was added, followed by the AP1 buffer and RNase A, and the incubation time was increased to 1–3 h. Samples were eluted in Nuclease-Free Water. The isolated DNA was tested for quality by gel electrophoresis (BioRad, Hercules, CA, USA) on a 1% agarose gel and quantity using spectrophotometric analysis (Denovix, Wilmington, DE, USA).
2.3. PCR Amplification and Purification
Two plastid barcode regions, rbcL and matK, and one nuclear ribosomal barcode regions, Internal Transcribed Spacer (ITS2), were amplified via Polymerase Chain Reaction (PCR) (Biorad, Biorad Laboratories, Singapore and Applied Biosystems Veriti Thermal Cycler, Life Technologies Holdings Pte. Ltd., Singapore) using forward and reverse primers of rbcL [35,66], matK (proposed by Ki-Joong Kim, see [67]), and ITS2 [68,69] (Table S3). The 25 μL PCR reaction using a 5× FIREPol master mix was prepared to amplify the respective barcode region. The standard thermal profile of all primers used is shown in Table S3. Modification in the annealing temperature was performed wherever required. PCR products were then verified through gel electrophoresis on a 2% agarose gel. Amplified products were purified using the MEGAquick-spinTM plus total fragment DNA Purification Kit (Intron Biotechnology) and then sequenced commercially.
2.4. Sequence Analysis
Bidirectional sequencing was performed for rbcL, matK, and ITS2 barcode markers. The obtained sequences were assembled and aligned in Geneious Prime v2021 (geneious.com (accessed on 27 December 2021).) and MEGA X. [70] using the Muscle algorithm. The sequences were then submitted to NCBI GenBank through a web-based sequence submission tool ‘BankIt,’ and accessions numbers were obtained for all the studied barcode markers (Table S1).
Further, the sequences were subjected to the taxonomic evaluation using the NCBI GenBank BLASTn to obtain homologies between the fragments [71]. In addition, unsupervised OTU picking methods were employed, where the phylogenetic analysis was performed using MEGA, and the assessment of OTUs was performed using ABGD and ASAP.
Along with the unsupervised OTU picking methods, Supervised Machine Learning methods (SML) were also implemented to recognize divergent taxa. The aligned datasets were formatted to the WEKA’s required file format using the FASTA to WEKA converter [54]. Further, in WEKA machine learning, the random forest and sequential minimal optimization classifiers were used through the 10-folds of cross-validation [72].
Phylogenetic tree construction was carried out in MEGAX. Initially, the phylogenetic model test was performed to determine the best-fit nucleotide substitution model with the lowest BIC scores (Bayesian Information Criterion). Accordingly, Maximum Likelihood (ML) phylogeny was inferred using the Kimura-2-Parameter (K2P) model with discrete gamma distribution was selected for the rbcL dataset. For the matK dataset, a ML phylogenetic tree was constructed using the General Time Reversible model (GTR) with discrete Gamma distribution (G). For the ITS2 dataset, ML phylogeny was achieved using the K2P model with discrete gamma distribution and invariant sites (G + I). All phylogenetic trees were given a bootstrap support of 1000. Moreover, for the phylogenetic tree annotation, the Interactive Tree of Life webserver was used. In addition, the ASAP webserver was used to build the partitions for species delimitation.
3. Results
3.1. Barcode Amplification and Sequencing Success
The core plant barcode markers rbcL, matK, and ITS2 were amplified at various temperature gradients and sequenced successfully (Figure 2).
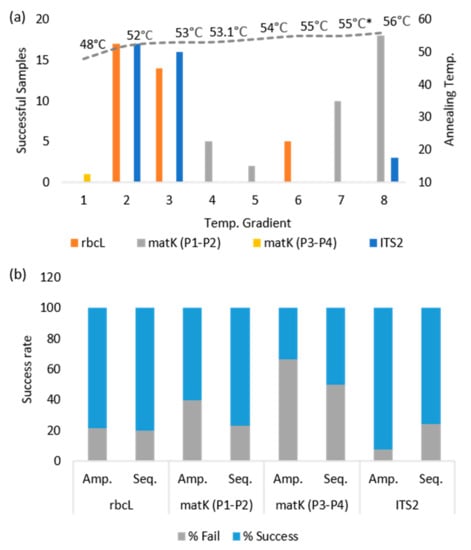
Figure 2.
PCR amplification and sequencing success rate for the DNA barcode markers. (a) Annealing temperature gradients used for the barcode amplification, (* represents the dual annealing temperature 50|55 °C), (b) PCR amplification and sequencing success of the attempted samples.
The rbcL and ITS2 markers showed significant success rates in PCR amplification ranging from 70% to 95%, while matK exhibited a lower success rate of about 60% (Figure 2). In sequencing, the rbcL marker showed the highest (80%) success rate, followed by the matK (76%) and ITS2 (75%) (Figure 2). In addition, another pair of matK markers was used for the amplicon recovery, where only one sample was successfully amplified. Overall, 99 sequences were obtained belonging to the rbcL (35), matK (34), and ITS2 (30) markers, representing about 13 species.
3.2. Taxonomic Assignment
The taxonomic identification of the collected specimens was initially made based on their key morphological characters. Further, the taxonomic evaluation was performed at the NCBI GenBank’s nucleotide database using the NCBI-BLASTn tool.
