Abstract
Although the existence of two cetacean species in and around Thai Seas from within the subfamily Globicephalinae, Risso’s dolphin Grampus griseus G. Cuvier, 1812, and false killer whale Pseudorca crassidens Owen, 1846, has been known for decades, current knowledge on the abundance, genetic diversity, and conservation status of these marine mammals is limited as these are rare oceanic species for Thailand’s territorial waters. Frozen skin tissue samples taken from six cetaceans (four Risso’s dolphins: two false killer whales) stranded along Thai coastlines were investigated. We aimed to identify the maternal lineage and connection of our samples throughout their distribution range. Accordingly, we analyzed the dataset of 110 and 50 mtDNA control region sequences of Risso’s dolphins and false killer whales, respectively. This dataset was retrieved from the online database of the National Center for Biotechnology Information (NCBI) and included six mtDNA sequences obtained from Thai Seas. Two unique haplotypes of Risso’s dolphins were found in the Thai Andaman Sea, whereas one haplotype identified as being from the Gulf of Thailand is a common haplotype shared with other regions of the Pacific Ocean. Two haplotypes were found for false killer whales from the Thai Andaman Sea, and these were also in common with other regions of the Indo Pacific Ocean. While shared haplotypes with other regions may imply inheritance from the same female ancestor, we speculate that distinct populations with unique genetic structures also exist in Thai Seas. Beneficially, our results could be used to monitor alterations of haplotypes or to assess the maternal genetic diversity of designated species in the future to establish baseline information for Thai Seas and adjacent waters.
1. Introduction
The Thai Seas, which consist of the Thai Andaman Sea and the Gulf of Thailand, are known to be the natural habitat of 27 cetacean species including whales, dolphins, and porpoises [1,2,3,4,5]. Risso’s dolphin (Grampus griseus G. Cuvier, 1812) and false killer whale (Pseudorca crassidens Owen, 1846), closely related species within the subfamily Globicephalinae based on mitogenomic phylogenetic analysis, are also known to occupy the Thai Seas [1,5,6,7,8]. There is a similarity in the distribution range that extends from coastal areas to oceanic depths in tropical and temperate waters; however, the cooler-temperate waters found between latitudes of 30–45° are preferred by Risso’s dolphins [9,10,11,12]. Water depth clearly affects the occurrence of this species as they exhibited a clear preference for the outer continental shelf at depths within a range of 300–1500 m in eastern Taiwanese waters [10,13,14,15]. It is unusual for this species to be found in the Gulf of Thailand where the average depth of the water is limited to only 45 m [10,16]. In contrast, the false killer whale, which is recognized as a warm temperate oceanic species, is more often sighted throughout the Thai Seas and adjacent waters when compared with Risso’s dolphin [1,8,17]. However, the number of stranded animals of both species along Thai coasts has been minimal since the first record in 1996 [1,18,19].
Currently, both species are listed in Appendix II of the Convention on International Trade in Endangered Species (CITES). According to the assessment of the International Union for Conservation of Nature (IUCN) Red List, Risso’s dolphin is now considered a species of least concern, while the status of the false killer whale is near threatened [20,21]. In fact, the populations of both species in many regions have been affected by a range of human activities such as fishery bycatch, intentional killings, loud anthropogenic sounds, and disturbance by the whale-watching boats of tourists [22,23,24,25,26,27,28,29]. Accordingly, high-intensity sonar and chemical contamination have been recognized as significant threats to the Risso’s dolphin population in the western Ligurian Sea. This has resulted in a decrease in the population size of these cetaceans [30]. Recently, the subpopulation of the false killer whale inhabiting the waters around the Hawaiian Islands has significantly declined as a result of the commercial fishing industry [31,32]. A high proportion of fishery-related scaring on the dorsal fins of false killer whales inhabiting the coastal waters of the main Hawaiian Islands also confirm the ongoing negative impacts of the fisheries located around this area [33]. The assessment of the regional conservation status of each species and an understanding of the potential threats to their existence are extremely important for the conservation of these species. Certain regional animal protection acts have been established to support the protection of these species, such as the Wildlife Conservation Act in Taiwan for Risso’s dolphin [34] and the Marine Mammals Protection Act in the United States for the false killer whale [20]. For Thailand, most of the cetaceans inhabiting Thai Seas are now being protected by the establishment of the Wild Animal Reservation and Protection Act (WARPA) of 1992 [35]. In recent decades, around 22 species of cetaceans, including the false killer whale, have been listed as protected marine animals by WARPA, while only two species of baleen whales are listed as conserved animals [35]. However, these lists do not cover all cetacean species that exist in Thailand’s waters. This is because there is insufficient data pertaining to some species that have recently been found in these waters, while there have also been very few records of stranded animals along Thai coasts, such as the Risso’s dolphins and humpback whales (Megaptera novaeangliae) [18].
Biological data related to the abundance and dispersal of each species, as well as the population structure, genetic variations, and the identification of specific threats for cetacean populations, will be needed to effectively evaluate their present conservation status and improve relevant conservation management strategies going forward [36,37,38]. Several studies on the local populations of Risso’s dolphins and false killer whales have been conducted using nuclear DNA microsatellite and mitochondrial DNA (mtDNA) markers to gain in-depth biological information. In these studies, information on the significant genetic differentiations in the populations of Risso’s dolphins inhabiting UK waters and the Mediterranean Sea was established without sharing mtDNA control region haplotypes between the populations of these animals, while a lower degree of genetic diversity was recorded for the population of UK dolphins [39]. Later, Chen, et al. [40] reported on the genetic differentiations among three populations of this species inhabiting the North Pacific Ocean. Based on mtDNA control region sequence analysis, these populations were also found to be genetically different from the populations inhabiting UK waters and the Mediterranean Sea in a previous study conducted by Gaspari et al. (2007) [39]. The phylogeographic structure of false killer whales inhabiting the North Pacific Ocean, and specifically the coastal waters of the main Hawaiian Islands, revealed a unique set of haplotypes [41]. A further investigation on the false killer whale population structure that inhabit the Hawaiian Archipelago was conducted by Martien, et al. [42]. Three unique haplotypes were identified throughout the coastal waters of the main Hawaiian Islands and the northwestern waters of the Hawaiian Islands that did not share haplotypes with other locations. This phylogeographic pattern of mtDNA also suggests that they have a common history of colonization.
Importantly, the populations of Risso’s dolphins and the false killer whales that inhabit Thai waters have never been studied. These marine mammals are rare oceanic species for the Thai Seas; although, some sightings and incidences of stranding have been documented [1,18,19]. Since 1994, pods of false killer whales have often been sighted around the Similan Islands and the Racha Islands in the Thai Andaman Sea, as well as around Kho Tao in the Gulf of Thailand [1,19]. In contrast, there have been no recorded sightings for free-ranging Risso’s dolphins in these areas. While this species is thought to be a rare species for the Thai Seas, there have been only a few reports of stranded individual dolphins along Thailand’s coasts, such as in the coastal areas of Nakhon Si Thammarat Province, Prachuap Khiri Khan Province in the Gulf of Thailand, and Phuket Province in the Thai Andaman Sea [18]. Currently, published data pertaining to the abundance, population, genetic diversity, and conservation status of Risso’s dolphins and false killer whales inhabiting Thailand’s waters is considered insufficient, while any connection between the populations of these species inhabiting the Thai Seas and the adjacent waters remains unclear. Thus, a lack of information on these species has led to difficulties in setting up effective conservation strategies and protective regulations. Therefore, in order to collect relevant biological data and understand how the cetaceans found stranded along Thai coasts may be connected to others around the world, we have investigated the mtDNA control region haplotypes of four Risso’s dolphin and two false killer whale sequences based on the limited stranded samples provided by the Phuket Marine Biological Center, Thailand. The mtDNA control regions obtained from six specimens were sequenced in order to determine their matrilineally genetic relationship to cetaceans inhabiting other oceanic waters by retrieving the available sequences deposited in the National Center for Biotechnology Information (NCBI) GenBank. This was done in order to observe the haplotype distribution of both species on a global scale.