Overall, we obtained 99 barcodes belonging to 36 specimens and 13 species in the present study. In addition, we retrieved 18 more barcodes belonging to 18 specimens and 11 species from the NCBI GenBank based on the records from previous studies performed on the flora of the UAE. Altogether, the dataset comprised about 117 barcodes, 54 specimens, and 20 species in common, viz., rbcL (n = 49), matK (n = 38), and ITS2 (n = 30).
Those barcode datasets were further analyzed using the unsupervised OTU picking methods, viz., ABGD and ASAP. The ABGD recognized groups of about 10 to 16 species only using J69 and K80 metrics. In addition, the initial partition exhibited lower accuracy in the species resolution than the recursive partition. Thus, the recursive partitions were further taken into consideration. The rbcL showed 6 partitions of which the fifth recursive partition resolved about 28 specimens and 7 species correctly (a priori intraspecific divergence of (P) = 0.0077, relative gap width (X) = 1.0) (Figure 3a and Table 1). In the case of matK, about 9 partitions were recognized, of which the eighth recursive partition was able to successfully resolve 29 specimens and 9 species (at p = 0.035 and X = 1) (Figure 3b and Table 1). In the ITS2 dataset, about 10 partitions were recognized, of which the seventh recursive partition was found to resolve 29 specimens and 10 species (at p = 0.0215 and X = 1) (Figure 3c and Table 1). The simple distance metric showed the lowest accuracy compared to JC69 and K80. Thus, it was not considered.
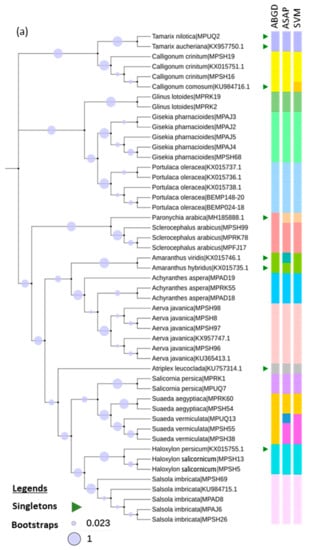
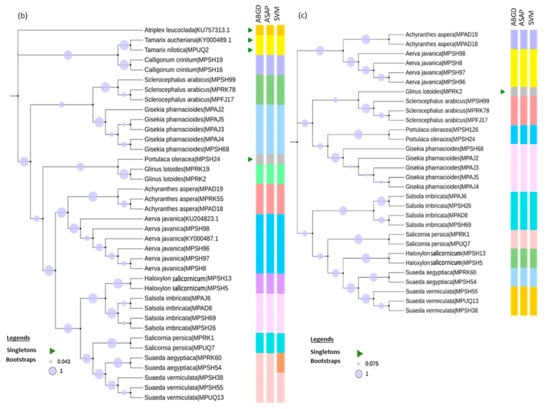
Figure 3.
Taxonomic evaluation using unsupervised (ABGD and ASAP) and supervised learning (SVM) methods. (a) RbcL maximum likelihood (ML) phylogeny inferred using Kimura-2-parameter (K2P) model and discrete gamma distribution with 100 bootstrap support. (b) MatK ML phylogeny inferred using the General Time Reversible model and discrete gamma distribution with 1000 bootstrap support. (c) ITS2 ML phylogeny inferred by K2P model along using discrete gamma distribution and invariant sites with 1000 bootstrap support.

Table 1.
Summary of species identification using unsupervised and supervised learning methods.
As seen in the ABGD analysis, the algorithm identified several species partitions for each p-value (priori), which might derive uncertainty from the data [50]. Therefore, it is recommended to implement an integrative taxonomic approach to evaluate the relevance of the ABGD partitions [50]. Thus, the species assignment was further validated using the ASAP, followed by the supervised machine learning approach. In the case of ASAP, it appeared to provide a gap-width score, p-value, threshold distance dT, and the number of species corresponding to each defined partition, and thus overcame the challenge of a priori defined by ABGD. The partition could then be prioritized by considering the smallest ASAP score and the asterisk marks that represent the overall best scores.
Accordingly, the partitions with the highest species resolution were discovered for the matK and ITS2 datasets at the threshold distance of 0.029 and 0.0134, respectively (Figure 4a,b). In the matK dataset, about 29 specimens and 9 species were resolved, while in the ITS2, about 29 specimens and 10 species were resolved successfully (Figure 4a,b and Table 1). However, for the rbcL dataset, the second successive partition at the threshold distance of 0.0045 with lower ASAP scores was found to be the best partition showing a higher resolution (Figure 4c), further accurately discriminating 33 specimens and 9 species, and was thus taken into consideration (Figure 3a, and Table 1).
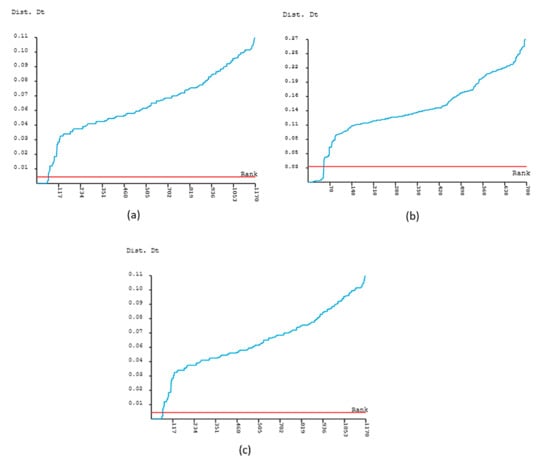
Figure 4.
Threshold distance ranking the best partition for species delimitation. (a) matK 2nd partition (ASAP score = 2.50, Nb = 13, p = 0.0027); (b) ITS2 2nd partition (ASAP score= 2.50, Nb = 11, p = 0.042); (c) rbcL 2nd partition (ASAP score = 4.50, number of species (Nb) = 18, p = 0.0052).
Following the unsupervised approach, the analysis through the SML approach exhibited the highest species resolution in all three markers, rbcL, matK, and ITS2. SML appeared to resolve about 39 specimens and 10 species in rbcL, 34 specimens and 10 species in matK, and about 29 specimens and 10 species in ITS2 (Figure 3a–c and Table 1).
Overall, in the rbcL dataset, the ASAP and SVM methods successfully differentiated Paronychia arabica from Scelerocephalus arabicus, and resolved the Suaeda genus (Suaeda aegyptiaca from Suaeda vermiculata, whereas Haloxylon persicum and Haloxylon salicornicum could not be discriminated using all three methods (ABGD, ASAP, and SVM) (Figure 3a). Moreover, in the rbcL dataset, ASAP alone was able to differentiate the Amaranthus genus (Amaranthus viridis and Amaranthus hybridus), while SVM alone was able to delimit the Calligonum genus (Calligonum crinitum and Calligonum comosum) (Figure 3a).
In the matK dataset, Suaeda aegyptiaca and Suaeda vermiculata were only resolved by SVM (Figure 3b), while in the ITS2 dataset, both the species seemed to be accurately differentiated using all three methods (ABGD, ASAP, and SVM) (Figure 3c). Altogether, 17 species were successfully resolved from the 20 barcoded species using the rbcL, matK, and ITS2 markers, though the matK and ITS2 datasets lacked enough species memberships for all 20 species.
3.3. Genetic Divergence
The genetic divergence analysis was conducted for the rbcL, matK and ITS2 datasets using the TaxonDNA (Table 2). The highest intraspecific distance of 2.45% was observed in the ITS2 dataset among the individuals of Salsola imbricata, while the highest interspecific divergence of 2.58% was observed between the species of the genus Suaeda. Similarly, the Suaeda genus seemed to exhibit maximum interspecific distances for the rbcL dataset, wherein the matK Tamarix genus showed higher interspecific divergence, followed by Suaeda. In the case of the species from the genus Suaeda, the genetic divergence in the rbcL (1.55%) and matK (1.21%) was not enough for the ABGD (rbcL and matK datasets) and ASAP (matK datasets) to discriminate the S. vermiculata from the S. aegyptiaca. However, all methods employed successfully resolved species from the Suaeda genus using the ITS2 dataset at the genetic divergence of 2.58%.

Table 2.
Intra- and interspecific genetic divergence.
4. Discussion
The use of herbal medicine traditionally for disease treatment and as a precursor for developing several important drugs [2,73] necessitates the accurate identification of medicinal plants. The results of our study suggest that the applications of DNA barcoding techniques can enhance the accurate identification of medicinally important species. Our study is among the first to utilize different DNA barcode markers and confirms the potential of the barcoding approach for the accurate identification of medicinally useful plants from the UAE that will help generate a reference dataset for research and other applications.
We investigated the efficacy of the three DNA barcode regions (rbcL, matK, and ITS2) for discriminating selected medicinal plant species belonging to the order Caryophyllales. The first step in assessing the potential candidate barcodes was to estimate the universality of the amplification and sequencing success rate across the studied taxa. The matK region showed a lower amplification rate (60%) than rbcL and ITS2, although two matK primers pairs were used with several attempts under different conditions (Figure 2a). The MatK (P1 and P2) pair was highly effective in the amplification success. However, the matK (P3 and P4) pair resulted in low recovery (only one sample was amplified successfully). The inconsistent success rate has been reported for matK. Several studies have indicated that the matK region was less amplified than other regions in different angiosperms and gymnosperms, including some arid desert plants [74,75,76]. The universality issues of the matK primer could be attributed to the nucleotide variations in the respective binding site that could inhibit the PCR amplification [74], or to the large amplified product size (≈900 bps) that could be susceptible to the degradation [77]. Cräutlein et al. [78] suggested the need for further efforts to improve primer design in matK to achieve higher efficiency. For sequencing, a higher number of good-quality sequences (80%) were obtained for rbcL than the other two regions. This result is aligned with previous studies that compared the three barcode loci for the coding genes (matK, rbcL, and rpoC1) for the discrimination of different plants of the UAE and concluded that the rbcL was more effective in discrimination between species [15,56,57].
Several different approaches based on the DNA barcoding technique have been advised for assigning species to their relevant taxa [52,54,79,80]. Our analysis applied an integrative approach for the delimitation of species using unsupervised “OTU picking” methods, viz., ABGD and ASAP that use only pairwise genetic distances, along with supervised methods for more data reliability. The ABGD method automatically identifies where the barcode gap is located in their distribution. This gap marks the limit between minimum interspecific and maximum intraspecific divergence. Thus, it is crucial to ensure the distance-based method’s effectiveness [51,81]. Our results showed that the recursive partitions in ABGD recognized more OTUs than primary ones, exhibiting a higher accuracy in species resolution under the analysis, which corroborates with previous observations [51,82,83]. Further, ASAP was performed to evaluate the relevance of the ABGD partitions, as any species partition must be subsequently tested against other evidence as recommended in an integrative taxonomy approach [50].