2. Materials and Methods
2.1. Samples and DNA Extraction
In this study, six skin tissue samples of stranded cetaceans were provided by the Phuket Marine Biological Center, Phuket 83000, Thailand. This center has collected and stored tissue samples of all stranded cetaceans along Thailand’s coasts since 1990 (Table 1). All samples were preserved in 95% ethanol and stored at −20 °C. Four tissue samples of Risso’s dolphins were collected from both the Thai Andaman Sea (ADM) and the Gulf of Thailand (GOT), whereas tissue samples of only two false killer whales were collected from ADM. The skin samples were extracted using a DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany) according to the method previously described [43,44] at the Faculty of Veterinary Medicine, Chiang Mai University, Thailand. Extracted DNA samples were prepared for purity evaluation using 2% agarose gel electrophoresis and were then visualized under ultraviolet light. DNA quantity was determined using a spectrophotometer in order to measure the absorbance ratio at 260–280 nm. Diluted samples (10 ng/μL) were stored at −20 °C for subsequent analyses. This study was approved of by the Animal Use Committee of the Faculty of Veterinary Medicine, Chiang Mai University, Thailand, in 2020 (S14/2563).

Table 1.
Cetacean samples used in this study obtained from Thailand’s territorial waters.
2.2. mtDNA Control Regions
The mtDNA control regions of six samples (Table 1) were amplified using PCR primers: forword, 5′-CAT ATT ACA ACG GTC TTG TAA ACC-3′; and reverse, 5′-GTC ATA AGT CCA TCG AGA TGT C-3′ [45]. PCR reactions were conducted in 25 μL reaction volumes consisting of 1X reaction buffer, 2 mM MgCl2, 0.4 mg/mL bovine serum albumin, 0.25 mM dNTPs, 0.4 μM of both forward and reverse primers, 5 U/μL Platinum Taq DNA polymerase (Invitrogen), and 10 ng/μL of the DNA sample. The PCR conditions were performed as follows: 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 50 °C for 45 s, 72 °C for 1 min, and 72 °C for 10 min. The amplicon size of all samples at 600 bp were visualized on 2% agarose gel electrophoresis. All PCR products obtained from the amplification were then sequenced by ATGC Co., Ltd., Pathum Thani, Thailand. The mtDNA control region sequences obtained from this study were visually prepared and edited manually using the MEGA-X program version 10.2.2 [46].
The mtDNA control region sequences of Risso’s dolphins and false killer whales available in the GenBank database were obtained to generate two datasets according to each species. These datasets were established using the following keyword searches: “mitochondrion/control region mtDNA/d-loop” + “Grampus griseus/Pseudorca crassidens”. The mtDNA sequences of both full length and partial length were presented through keyword searches, although only the D-loop Feature in the “Graphics” section was exported as a FASTA record. A total of 110 sequences of Risso’s dolphins and 50 sequences of false killer whales were obtained through designated keyword searches. The mtDNA control sequences of six studied samples were included in these datasets for subsequent analysis. The final two datasets and the accession numbers of all sequences are presented in Tables S1 and S2.
2.3. mtDNA Sequence Analysis and Phylogeographic Relationships
In our sequence samples, each species was aligned to the worldwide sequences in each dataset as implemented by Clustal W in MEGA X version 10.2.2 [46,47]. The haplotypes of each dataset were determined using DnaSP program version 6.12.3 [48]. Geographic maps of haplotype distribution were generated using the tmap function implemented in R studio [49]. The Median Joining Networks (MJNs) of each species were constructed using PopART program version 1.7 [50].
2.4. Phylogenetic Tree Construction
Phylogenetic trees of mtDNA control region sequences for each species were constructed using Bayesian Analysis implemented in the MrBayes program version 3.2.7 [51]. To identify the best tree evolutionary models for Risso’s dolphins and false killer whales, jModelTest version 2.1.10 was used to achieve a result of the best fit model established as HKY+G and GTR+I+G, respectively [52,53]. The total run length of Markov Chain Monte Carlo (MCMC) sampling at 10,000,000 iterations for each tree was performed, while samples were drawn for every 5000 iterations. Convergence diagnostics with an average standard deviation of the split frequencies below 0.01 were used. The first 2,500,000 iterations were discarded in the burn-in step. Mitochondrial control region DNA sequences of other cetacean species were used to serve as an out group for each phylogenetic tree: white-beaked dolphin (Lagenorhynchus albirostris AJ554061), rough-tooth dolphin (Steno bredanensis NC 042716), and killer whale (Orcinus orca NC 023889). The phylogenetic trees obtained were then visualized by iTOL version 6.1.1 [54].
3. Results
3.1. mtDNA Control Region
A total of six mtDNA control region sequences were successfully amplified from the Thai samples (4 Risso’s dolphins: 2 false killer whales). Maximum sequence lengths of 491 bp for Risso’s dolphins and 484 bp for false killer whales were obtained in this study and deposited in GenBank (MZ401232, MZ401234-36, and MZ401168-69). The datasets of each species obtained from GenBank, including our new sequences, were of different lengths; however, for the purposes of comparison with all available sequences in each species, the length of each dataset was reduced and aligned with the consensus fragments of 390 bp for Risso’s dolphins and 323 bp for false killer whales.
3.2. Haplotypes and Phylogeographic Relationships
Overall, with regard to the global scale of mtDNA control region haplotypes, we identified a total of 58 variable sites: 81 haplotypes for Risso’s dolphins at 22 variable sites and 23 haplotypes for false killer whales. The haplotype distribution of each species is shown in Figure 1 and Figure 2. For our samples, three haplotypes of Risso’s dolphins (haplotypes 2–4, Figure 1) and two haplotypes of false killer whales (haplotypes 1 and 2, Figure 2) were detected.
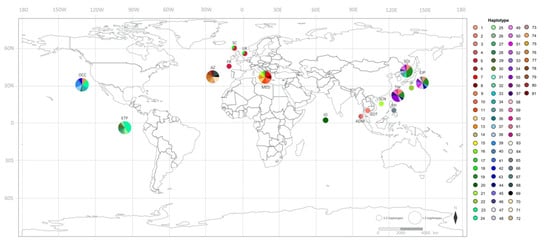
Figure 1.
Map showing the geographic distribution of Risso’s dolphin mtDNA control region haplotypes. Each circle represents the haplotypes of Risso’s dolphins in each location. The two sizes of circles indicate the number of haplotypes: smaller, 1–2 haplotypes; larger, >2 haplotypes. The color indicating the haplotype and the proportion of color in each circle are related to haplotype frequency. Abbreviations for location are as follows: AZ, Azores Island; MED, Mediterranean; FR, France; SC, Scotland; UK, United Kingdom; IO, Indian Ocean; ADM, Thai Andaman Sea; GOT, Gulf of Thailand; PH, Philippines; TW, Taiwan; SCN, Southeastern China Sea; SOJ, Sea of Japan; JP, Japan; EJP, Eastern Japan; ETP, Eastern Tropical Pacific Ocean; OCC, Oregon-California Coastal.