Our results indicated that the unsupervised ABGD method showed taxonomic conflicts in rbcL between Amaranthus species (A. hybridus and A. viridis), and between Paronychia arabica and Sclerocephalus arabicus. Interestingly, these species differed morphologically and could be discriminated easily (Figure 3a). Moreover, merged taxa were observed for the genus Suaeda (S. aegyptiaca and S. vermiculata) in the rbcL dataset using ABGD, as well as in the matK datasets using both the ABGD and ASAP methods (Figure 3a,b). Moreover, a low pairwise interspecific divergence of rbcL (=1.55%) and matK (=1.21%) was observed between the species of Suaeda, thereby exhibiting a monophyletic relationship. A similar result was observed by Kapralov et al. [80], who provided strong statistical support for the monophyly. The taxonomic relationships might be confusing due to the absence of a barcode gap, which can result from a limited number of sequences per species (i.e., <3–5) [51].
Following the ABGD and ASAP methods, species delimitation through character-based supervised machine learning methods was utilized to understand better the confirmation of the initial identification [84]. So far, several studies have performed the character-based barcoding approach, which has proved its usefulness in identifying plant species better than the conventional unsupervised methods [52,53,54,85]. In our analysis, the unsupervised ASAP method tended to provide a better resolution potential for the rbcL dataset than its neighboring ABGD method (Table 1). In addition, ASAP was able to resolve two singleton species in the rbcL dataset that were not even recognized using the ABGD method (Figure 3a). Moreover, when compared with the supervised learning approach, the SVM method stood out as the more efficient method to provide an accurate identification than the unsupervised approach with the higher number of species, as observed in the rbcL and matK datasets (Table 1 and Figure 3a,b). In addition, S. aegyptiaca and S. vermiculata were also recovered as separate clades, which indicates that the intraspecific diversity could be hidden [34,86].
It has been reported that OTUs proposed by one or more methods could be inconsistent in distinguishing between the members of closely related genera [49]. In our study, we observed that the members of genus Amaranthus (A. viridis and A. hybridus) were only discriminated through ASAP, but members of Calligonum (C. crinitum and C. comosum) were distinguished only by SVM. This supports the importance of using more than one method, especially for closely related species that are difficult to discriminate morphologically, such as C. crinitum and C. comosum. The use of more than one method can maximize the probability of identifying morphologically similar species and overcoming the limitation associated with each method [50,87].
Overall, the taxonomic performance of SVM was stronger than that of ABGD and ASAP in the rbcL dataset. The SVM delivered the highest incidence of correct matches (55.0%) across the 20 species compared to 35.0% and 45.0% for ABGD and ASAP, respectively (Table 1). In the matK dataset, the performance of ABGD was similar to ASAP (60.0%) and was improved to 73.33% using supervised learning methods. However, all the methods delivered a similar percentage of correct matches in the ITS2 dataset (Table 1). Considerably, it is now a well-known fact that the combination of the two plastid markers, ribulose 1,5-bisphosphate carboxylase gene (rbcL) and maturase K (matK), that were accepted as the core barcoding regions [33], do not grant a suitable coverage of plant species. Thus, they must often be implemented along with the other hypervariable sequences, such as nuclear ITS or the plastid interspacer region trnH-psbA [88].
Moreover, the efficiency of the utilized markers and methods depends on the sample size, as the singleton species or small sample size could lead to skewed results [21]. In our study, we had about 10 singleton species, which were considered as singletons and not independent OTUs to reduce the probability of biased identification. Thus, an adequate sample size and proper implementation of the DNA barcoding technique can provide a scientific basis for the molecular identification and conservation of valuable medicinal species. Our study is among the first to utilize different DNA barcode markers and to confirm the potential of DNA barcoding in the accurate identification of medicinally important plants from the UAE. The dataset generated through this study will assist in developing the reference library, and allows others to contribute and explore the genetic potential of the available germplasm for various applications.
5. Conclusions
The results support the potential use of DNA barcoding in discriminating closely related taxa of Caryophyllales. The ITS2 was more effective in the discrimination between studied species, indicating its potential for distinguishing between Caryophyllales medicinally important plants and non-medicinal plants or other undesirable plant tissues. However, due to the inability of one DNA barcoding analysis method to discriminate between members of closely related genera, we propose combining two or more methods. The results of this study could fill a small gap of generating DNA barcodes for local (i.e., the UAE), regional (i.e., the Arab Gulf region), and global libraries of vascular plant flora.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14040262/s1, Table S1: List of specimen samples used in this study. Table S2: Information on ecological characteristics and medicinal benefits of studied plant species, Table S3: List of Primers with their thermal profile used in this study.
Author Contributions
K.A.M., A.E.-K. and R.J. conceived the ideas and designed the experiment; R.J., S.G., H.S. and T.M. collected the samples; R.J., K.A.S., E.A.H., M.A.S. and M.A.J. performed the Laboratory analysis; R.J. conducted the bioinformatics analysis; R.J., K.A.S., E.A.H., M.A.S., M.A.J., S.G., H.S. and T.M. were involved in data validation and curation; R.J., K.A.S., E.A.H., M.A.S., M.A.J., S.G., H.S. and T.M. prepared the original draft; K.A.M. and A.E.-K. reviewed and edited the manuscript; K.A.M. and A.E-K were involved in project supervision, administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a collaborative research grant from the University of Sharjah and Sharjah Research Academy, project # 1702145054-P.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in the NCBI GenBank (Table S1).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, A.; Singh, S.; Mohan Prasad, S. Role of Medicinal Plants for Health Perspective: Special Reference to Antioxidant Potential. J. Chem. Biol. Ther. 2016, 1, 106. [Google Scholar] [CrossRef]
- Hostettmann, K.; Marston, A.; Ndjoko, K.; Wolfender, J.-L. The Potential of African Plants as a Source of Drugs. Curr. Org. Chem. 2005, 4, 973–1010. [Google Scholar] [CrossRef]
- Ljubuncic, P.; Azaizeh, H.; Portnaya, I.; Cogan, U.; Said, O.; Saleh, K.A.; Bomzon, A. Antioxidant activity and cytotoxicity of eight plants used in traditional Arab medicine in Israel. J. Ethnopharmacol. 2005, 99, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Cope, T.A.; Miller, A.G.; Morris, M. Plants of Dhofar: The Southern Region of Oman. Traditional, Economic and Medicinal Uses. Geogr. J. 1990, 156, 89. [Google Scholar] [CrossRef]
- Ghazanfar, S.A. Handbook of Arabian Medicinal Plants; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Bussmann, R.W.; Malca, G.; Glenn, A.; Sharon, D.; Nilsen, B.; Parris, B.; Dubose, D.; Ruiz, D.; Saleda, J.; Martinez, M.; et al. Toxicity of medicinal plants used in traditional medicine in Northern Peru. J. Ethnopharmacol. 2011, 137, 121–140. [Google Scholar] [CrossRef]
- Chandra, S.; Rawat, D.S. Medicinal plants of the family Caryophyllaceae: A review of ethno-medicinal uses and pharmacological properties. Integr. Med. Res. 2015, 4, 123–131. [Google Scholar] [CrossRef]
- Phondani, P.C.; Bhatt, A.; Elsarrag, E.; Horr, Y.A. Ethnobotanical magnitude towards sustainable utilization of wild foliage in Arabian Desert. J. Tradit. Complement. Med. 2016, 6, 209–218. [Google Scholar] [CrossRef]
- Ullah, R.; Alqahtani, A.S.; Noman, O.M.A.; Alqahtani, A.M.; Ibenmoussa, S.; Bourhia, M. A review on ethno-medicinal plants used in traditional medicine in the Kingdom of Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 2706–2718. [Google Scholar] [CrossRef]
- Aati, H.; El-Gamal, A.; Shaheen, H.; Kayser, O. Traditional use of ethnomedicinal native plants in the Kingdom of Saudi Arabia. J. Ethnobiol. Ethnomed. 2019, 15, 2. [Google Scholar] [CrossRef]
- Sakkir, S. Medicinal plants diversity and their conservation status in the United Arab Emirates (UAE). J. Med. Plants Res. 2012, 6, 1304–1322. [Google Scholar] [CrossRef]
- Cybulska, I.; Brudecki, G.; Alassali, A.; Thomsen, M.; Jed Brown, J. Phytochemical composition of some common coastal halophytes of the United Arab Emirates. Emirates J. Food Agric. 2014, 26, 1046–1056. [Google Scholar] [CrossRef]
- Sajjad, A.; Syed, A.; Hasnain, A.; Mohamed, E. Ethno Botanical Study of Traditional Native Plants in Al Ain UAE. Int. J. Adv. Res. Biol. Sci. 2017, 4, 1–10. [Google Scholar] [CrossRef]
- Parvathy, V.A.; Swetha, V.P.; Sheeja, T.E.; Sasikumar, B. Detection of plant-based adulterants in turmeric powder using DNA barcoding. Pharm. Biol. 2015, 53, 1774–1779. [Google Scholar] [CrossRef]
- Mosa, K.A.; Soliman, S.; El-Keblawy, A.; Ali, M.A.; Hassan, H.A.; Tamim, A.A.B.; Al-Ali, M.M. Using DNA Barcoding to Detect Adulteration in Different Herbal Plant-Based Products in the United Arab Emirates: Proof of Concept and Validation. Recent Pat. Food. Nutr. Agric. 2018, 9, 55–64. [Google Scholar] [CrossRef]
- Abdel-Aziz, S.M.; Aeron, A.; Kahil, T.A.; Abdel-Aziz, S.M.; Kahil, T.A.; Aeron, A. Health Benefits and Possible Risks of Herbal Medicine. In Microbes in Food and Health; Garg, N., Abdel-Aziz, S., Aeron, A., Eds.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Pratiwi, R.; Dipadharma, R.H.F.; Prayugo, I.J.; Layandro, O.A. Recent Analytical Method for Detection of Chemical Adulterants in Herbal Medicine. Molecules 2021, 26, 6606. [Google Scholar] [CrossRef]
- Tungmunnithum, D.; Renouard, S.; Drouet, S.; Blondeau, J.P.; Hano, C. A Critical Cross-Species Comparison of Pollen from Nelumbo nucifera Gaertn. vs. Nymphaea lotus L. for Authentication of Thai Medicinal Herbal Tea. Plants 2020, 9, 921. [Google Scholar] [CrossRef]