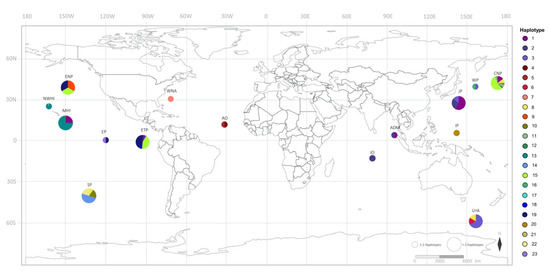
Figure 2.
Map showing the geographic distribution of false killer whale mtDNA control region haplotypes. Each circle represents the haplotypes of false killer whales in each location. The two sizes of circles indicate the number of haplotypes: smaller, 1–2 haplotypes; larger, >2 haplotypes. The color indicating the haplotype and the proportion of color in each circle are related to haplotype frequency. Abbreviations for location are as follows: ADM, Thai Andaman Sea; CNP, Central North Pacific Ocean; ENP, Eastern North Pacific Ocean; EP, East Pacific Ocean; ETP, Eastern Tropical Pacific Ocean; IP, Indo Pacific Ocean; JP, Japan; MHI, Main Hawaiian Islands; NWHI, Northwestern Hawaiian Islands; SP, South Pacific Ocean; WP, West Pacific Ocean; IO, Indian Ocean; AO, Atlantic Ocean; WNA, West Northern Atlantic Ocean; Unk, Unknown.
The MJNs of Risso’s dolphin indicated multiple alternative connections among haplotypes worldwide (Figure 3). Many haplotypes associated with a small degree of frequency were connected to each other through one to five mutation steps, whereas some clusters of haplotypes were present in a specific geographic region. For example, haplotypes 5 and 18 were restricted to the North Atlantic region, while haplotypes 6, 10, and 13 were restricted to the Mediterranean Sea. Shared haplotypes also occurred across many locations such as haplotypes 5 and 19 that were shared between Eastern Japan, the Sea of Japan, and Taiwan. However, haplotypes shared between oceanic regions were not observed. For our Risso’s dolphin samples, two unique Thai haplotypes, namely haplotypes 3 and 4 obtained from the Thai Andaman Sea, were reported, while haplotype 2 obtained from the Gulf of Thailand is shared with specimens obtained from Eastern Japan, the Sea of Japan, Taiwan, and the Oregon-California Coastal region (Figure 3). A simpler pattern of MJNs was observed for false killer whale mtDNA control region haplotypes (Figure 4). The haplotypes were also connected to each other with one to five mutation steps. The sharing of haplotypes between the Atlantic and Pacific Oceans was not detected. Instead, it appeared to occur between the Indian and Pacific Oceans, wherein haplotype 2 included our false killer whale sample obtained from the Thai Andaman Sea and others obtained from the Indian Ocean and the territorial waters of Japan. According to the analysis of another sample of the false killer whale in our study, cetaceans of the Thai Andaman Sea also shared haplotypes with several locations in the Pacific Ocean (haplotype 1), i.e., the Central North Pacific Ocean, the Eastern Tropical Pacific Ocean, the territorial waters of Japan, and the coastal waters of the main Hawaiian Islands. This high frequency haplotype is likely to be the ancestral haplotype for the Indo Pacific Ocean as well as the haplotype 13 for the main Hawaiian and Northwestern Hawaiian Islands (Figure 4).
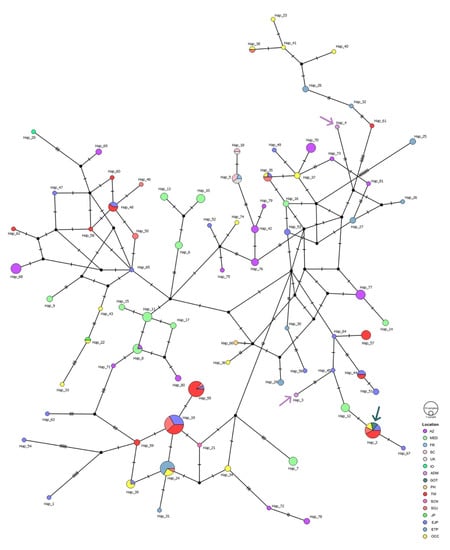
Figure 3.
Median joining network (MJN) of all Risso’s dolphin mtDNA control region haplotypes. The MJN was generated from 390 base pairs of Risso’s dolphin mtDNA control region sequences and indicated that 81 haplotypes were found in 16 locations. Each haplotype is colored by location and the small black circles represent inferred haplotypes (not sampled). The size of each circle is proportional to its haplotype frequency, while nucleotide substitutions are shown at the branches as small transverse bars. Purple arrows indicate the haplotypes from the Thai Andaman Sea and the green arrow indicates the haplotype from the Gulf of Thailand. Abbreviations for location are as follows: AZ, Azores Island; MED, Mediterranean; FR, France; SC, Scotland; UK, United Kingdom; IO, Indian Ocean; ADM, Thai Andaman Sea; GOT, Gulf of Thailand; PH, Philippines; TW, Taiwan; SCN, Southeastern China Sea; SOJ, Sea of Japan; JP, Japan; EJP, Eastern Japan; ETP, Eastern Tropical Pacific Ocean; OCC, Oregon-California Coastal.
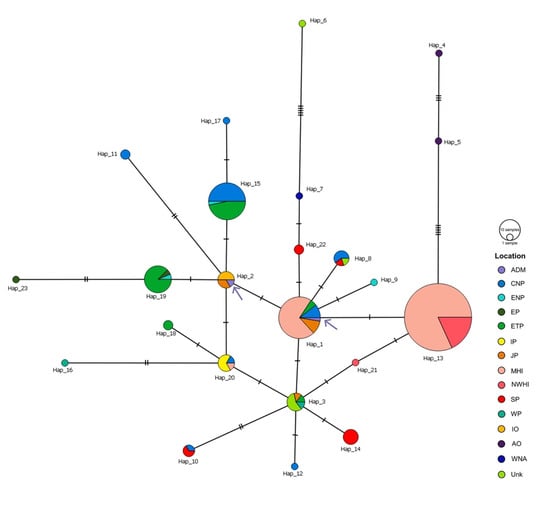
Figure 4.
Median joining network (MJN) of all false killer whale mtDNA control region haplotypes. The MJN was generated from 323 base pairs of false killer whale mtDNA control region sequences and indicated that 23 haplotypes were found in 15 locations. Each haplotype is colored by location. The size of each circle is proportional to its haplotype frequency, while nucleotide substitutions are shown at the branches as small transverse bars. Purple arrows indicate haplotypes from the Thai Andaman Sea. Abbreviations of location are as follows: ADM, Thai Andaman Sea; CNP, Central North Pacific Ocean; ENP, Eastern North Pacific Ocean; EP, East Pacific Ocean; ETP, Eastern Tropical Pacific Ocean; IP, Indo Pacific Ocean; JP, Japan; MHI, Main Hawaiian Islands; NWHI, Northwestern Hawaiian Islands; SP, South Pacific Ocean; WP, West Pacific Ocean; IO, Indian Ocean; AO, Atlantic Ocean; WNA, West Northern Atlantic Ocean; Unk, Unknown.
3.3. Phylogenetic Reconstruction
Bayesian phylogenetic reconstructions revealed a monophyletic taxon of mtDNA lineages for Risso’s dolphins and false killer whales from worldwide samples with a posterior probability of 1.00 for both species (Figure 5 and Figure 6). In terms of Risso’s dolphin phylogeny, many clades of haplotypes were observed. Our two samples obtained from the Gulf of Thailand (haplotype 2) were clustered in a monophyletic clade with other samples obtained from the waters of Eastern Japan and Taiwan in the Pacific Ocean (Figure 5). This clade is also paraphyly to the unique haplotypes of Risso’s dolphins obtained from the Thai Andaman Sea (haplotype 3 and haplotype 4). With regard to false killer whales, there was only one large clade that contained all haplotypes. This included our two samples obtained from the Thai Andaman Sea (haplotype 1 and haplotype 2, Figure 6).
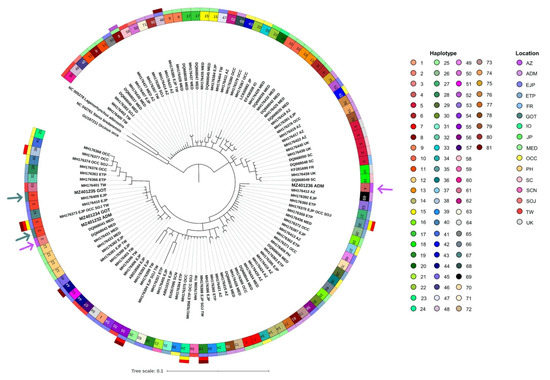
Figure 5.
Bayesian phylogenetic tree of Risso’s dolphin based on 390 bp alignment of mitochondrial DNA control region. Accordingly, 96 sequences of Risso’s dolphins were acquired from the NCBI database, while four sequences were obtained from this study. The white-beaked dolphin (Lagenorhynchus albirostris AJ554061), rough-tooth dolphin (Steno bredanensis NC 042716), and killer whale (Orcinus orca NC 023889) were utilized as out groups. The circular bands of color around the tree indicate haplotype: first layer and location, second, third, fourth, and fifth layers (inner to outer). Purple arrows indicate samples from the Thai Andaman Sea and green arrows indicate samples from the Gulf of Thailand. Abbreviations of location are as the follows: AZ, Azores Island; MED, Mediterranean; FR, France; SC, Scotland; UK, United Kingdom; IO, Indian Ocean; ADM, Thai Andaman Sea; GOT, Gulf of Thailand; PH, Philippines; TW, Taiwan; SCN, Southeastern China Sea; SOJ, Sea of Japan; JP, Japan; EJP, Eastern Japan; ETP, Eastern Tropical Pacific Ocean; OCC, Oregon-California Coastal.
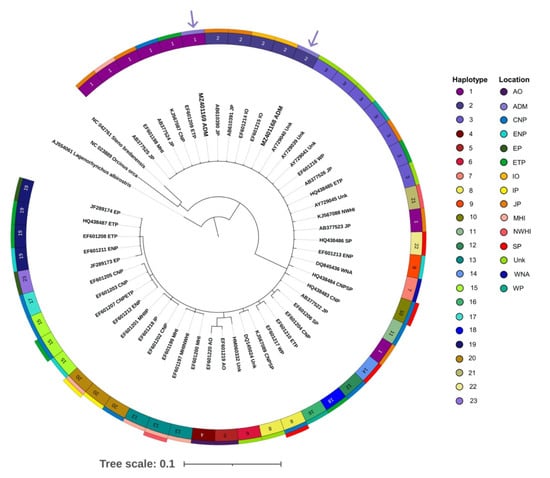
Figure 6.
Bayesian phylogenetic tree of false killer whales based on 323 bp alignment of mitochondrial DNA control regions. Accordingly, 48 sequences of false killer whales were obtained from the NCBI database, while two sequences were obtained from this study. The white-beaked dolphin (Lagenorhynchus albirostris AJ554061), rough-tooth dolphin (Steno bredanensis NC 042716), and killer whale (Orcinus orca NC 023889) were utilized as out groups. The circular bands of color around the tree indicate haplotype: first layer and location, second and third layers (inner to outer). Purple arrows indicate samples from Thai Andaman Sea. Abbreviations of location are as follows: ADM, Thai Andaman Sea; CNP, Central North Pacific Ocean; ENP, Eastern North Pacific Ocean; EP, East Pacific Ocean; ETP, Eastern Tropical Pacific Ocean; IP, Indo Pacific Ocean; JP, Japan; MHI, Main Hawaiian Islands; NWHI, Northwestern Hawaiian Islands; SP, South Pacific Ocean; WP, West Pacific Ocean; IO, Indian Ocean; AO, Atlantic Ocean; WNA, West Northern Atlantic Ocean; Unk, Unknown.
4. Discussion
4.1. Unique Haplotypes of Risso’s Dolphins and Potential Distribution in Thai Seas
According to our assessment of the unique mtDNA control region haplotypes of Risso’s dolphins inhabiting the Thai Andaman Sea, we found the potential presence of a different genetic unit when compared to other studied areas. These haplotypes were not included in previous genetic studies or other established databases; however, they did appear to be present in different habitats that are known to be home to a wide range of prey species. The topography of the Andaman Sea, including continental shelves with the preferred depth for this species, also support the possibility of the occurrence for a distinct Risso’s dolphin population in this region where the water depth is around 200–2000 m and deeper than the Gulf of Thailand [55]. Within a short geographic distance, many factors, such as divergent habitats, foraging specializations, site fidelity, and social structure, can be involved in shaping the population structure of cetaceans and specifically odontocetes [56,57,58]. This would be particularly true for Risso’s dolphin populations in eight regions according to the outcomes of a number of previous studies. These regions included the territorial waters of the UK, the Mediterranean Sea, the Azores Islands, Taiwanese waters, the Sea of Japan, eastern Japan, Eastern Tropical Pacific waters, and the Oregon-California coastal region, all of which have indicated significant differentiations in mtDNA control regions (Fst, p value < 0.05) across these differing habitats [39,40].
Differences in prey specialization and the foraging habitats of distinct populations have also been suggested as potential mechanisms for the genetic differentiations that have occurred among other cetaceans, i.e., killer whales and bottlenose dolphins [56,59,60]. Similarly, it is generally known that Risso’s dolphins feed primarily on a wide variety of cephalopod species that inhabit both the water column and the ocean floor [26,61]; however, differences in the main prey species within each region have been observed. A study of the stomach contents of Risso’s dolphins obtained from the Northern Atlantic region and the Faroe Islands revealed different types of squid as the main prey species during different periods of the year i.e., Todarodes sagittatus in September and Eledona cirrhosa in April [26]; whereas in the Mediterranean Sea, two types of squid in the family Histioteuthidae, i.e., Histioteuthis bonnellii and Histioteuthis reversa, were reported as being representative of the primary diet of this dolphin species [62]. Unfortunately, the prey species found in the stomach contents of our samples obtained from the Thai Seas have never been identified. Therefore, we suggest that, in the future, the stomach contents of dead stranded cetaceans should be investigated in further studies. This information could provide even more supportive evidence for the existence of a distinct dolphin populations in relation to prey specialization.
Although the sighting and capture records of Risso’s dolphins in worldwide oceans have been reported by collecting all available data from 1950 to 2012 [1,10], the records from the Thai Andaman Sea and the Gulf of Thailand have not been included in any of the previous studies. In the last decade, there has been a lack of evidence to prove the existence of this species in Thai Seas, particularly in the waters of the Gulf of Thailand where the maximum water depth is limited to only 80 m [16]. Consequently, the Gulf of Thailand is thought to be an unfavorable area for Risso’s dolphins, which are known to prefer deeper waters [10]. In addition to the shallow waters of the gulf and the absence of reported sightings of free-ranging cetaceans in this area, our results also support the contention that the Gulf of Thailand might not be the core habitat of the Risso’s dolphin as the haplotype we found (haplotype 2) from two stranded dolphins in this area was also found in other regions of the Pacific Ocean, i.e., the waters of Eastern Japan, the Sea of Japan, Taiwan, and the Oregon-California Coastal region. Similarly, it is unusual for this species to be observed in western Taiwanese waters, where the water is much shallower than it is in eastern Taiwanese waters. Only four sightings of Risso’s dolphin groups and a few incidences of stranding were recorded in this area [34,63], while there was a significant number of sightings, at 1141, recorded in a survey conducted in the deeper waters of the Hualien and Shirti ports of east-central Taiwanese waters over a 17-year period [15]. Despite this species’ preference for water-depth [6,10,64,65], it is likely that the Risso’s dolphins found stranded in the Gulf of Thailand were transient individuals that may have strayed far from the pods that inhabit the waters outside the gulf in the South China Sea, where the water is known to be deeper than 1000 m [66,67].