- Wu, L.; Wu, M.; Cui, N.; Xiang, L.; Li, Y.; Li, X.; Chen, S. Plant super-barcode: A case study on genome-based identification for closely related species of Fritillaria. Chinese Med. 2021, 16, 1–11. [Google Scholar] [CrossRef]
- Mosa, K.A.; Gairola, S.; Jamdade, R.; El-Keblawy, A.; Al Shaer, K.I.; Al Harthi, E.K.; Shabana, H.A.; Mahmoud, T. The Promise of Molecular and Genomic Techniques for Biodiversity Research and DNA Barcoding of the Arabian Peninsula Flora. Front. Plant. Sci. 2019, 9, 1929. [Google Scholar] [CrossRef]
- Jamdade, R.; Upadhyay, M.; Al Shaer, K.; Al Harthi, E.; Al Sallani, M.; Al Jasmi, M.; Ketbi, A. Al Evaluation of Arabian Vascular Plant Barcodes (rbcL and matK): Precision of Unsupervised and Supervised Learning Methods towards Accurate Identification. Plants 2021, 10, 2741. [Google Scholar] [CrossRef]
- Jamdade, R.A.; Mahmoud, T.; Gairola, S. Prospects of genomic resources available at the global databases for the flora of United Arab Emirates. 3 Biotech. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Urumarudappa, S.K.J.; Tungphatthong, C.; Prombutara, P.; Sukrong, S. DNA metabarcoding to unravel plant species composition in selected herbal medicines on the National List of Essential Medicines (NLEM) of Thailand. Sci. Rep. 2020, 10, 18259. [Google Scholar] [CrossRef]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Aghayeva, P.; Cozzolino, S.; Cafasso, D.; Ali-zade, V.; Fineschi, S.; Aghayeva, D. DNA barcoding of native Caucasus herbal plants: Potentials and limitations in complex groups and implications for phylogeographic patterns. Biodivers. Data J. 2021, 9, 1–28. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Burns, J.M.; Janzen, D.H.; Hajibabaei, M.; Hallwachs, W.; Hebert, P.D.N. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservación Guanacaste, Costa Rica. Proc. Natl. Acad. Sci. USA 2008, 105, 6350–6355. [Google Scholar] [CrossRef]
- Dick, C.W.; Webb, C.O. Plant DNA Barcodes, Taxonomic Management, and Species Discovery in Tropical Forests. Methods Mol. Biol. 2012, 858, 379–393. [Google Scholar] [CrossRef]
- Celiński, K.; Kijak, H.; Wojnicka-Półtorak, A.; Buczkowska-Chmielewska, K.; Sokołowska, J.; Chudzińska, E. Effectiveness of the DNA barcoding approach for closely related conifers discrimination: A case study of the Pinus mugo complex. Comptes Rendus Biol. 2017, 340, 339–348. [Google Scholar] [CrossRef]
- Yu, J.; Wu, X.; Liu, C.; Newmaster, S.; Ragupathy, S.; Kress, W.J. Progress in the use of DNA barcodes in the identification and classification of medicinal plants. Ecotoxicol. Environ. Saf. 2021, 208, 111691. [Google Scholar] [CrossRef]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef]
- Nazar, N.; Howard, C.; Slater, A.; Sgamma, T. Challenges in Medicinal and Aromatic Plants DNA Barcoding—Lessons from the Lamiaceae. Plants 2022, 11, 137. [Google Scholar] [CrossRef]
- CBOL Plant Working Group; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding rbcL Gene Complements the Non-Coding trnH-psbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed]
- Chase, M.W.; Salamin, N.; Wilkinson, M.; Dunwell, J.M.; Kesanakurthi, R.P.; Haidar, N.; Savolainen, V. Land plants and DNA barcodes: Short-term and long-term goals. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1889–1895. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef]
- Mishra, P.; Kumar, A.; Nagireddy, A.; Mani, D.N.; Shukla, A.K.; Tiwari, R.; Sundaresan, V. DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant. Biotechnol. J. 2016, 14, 8–21. [Google Scholar] [CrossRef]
- Hashim, A.M.; Alatawi, A.; Altaf, F.M.; Qari, S.H.; Elhady, M.E.; Osman, G.H.; Abouseadaa, H.H. Phylogenetic relationships and DNA barcoding of nine endangered medicinal plant species endemic to Saint Katherine protectorate. Saudi J. Biol. Sci. 2021, 28, 1919–1930. [Google Scholar] [CrossRef]
- Gao, T.; Yao, H.; Song, J.; Liu, C.; Zhu, Y.; Ma, X.; Pang, X.; Xu, H.; Chen, S. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol. 2010, 130, 116–121. [Google Scholar] [CrossRef]
- Jiao, J.; Huang, W.; Bai, Z.; Liu, F.; Ma, C.; Liang, Z. DNA barcoding for the efficient and accurate identification of medicinal polygonati rhizoma in China. PLoS ONE 2018, 13, e0201015. [Google Scholar] [CrossRef]
- Pathak, M.R.; Mohamed, A.A.M.; Farooq, M. DNA Barcoding and Identification of Medicinal Plants in the Kingdom of Bahrain. Am. J. Plant. Sci. 2018, 9, 2757–2774. [Google Scholar] [CrossRef]
- Tahir, A.; Hussain, F.; Ahmed, N.; Ghorbani, A.; Jamil, A. Assessing universality of DNA barcoding in geographically isolated selected desert medicinal species of Fabaceae and Poaceae. Peer J. 2018, 6, e4499. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Dong, X.; Lin, R.; Fan, J.; Chen, Z. Evaluation of four commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae. PLoS ONE 2015, 10, e0125574. [Google Scholar] [CrossRef]