Sightings of Risso’s dolphins in and around the Andaman Sea and adjacent waters, i.e., the Bay of Bengal and the southeastern coasts of Indonesia and Australia, have been scarcely noted [10]; however, samples obtained from these areas are rare and hard to obtain. In our study, although unique haplotypes were observed from the Thai Andaman Sea, more information is needed to confirm the existence of distinct populations inhabiting these areas. In the last decade, the recent emergence of this species in the Thai Seas and the lack of other biological information could lead to hard decisions being made on the establishment of accurate conservation status; thus, this species has not yet been considered for acceptance on WARPA’s animal protected lists of Thailand. We suggest that additional genetic studies involving even more samples of this species in these areas, particularly from the Thai Andaman Sea, would be beneficial in revealing further information of evidentiary value.
4.2. Maternal Lineage of False Killer Whales Inhabiting the Thai Andaman Sea and Other Regions
The existence of shared haplotypes of false killer whales between the Thai Andaman Sea and other areas within the Pacific Ocean was observed in our study. Haplotype 1 in our study is an ancestral haplotype that is widely distributed across the Pacific Ocean, including MHI, as it appears to exhibit the greatest degree of frequency and is located in the middle of the MJNs. In the previous study, three unique haplotypes of this species were thought to be found only in MHI and NWHI as demographical isolated populations, while these haplotypes were not found elsewhere in the Pacific Ocean [42]. However, in that study, not only were three unique haplotypes recorded around the Hawaiian Archipelago, but another haplotype was also shared between Northern Australia and MHI. It may also be possible that the false killer whale population around the Thai Andaman Sea has shared female ancestors with other regions. Apparently, this haplotype has persisted and survived even in diverse habitats, though more samples should be investigated to confirm the common haplotype of this population within this area and perhaps the existence of even more haplotypes.
Although pods of free-ranging false killer whales have often been sighted in the Thai Seas [1,19], a connection of maternal lineage between the Thai Andaman Sea and the Gulf of Thailand has never been revealed. This is because no relationship could be established from samples obtained from the Gulf of Thailand and from stranded individual cetaceans along both coasts. Sightings and stranding incidences for this species were recorded around the Malay Peninsula, i.e., the Langkawi Archipelago, Tioman Island, and Sabha, Malaysia, all of which were close to our study area [68]. However, there is limited knowledge on the population of this species throughout these areas. In addition, their genetic information has rarely been investigated. Only two sequences of unknown location from the Indo Pacific Ocean have been deposited in the online database (NCBI), though this haplotype (haplotype 20) is closely related to our samples with differences in one to two nucleotide positions. With regard to the matrilineal social systems of this species [69], the remarkably low diversity of the mtDNA control region has been revealed across many regions, i.e., MHI (hd = 0.395 ± 0.043 to 0.554 ± 0.053), NWHI (hd = 0.105 ± 0.092), and ETP (hd = 0.676 ± 0.052) [42]. Thus, we speculate that very few haplotypes have occurred around the Thai Seas, while haplotype 20 in our study is also likely to be found in populations around these areas.
The number of deaths and stranded cetaceans along Thai coasts have been reported annually. Fishing equipment has been identified as posing the greatest potential threat to these mammals [70], though the specific threat for two species, namely Risso’s dolphins and the false killer whale, is poorly understood. We suggest that monitoring through active surveys, i.e., line transect surveys on abundance, sight effort records, and photo-identification studies, on free ranging cetaceans of both species in the Thai Seas should be initiated to evaluate the abundance, distribution pattern, and potential threats for each species. Studies such as these have previously been performed involving other species such as the Irrawaddy dolphin (Orcaella brevirostris), the Indo Pacific humpback dolphin (Sousa chinensis), and the Indo Pacific finless porpoise (Neophocaena phocaenoides) [2,3]. Additionally, further genetic studies involving potential molecular markers, such as nuclear DNA microsatellites, are urgently needed to assess their genetic diversity and population structure. This information will help to improve vital and relevant conservation measures for Thailand’s cetacean populations.
Evaluation of the conservation status and establishment of effective conservation strategies for both species have been a challenge for ecologists and conservation biologists, though the achievement of which may currently be impossible as little is known about these marine mammals in the Thai Seas. To support the conservation of these cetaceans in these areas, we are providing useful genetic information to enable researchers to better understand their distribution and the connection of maternal lineage in the Thai Seas and oceanic waters throughout the world. In the past, there has been limited published data on Risso’s dolphins in and around the Malay Peninsula. Consequently, findings on the unique haplotypes for our samples obtained from the Thai Andaman Seas indicate the presence of new genetic evidence for the potential distribution and existence of distinct populations in this area. Moreover, the haplotypes of both species include shared haplotypes of the false killer whales, which could be used to monitor alterations in haplotypes or maternal genetic diversity in the future. Importantly, our findings can contribute to the establishment of baseline genetic information for the Thai Seas and adjacent waters.
5. Conclusions
Our study highlights include valuable information on the mtDNA control region haplotype for both Risso’s dolphins and false killer whales in comparisons made between the Thai Seas and oceans throughout the world. We have also provided new genetic information for both species derived from cetaceans that were stranded along Thai coasts. Despite the limited number of samples in our study, two unique haplotypes of Risso’s dolphins obtained from the Thai Andaman Sea were observed. While the unique haplotypes of this species may infer the existence of local dolphin populations around the Thai Andaman Sea and adjacent waters, the shared haplotype of both species, including that of the false killer whales that inhabit the Thai Seas and other locations, also indicates a connection and potential distribution of the maternal lineage throughout the distribution range of this species. However, in order to identify the existence of local dolphin populations, more information pertaining to population structure, abundance, distribution range, and an understanding of the prey species derived from the stomach contents of the cetaceans in these areas will be needed. Furthermore, more samples from these areas will need to be obtained and investigated in order to gain in-depth population information and to develop and establish effective conservation strategies for Thai cetaceans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14040257/s1, Table S1: The accession number of Risso’s dolphins (Granpus griseus) used in this study; Table S2: The accession number of false killer whales (Pseudorca crassiden) used in this study. The references [7,39,40,41,42,71,72,73,74,75,76,77,78,79,80,81,82,83] are cited in the supplementary materials.
Author Contributions
Data curation, P.P.; funding acquisition, K.N.; investigation, P.P. and A.P.; methodology, P.P. and A.P.; resources, P.K. and K.K.; supervision, K.B., J.K., S.C. and K.N.; validation, K.B., J.K., S.C. and K.N.; visualization, J.K., S.C. and K.N.; writing—original draft, P.P. and K.B.; writing—review and editing, K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Excellence Center in Veterinary Bioscience, Chiang Mai University, Chiang Mai 50200, Thailand and the CMU Presidential Scholarship, Chiang Mai University, Thailand.
Institutional Review Board Statement
This study was approved of by the Animal Use Committee of the Faculty of Veterinary Medicine, Chiang Mai University, Thailand in 2020 (S14/2563).
Informed Consent Statement
Not applicable.
Data Availability Statement
The accession numbers presented in this study are available in Tables S1 and S2.
Acknowledgments
We dedicate the value of this research to the Phuket Marine Biological Center, Phuket, Thailand. To this center, we express our deepest gratitude for providing samples, data, and their helpful insight that allowed us to fully analyze the results of this study.
Conflicts of Interest
The authors declare no conflict of interest. Moreover, the funders of this research had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Chantrapornsyl, S.; Adulyanukosol, K.; Kittiwathanawong, K. Records of cetaceans in Thailand. Phuket Mar. Biol. Cent. Res. Bull. 1996, 61, 39–63. [Google Scholar]
- Jutapruet, S.; Intongcome, A.; Wang, X.; Kittiwattanawong, K.; Huang, S.-L. Distribution of Three Sympatric Cetacean Species Off the Coast of the Central-Western Gulf of Thailand. Aquat. Mamm. 2017, 43, 465–473. [Google Scholar] [CrossRef]
- Hines, E.; Strindberg, S.; Junchompoo, C.; Ponnampalam, L.S.; Ilangakoon, A.D.; Jackson-Ricketts, J.; Mananunsap, S. Line transect estimates of Irrawaddy dolphin abundance along the eastern Gulf Coast of Thailand. Front. Mar. Sci. 2015, 2, 63. [Google Scholar] [CrossRef]
- Jutapruet, S.; Huang, S.-L.; Li, S.; Lin, M.; Kittiwattanawong, K.; Pradit, S. Population Size and Habitat Characteristics of the Indo-Pacific Humpback Dolphin (Sousa chinensis) Off Donsak, Surat Thani, Thailand. Aquat. Mamm. 2015, 41, 129–142. [Google Scholar] [CrossRef]
- Department of Marine and Coastal Resources. Whale and Dolphin Species. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: http://km.dmcr.go.th/th/c_7/d_2448 (accessed on 17 August 2021).
- Hartman, K.L. Risso’s dolphin: Grampus griseus. In Encyclopedia of Marine Mammals; Elsevier: Amsterdam, The Netherlands, 2018; pp. 824–827. [Google Scholar]
- Vilstrup, J.T.; Ho, S.Y.; Foote, A.D.; Morin, P.A.; Kreb, D.; Krützen, M.; Parra, G.J.; Robertson, K.M.; de Stephanis, R.; Verborgh, P. Mitogenomic phylogenetic analyses of the Delphinidae with an emphasis on the Globicephalinae. BMC Evol. Biol. 2011, 11, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, M.; Kinze, C.C. Annotated checklist and identification key to the whales, dolphins, and porpoises (Order Cetacea) of Thailand and adjacent waters. Nat. Hist. Bull. Siam. Soc. 1999, 47, 27–62. [Google Scholar]
- Jefferson, T.; Webber, M.; Pitman, R. Marine Mammals of the World: A Comprehensive Guide to Their Identification; Academic Press: San Diego, CA, USA, 2015. [Google Scholar]
- Jefferson, T.A.; Weir, C.R.; Anderson, R.C.; Ballance, L.T.; Kenney, R.D.; Kiszka, J.J. Global distribution of R isso’s dolphin G rampus griseus: A review and critical evaluation. Mamm. Rev. 2014, 44, 56–68. [Google Scholar] [CrossRef]
- Evans, P.G.; Anderwald, P.; Baines, M.E. UK cetacean status review. In Report to English Nature and Countryside Council for Wales; Natural England: York, UK, 2003. [Google Scholar]
- Reid, J.B.; Evans, P.G.; Northridge, S.P. Atlas of Cetacean Distribution in North-West European Waters; Joint Nature Conservation Committee: Peterborough, UK, 2003.
- Azzellino, A.; Gaspari, S.; Airoldi, S.; Nani, B. Habitat use and preferences of cetaceans along the continental slope and the adjacent pelagic waters in the western Ligurian Sea. Deep Sea Res. Part I Oceanogr. Res. Pap. 2008, 55, 296–323. [Google Scholar] [CrossRef]
- Praca, E.; Gannier, A. Ecological niches of three teuthophageous odontocetes in the northwestern Mediterranean Sea. Ocean Sci. 2008, 4, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Hsin-Yi, Y.; Ching-Yi, L.; Wen-Jyh, Y.; Lien-Siang, C. Distribution of Risso’s dolphin (Grampus griseus) in the east-central coastal waters of Taiwan based on whale-watching records. Taiwania 2019, 64, 417–425. [Google Scholar]
- Wattayakorn, G. Environmental issues in the Gulf of Thailand. In The Environment in Asia Pacific Harbours; Springer: Berlin/Heidelberg, Germany, 2006; pp. 249–259. [Google Scholar]
- Jaaman, S.A.; Najib, R.; Syuhaime, A.; Yuhana, U. Records of marine mammals in Peninsular Malaysia: A review. In Tropical Marine Environment: Charting Strategies for the Millennium; FSES: Serdang, Malaysia, 2002; pp. 499–515. [Google Scholar]
- Department of Marine and Coastal Resources. Risso’s Dolphin. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: http://km.dmcr.go.th/th/c_1/s_96/d_4205 (accessed on 17 August 2021).
- Department of Marine and Coastal Resources. False Killer Whale. Central Database System and Data Standard for Marine and Coastal Resources. 2013. Available online: http://km.dmcr.go.th/th/c_1/s_96/d_4183 (accessed on 17 August 2021).
- Baird, R.W. Pseudorca crassidens (Errata Version Published in 2019). The IUCN Red List of Threatened Species 2018: E. T18596A145357488. 2018. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T18596A145357488.en (accessed on 20 August 2021).
- Kiszka, J.; Braulik, G. Grampus griseus. The IUCN Red List of Threatened Species 2018: E. T9461A50356660. 2019. Available online: https://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T9461A50356660.en (accessed on 20 August 2021).
- Visser, F.; Hartman, K.L.; Rood, E.J.; Hendriks, A.J.; Zult, D.B.; Wolff, W.J.; Huisman, J.; Pierce, G.J. Risso’s dolphins alter daily resting pattern in response to whale watching at the Azores. Mar. Mamm. Sci. 2011, 27, 366–381. [Google Scholar] [CrossRef] [Green Version]
- Kiszka, J. Marine mammals: A review of status, distribution and interaction with fisheries in the Southwest Indian Ocean. In Offshore Fisheries of the Southwest Indian Ocean: Their Status and the Impact on Vulnerable Species; Van der Elst, R.P., Everett, B.I., Eds.; Oceanographic Research Institute: Durban, South Africa, 2015; Chapter 8; pp. 303–323. [Google Scholar]
- Kasuya, T. Whaling, Japanese. In Encyclopedia of Marine Mammals; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1066–1070. [Google Scholar]
- Southall, B.L.; Nowacek, D.P.; Miller, P.J.; Tyack, P.L. Experimental field studies to measure behavioral responses of cetaceans to sonar. Endanger. Species Res. 2016, 31, 293–315. [Google Scholar] [CrossRef] [Green Version]
- Bloch, D.; Desportes, G.; Harvey, P.; Lockyer, C.; Mikkelsen, B. Life history of Risso’s dolphin (Grampus griseus) (G. Cuvier, 1812) in the Faroe Islands. Aquat. Mamm. 2012, 38, 250–266. [Google Scholar] [CrossRef]
- Thode, A.; Wild, L.; Straley, J.; Barnes, D.; Bayless, A.; O’Connell, V.; Oleson, E.; Sarkar, J.; Falvey, D.; Behnken, L. Using line acceleration to measure false killer whale (Pseudorca crassidens) click and whistle source levels during pelagic longline depredation. J. Acoust. Soc. Am. 2016, 140, 3941–3951. [Google Scholar] [CrossRef]
- Bradford, A.L.; Forney, K.A. Injury Determinations for Cetaceans Observed Interacting with Hawaii and American Samoa Longline Fisheries during 2007–2011; NOAA: Washington, DC, USA, 2014.