- Moon, B.C.; Kim, W.J.; Ji, Y.; Lee, Y.M.; Kang, Y.M.; Choi, G. Molecular identification of the traditional herbal medicines, Arisaematis Rhizoma and Pinelliae Tuber, and common adulterants via universal DNA barcode sequences. Genet. Mol. Res. 2016, 15, gmr7064. [Google Scholar] [CrossRef]
- Kim, W.J.; Ji, Y.; Choi, G.; Kang, Y.M.; Yang, S.; Moon, B.C. Molecular identification and phylogenetic analysis of important medicinal plant species in genus Paeonia based on rDNA-ITS, matK, and rbcL DNA barcode sequences. Genet. Mol. Res. 2016, 15, gmr.15038472. [Google Scholar] [CrossRef]
- Theodoridis, S.; Stefanaki, A.; Tezcan, M.; Aki, C.; Kokkini, S.; Vlachonasios, K.E. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çeşme-Karaburun Peninsula (Turkey). Mol. Ecol. Resour. 2012, 12, 620–633. [Google Scholar] [CrossRef]
- Schori, M.; Showalter, A.M. DNA barcoding as a means for identifying medicinal plants of Pakistan. Pakistan J. Bot. 2011, 43, 1–4. [Google Scholar]
- Ducasse, J.; Ung, V.; Lecointre, G.; Miralles, A. LIMES: A tool for comparing species partition. Bioinformatics 2020, 36, 2282–2283. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- He, T.; Jiao, L.; Wiedenhoeft, A.C.; Yin, Y. Machine learning approaches outperform distance- and tree-based methods for DNA barcoding of Pterocarpus wood. Planta 2019, 249, 1617–1625. [Google Scholar] [CrossRef]
- Emu, M.; Sakib, S. Species Identification using DNA Barcode Sequences through Supervised Learning Methods. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’sBazar, Bangladesh, 7–9 February 2019. [Google Scholar] [CrossRef]
- Weitschek, E.; Fiscon, G.; Felici, G. Supervised DNA Barcodes species classification: Analysis, comparisons and results. BioData Min. 2014, 7, 1–18. [Google Scholar] [CrossRef]
- Patil, T.S.; Jamdade, R.A.; Patil, S.M.; Govindwar, S.P.; Muley, D.V. DNA barcode based delineation of freshwater fishes from northern Western Ghats of India, one of the world’s biodiversity hotspots. Biodivers. Conserv. 2018, 27, 3349–3371. [Google Scholar] [CrossRef]
- Enan, M.R.; Palakkott, A.R.; Ksiksi, T.S. DNA barcoding of selected UAE medicinal plant species: A comparative assessment of herbarium and fresh samples. Physiol. Mol. Biol. Plants 2017, 23, 221–227. [Google Scholar] [CrossRef][Green Version]
- Maloukh, L.; Kumarappan, A.; Jarrar, M.; Salehi, J.; El-wakil, H.; Rajya Lakshmi, T.V. Discriminatory power of rbcL barcode locus for authentication of some of United Arab Emirates (UAE) native plants. 3 Biotech. 2017, 7, 144. [Google Scholar] [CrossRef]
- Sukhorukov, A.P.; Mavrodiev, E.V.; Struwig, M.; Nilova, M.V.; Dzhalilova, K.K.; Balandin, S.A.; Erst, A.; Krinitsyna, A.A. One-seeded fruits in the core caryophyllales: Their origin and structural diversity. PLoS ONE 2015, 10, e0130783. [Google Scholar] [CrossRef]
- Kool, A.; de Boer, H.J.; Krüger, Å.; Rydberg, A.; Abbad, A.; Björk, L.; Martin, G. Molecular identification of commercialized medicinal plants in Southern Morocco. PLoS ONE 2012, 7, e39459. [Google Scholar] [CrossRef]
- Hernández-Ledesma, P.; Berendsohn, W.G.; Borsch, T.; Von Mering, S.; Akhani, H.; Arias, S.; Castañeda-Noa, I.; Eggli, U.; Eriksson, R.; Flores-Olvera, H.; et al. A taxonomic backbone for the global synthesis of species diversity in the angiosperm order caryophyllales. Willdenowia 2015, 45, 281–383. [Google Scholar] [CrossRef]
- Cuénoud, P.; Savolainen, V.; Chatrou, L.W.; Powell, M.; Grayer, R.J.; Chase, M.W. Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB, and matK DNA sequences. Am. J. Bot. 2002, 89, 132–144. [Google Scholar] [CrossRef]
- Jongbloed, M.V.D.; Feulner, G.R.; Böer, B.B.; Western, A.R. The Comprehensive Guide to the Wild Flowers of the United Arab Emirates; Environmental Research and Wildlife Development Agency: Abu Dhabi, United Arab Emirates, 2003; ISBN 978-9948408246. [Google Scholar]
- Karim, F.M.; Fawzi, N.M. Flora of the United Arab Emirates; Publications Department; United Arab Emirates University: Al Ain, United Arab Emirates, 2007; ISBN 9789948021407. [Google Scholar]
- Feulner, G.R. The Flora of the Ru’us al-Jibal—the Mountains of the Musandam Peninsula: An Annotated Checklist and Selected Observations. Available online: http://www.enhg.org/Portals/1/trib/V19/TribulusV19.pdf (accessed on 6 July 2021).
- Feulner, G.R. The Olive Highlands: A Unique “Island” of Biodiversity within the Hajar Mountains of the United Arab Emirates. Available online: http://www.enhg.org/Portals/1/trib/V22/TribulusV22.pdf (accessed on 6 July 2021).
- Levin, R.A.; Wagner, W.L.; Hoch, P.C.; Nepokroeff, M.; Pires, J.C.; Zimmer, E.A.; Sytsma, K.J. Family-level relationships of Onagraceae based on chloroplast rbc L and ndh F data. Am. J. Bot. 2003, 90, 107–115. [Google Scholar] [CrossRef]
- Lee, H.L.; Yi, D.K.; Kim, J.S. Development of plant DNA barcoding markers from the variable noncoding regions of chloroplast genome. In Proceedings of the Abstract Presented at the Second International Barcode of Life Conference, Academia Sinica, Taipei, Taiwan, 18–20 September 2007. [Google Scholar]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. No Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; MA, W.T., Gelfand, D., Sninsky, J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Balunas, M.J.; Kinghorn, A.D. Drug discovery from medicinal plants. Life Sci. 2005, 78, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Bafeel, S.O.; Arif, I.A.; Bakir, M.A.; Khan, H.A.; Al Farhan, A.H.; Al Homaidan, A.A.; Ahamed, A.; Thomas, J. Comparative Evaluation of PCR Success with Universal Primers of Maturase K (matK) and Ribulose-1, 5-Bisphosphate Carboxylase Oxygenase Large Subunit (rbcL) for Barcoding of Some Arid Plants. Plant. Omics 2011, 4, 195–198. [Google Scholar]
- Costion, C.; Ford, A.; Cross, H.; Crayn, D.; Harrington, M.; Lowe, A. Plant DNA Barcodes Can Accurately Estimate Species Richness in Poorly Known Floras. PLoS ONE 2011, 6, e26841. [Google Scholar] [CrossRef] [PubMed]
- YU, J.; XUE, J.-H.; ZHOU, S.-L. New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Fazekas, A.J.; Kuzmina, M.L.; Newmaster, S.G.; Hollingsworth, P.M. DNA barcoding methods for land plants. Methods Mol. Biol. 2012, 858, 223–252. [Google Scholar] [CrossRef]
- Von Cräutlein, M.; Korpelainen, H.; Pietiläinen, M.; Rikkinen, J. DNA barcoding: A tool for improved taxon identification and detection of species diversity. Biodivers. Conserv. 2011, 20, 373–389. [Google Scholar] [CrossRef]
- Casiraghi, M.; Labra, M.; Ferri, E.; Galimberti, A.; de Mattia, F. DNA barcoding: A six-question tour to improve users’ awareness about the method. Brief. Bioinform. 2010, 11, 440–453. [Google Scholar] [CrossRef]
- Kapralov, M.V.; Akhani, H.; Voznesenskaya, E.V.; Edwards, G.; Franceschi, V.; Roalson, E.H. Phylogenetic Relationships in the Salicornioideae / Suaedoideae / Salsoloideae s.l. (Chenopodiaceae) Clade and a Clarification of the Phylogenetic Position of Bienertia and Alexandra Using Multiple DNA Sequence Datasets. Syst. Bot. 2006, 31, 571–585. [Google Scholar] [CrossRef]
- Wyler, S.C.; Naciri, Y. Evolutionary histories determine DNA barcoding success in vascular plants: Seven case studies using intraspecific broad sampling of closely related species. BMC Evol. Biol. 2016, 16, 103. [Google Scholar] [CrossRef]
- Liu, X.F.; Yang, C.H.; Han, H.L.; Ward, R.D.; Zhang, A. Identifying species of moths (Lepidoptera) from Baihua Mountain, Beijing, China, using DNA barcodes. Ecol. Evol. 2014, 4, 2472–2487. [Google Scholar] [CrossRef]
- Yang, Z.; Landry, J.-F.; Hebert, P.D.N. A DNA Barcode Library for North American Pyraustinae (Lepidoptera: Pyraloidea: Crambidae). PLoS ONE 2016, 11, e0161449. [Google Scholar] [CrossRef]
- Zou, S.; Fei, C.; Song, J.; Bao, Y.; He, M.; Wang, C. Combining and Comparing Coalescent, Distance and Character-Based Approaches for Barcoding Microalgaes: A Test with Chlorella-Like Species (Chlorophyta). PLoS ONE 2016, 11, e0153833. [Google Scholar] [CrossRef]
- Jaén-Molina, R.; Marrero-Rodríguez, Á.; Reyes-Betancort, J.A.; Santos-Guerra, A.; Naranjo-Suárez, J.; Caujapé-Castells, J. Molecular taxonomic identification in the absence of a ‘barcoding gap’: A test with the endemic flora of the Canarian oceanic hotspot. Mol. Ecol. Resour. 2015, 15, 42–56. [Google Scholar] [CrossRef]
- Jiang, K.W.; Zhang, R.; Zhang, Z.F.; Pan, B.; Tian, B. DNA barcoding and molecular phylogeny of Dumasia (Fabaceae: Phaseoleae) reveals a cryptic lineage. Plant. Divers. 2020, 42, 376–385. [Google Scholar] [CrossRef]
- Barley, A.J.; Thomson, R.C. Assessing the performance of DNA barcoding using posterior predictive simulations. Mol. Ecol. 2016, 25, 1944–1957. [Google Scholar] [CrossRef]
- Morello, L.; Braglia, L.; Gavazzi, F.; Gianì, S.; Breviario, D. Tubulin-Based DNA Barcode: Principle and Applications to Complex Food Matrices. Genes 2019, 10, 229. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).