- Dai, X.; Wu, F.; Wang, X. Annual Report to the Commission Part 1: Information on Fisheries, Research and Statistics; WCPFC: Kolonia, Federated States of Micronesia, 2020. [Google Scholar]
- Azzellino, A.; Airoldi, S.; Gaspari, S.; Lanfredi, C.; Moulins, A.; Podestà, M.; Rosso, M.; Tepsich, P. Risso’s dolphin, Grampus griseus, in the Western Ligurian Sea: Trends in population size and habitat use. Adv. Mar. Biol. 2016, 75, 205–232. [Google Scholar]
- Reeves, R.R.; Leatherwood, S.; Baird, R.W. Evidence of a Possible Decline since 1989 in False Killer Whales (Pseudorca crassidens) around the Main Hawaiian Islands1. Pac. Sci. 2009, 63, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Baird, R.W.; Gorgone, A.M. False Killer Whale Dorsal Fin Disfigurements as a Possible Indicator of Long-Line Fishery Interactions in Hawaiian Waters1. Pac. Sci. 2005, 59, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Baird, R.W.; Mahaffy, S.D.; Gorgone, A.M.; Cullins, T.; McSweeney, D.J.; Oleson, E.M.; Bradford, A.L.; Barlow, J.; Webster, D.L. False killer whales and fisheries interactions in Hawaiian waters: Evidence for sex bias and variation among populations and social groups. Mar. Mamm. Sci. 2015, 31, 579–590. [Google Scholar] [CrossRef]
- Yu, H.-Y.; Li, W.-T.; Chou, L.-S. Ecological and biological characteristics for the Risso’s dolphins (Grampus griseus) off Taiwan, with conservation evaluations on potential anthropogenic threats. Mammal. Study 2019, 44, 77–89. [Google Scholar] [CrossRef]
- Department of National Parks, Wildlife and Plant Conservation. Wildlife Conservation and Protection Act, B.E. 2562. 2019. Available online: http://portal.dnp.go.th/Content/LegalAffairs?contentId=22540 (accessed on 20 August 2021).
- Braulik, G.T.; Kasuga, M.; Wittich, A.; Kiszka, J.J.; MacCaulay, J.; Gillespie, D.; Gordon, J.; Said, S.S.; Hammond, P.S. Cetacean rapid assessment: An approach to fill knowledge gaps and target conservation across large data deficient areas. Aquat. Conserv. 2018, 28, 216–230. [Google Scholar] [CrossRef]
- Freeman, M.M. Challenges of assessing cetacean population recovery and conservation status. Endanger. Species Res. 2008, 6, 173–184. [Google Scholar] [CrossRef]
- Hoelzel, A. Genetic structure of cetacean populations in sympatry, parapatry, and mixed assemblages: Implications for conservation policy. J. Hered. 1998, 89, 451–458. [Google Scholar] [CrossRef] [Green Version]
- Gaspari, S.; Airoldi, S.; Hoelzel, A.R. Risso’s dolphins (Grampus griseus) in UK waters are differentiated from a population in the Mediterranean Sea and genetically less diverse. Conserv. Genet. 2007, 8, 727–732. [Google Scholar] [CrossRef]
- Chen, I.; Nishida, S.; Chou, L.S.; Tajima, Y.; Yang, W.C.; Isobe, T.; Yamada, T.K.; Hartman, K.; Hoelzel, A.R. Concordance between genetic diversity and marine biogeography in a highly mobile marine mammal, the Risso’s dolphin. J. Biogeogr. 2018, 45, 2092–2103. [Google Scholar] [CrossRef]
- Chivers, S.J.; Baird, R.W.; McSweeney, D.J.; Webster, D.L.; Hedrick, N.M.; Salinas, J.C. Genetic variation and evidence for population structure in eastern North Pacific false killer whales (Pseudorca crassidens). Can. J. Zool. 2007, 85, 783–794. [Google Scholar] [CrossRef] [Green Version]
- Martien, K.K.; Chivers, S.J.; Baird, R.W.; Archer, F.I.; Gorgone, A.M.; Hancock-Hanser, B.L.; Mattila, D.; McSweeney, D.J.; Oleson, E.M.; Palmer, C. Nuclear and mitochondrial patterns of population structure in North Pacific false killer whales (Pseudorca crassidens). J. Hered. 2014, 105, 611–626. [Google Scholar] [CrossRef] [Green Version]
- Cherdsukjai, P.; Buddhachat, K.; Brown, J.; Kaewkool, M.; Poommouang, A.; Kaewmong, P.; Kittiwattanawong, K.; Nganvongpanit, K. Age relationships with telomere length, body weight and body length in wild dugong (Dugong dugon). PeerJ 2020, 8, e10319. [Google Scholar] [CrossRef]
- Poommouang, A.; Kriangwanich, W.; Buddhachat, K.; Brown, J.L.; Piboon, P.; Chomdej, S.; Kampuansai, J.; Mekchay, S.; Kaewmong, P.; Kittiwattanawong, K. Genetic diversity in a unique population of dugong (Dugong dugon) along the sea coasts of Thailand. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Blair, D.; McMahon, A.; McDonald, B.; Tikel, D.; Waycott, M.; Marsh, H. Pleistocene sea level fluctuations and the phylogeography of the dugong in Australian waters. Mar. Mamm. Sci. 2014, 30, 104–121. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tennekes, M. tmap: Thematic Maps in R. J. Stat. Softw. 2018, 84, 1–39. [Google Scholar] [CrossRef] [Green Version]
- Leigh, J.W.; Bryant, D. popart: Full—feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.; Teslenko, M.; Nylander, J. Draft MrBayes version 3.2 manual: Tutorials and model summaries. 2011. Available online: http://41.204.190.30/resources_2/mb3.2_manual.pdf (accessed on 15 August 2021).
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Gibson, R.; Atkinson, R.; Gordon, J. Coral reefs of the Andaman Sea—an integrated perspective. Oceanogr. Mar. Biol. 2007, 45, 173–194. [Google Scholar]
- Bowen, B.W.; Gaither, M.R.; DiBattista, J.D.; Iacchei, M.; Andrews, K.R.; Grant, W.S.; Toonen, R.J.; Briggs, J.C. Comparative phylogeography of the ocean planet. Proc. Natl. Acad. Sci. USA 2016, 113, 7962–7969. [Google Scholar] [CrossRef] [Green Version]
- Hoelzel, A. Evolution of population genetic structure in marine mammal species. In Population Genetics for Animal Conservation; Cambridge University Press: Cambridge, UK, 2009; pp. 294–318. [Google Scholar]
- Möller, L.M.; Wiszniewski, J.; Allen, S.J.; Beheregaray, L.B. Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar. Freshw. Res. 2007, 58, 640–648. [Google Scholar] [CrossRef] [Green Version]
- Sellas, A.B.; Wells, R.S.; Rosel, P.E. Mitochondrial and nuclear DNA analyses reveal fine scale geographic structure in bottlenose dolphins (Tursiops truncatus) in the Gulf of Mexico. Conserv. Genet. 2005, 6, 715–728. [Google Scholar] [CrossRef]
- Hoelzel, A.; Dahlheim, M.; Stern, S. Low genetic variation among killer whales (Orcinus orca) in the eastern North Pacific and genetic differentiation between foraging specialists. J. Hered. 1998, 89, 121–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cockcroft, V.; Haschick, S.; Klages, N.W. The diet of Risso’s dolphin, Grampus griseus (Cuvier, 1812), from the east coast of South Africa. Z. Saugetierkd. 1993, 58, 286–293. [Google Scholar]
- Luna, A.; Sánchez, P.; Chicote, C.; Gazo, M. Cephalopods in the diet of Risso’s dolphin (Grampus griseus) from the Mediterranean Sea: A review. Mar. Mamm. Sci. 2021, 1–17. [Google Scholar] [CrossRef]
- Huang, C. Fauna and Distribution of Cetaceans in Taiwan and Abundance Estimate of Small Cetaceans in South-Western Taiwan Waters. Master’s Thesis, National Taiwan Ocean University, Keelung, Taiwan, 1996; p. 88. [Google Scholar]
- Baumgartner, M.F. The distribution of Risso’s dolphin (Grampus griseus) with respect to the physiography of the northern Gulf of Mexico. Mar. Mamm. Sci. 1997, 13, 614–638. [Google Scholar] [CrossRef]
- Kruse, S.L. Aspects of the Biology, Ecology, and Behavior of Risso’s Dolphins (Grampus griseus) off the California Coast; University of California: Santa Cruz, CA, USA, 1989. [Google Scholar]
- Hsu, S.-K.; Yeh, Y.-C.; Doo, W.-B.; Tsai, C.-H. New bathymetry and magnetic lineations identifications in the northernmost South China Sea and their tectonic implications. Mar. Geophys. Res. 2004, 25, 29–44. [Google Scholar] [CrossRef]
- Qu, T.; Mitsudera, H.; Yamagata, T. Intrusion of the north Pacific waters into the South China Sea. J. Geophys. Res. Oceans 2000, 105, 6415–6424. [Google Scholar] [CrossRef]
- Ponnampalam, L.S. Opportunistic observations on the distribution of cetaceans in the Malaysian South China, Sulu and Sulawesi Seas and an updated checklist of marine mammals in Malaysia. Raffles Bull. Zool. 2012, 60, 221–231. [Google Scholar]
- Baird, R.W.; Gorgone, A.M.; McSweeney, D.J.; Webster, D.L.; Salden, D.R.; Deakos, M.H.; Ligon, A.D.; Schorr, G.S.; Barlow, J.; Mahaffy, S.D. False killer whales (Pseudorca crassidens) around the main Hawaiian Islands: Long-term site fidelity, inter-island movements, and association patterns. Mar. Mamm. Sci. 2008, 24, 591–612. [Google Scholar] [CrossRef] [Green Version]
- Department of Marine and Coastal Resources. The Report of Rare Marine Animals 2017. Central Database System and Data Standard for Marine and Coastal Resources. 2017. Available online: https://dmcrth.dmcr.go.th/attachment/dw/download.php?WP=rUqjMT02qmWZG22DM7y04TyerPMjZT0lqmWZZz1CM5O0hJatrTDo7o3Q (accessed on 9 October 2021).
- Xiong, Y.; Brandley, M.C.; Xu, S.; Zhou, K.; Yang, G. Seven new dolphin mitochondrial genomes and a time-calibrated phylogeny of whales. BMC Evol. Biol. 2009, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Jayasankar, P.; Anoop, B.; Rajagopalan, M.; Yousuf, K.; Reynold, P.; Krishnakumar, P.; Afsal, V.; Anoop, A. Indian efforts on the inventorization of marine mammal species for their conservation and management. Asian Fish. Sci. 2009, 22, 143–155. [Google Scholar] [CrossRef]
- Alfonsi, E.; Méheust, E.; Fuchs, S.; Carpentier, F.-G.; Quillivic, Y.; Viricel, A.; Hassani, S.; Jung, J.-L. The use of DNA barcoding to monitor the marine mammal biodiversity along the French Atlantic coast. Zookeys 2013, 5. [Google Scholar]
- Senevirathna, J.D.M.; Asakawa, S.; Yoshitake, K. (The University of Tokyo, Aquatic Bioscience, Yayoi, Bunkyo-Ku, Tokyo, Japan); Grampus Griseus, Mitochondrial DNA, Complete Genome, Accession Number LC630882. 2021. Available online: https://www.ncbi.nlm.nih.gov/nuccore/LC630882 (accessed on 7 July 2021).
- Yamagiwa, D. (Department of Veterinary Anatomy, Graduate School of Agricultural & Life Sciences, The University of Tokyo, 1-1-1 Yayoi Bunkyo-Ku, Tokyo, Japan); Grampus Griseus Mitochondrial DNA, D-Loop Region, Accession Number AB018584. 1998. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AB018584.1/ (accessed on 7 July 2021).
- Kitamura, S.; Abe, S. (Hokkaido University, Graduate School of Fisheries Sciences, Minato 3-1-1, Hakodate, Hokkaido 041-8611, Japan); Grampus Griseus Mitochondrial DNA, D-Loop, Partial Sequence, Accession Number AB610375. 2011. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AB610375 (accessed on 7 July 2021).
- Morin, P.A.; Archer, F.I.; Foote, A.D.; Vilstrup, J.; Allen, E.E.; Wade, P.; Durban, J.; Parsons, K.; Pitman, R.; Li, L. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 2010, 20, 908–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, K.; Takashi, Y.; Sano, A.; Miyahara, H. (Okinawa Churaumi Aquarium, Marinemammal Section, 424 Ishikawa, Motobu, Okinawa 905-0206, Japan); Pseudorca Crassidens Mitochondrial D-Loop, Partial Sequence, Accession Number AB377522-26. 2008. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AB377522 (accessed on 7 July 2021).
- Kitamura, S.; Abe, S. (Hokkaido University, Graduate School of Fisheries Sciences, Minato 3-1-1, Hakodate, Hokkaido 041-8611, Japan); Pseudorca Crassidens Mitochondrial DNA, D-Loop, Partial Sequence, Accession Number AB610390-91. 2011. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AB610390 (accessed on 7 July 2021).
- Zhang, T.; Yang, G.; Zhou, K.; Wei, F. (Institute of Genetic Resources, Nanjing Normal University, Ninghai Road 122#, Nanjing, Jiangsu 210097, China); Pseudorca Crassidens Isolate NJNU0072 Mitochondrial Control Region, Partial Sequence, Accession Number AY729039-41,45. 2004. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AY729039 (accessed on 7 July 2021).
- Baker, C.S. (School of Biological Sciences, University of Auckland, 3a Symonds Street, Auckland 1001, New Zealand); Pseudorca Crassidens Voucher PcraNZ1.MD Mitochondrial Control Region, Partial Sequence, Accession Number DQ145024. 2005. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ145024 (accessed on 7 July 2021).
- Rosel, P.E. (National Marine Fisheries Service, 646 Cajundome Blvd., Lafayette, LA 70506, USA); Pseudorca Crassidens HAPLOTYPE Pcra001 Mitochondrial Control Region, Partial Sequence, Accession Number DQ845436. 2006. Available online: https://www.ncbi.nlm.nih.gov/nuccore/DQ845436 (accessed on 7 July 2021).
- Chivers, S.J.; Baird, R.W.; Martien, K.M.; Taylor, B.L.; Archer, E.; Gorgone, A.M.; Hancock, B.L.; Hedrick, N.M.; Matilla, D.; McSweeney, D.J. Evidence of genetic differentiation for Hawaii insular false killer whales (Pseudorca crassidens); Cascadia Research Collective: Olympia, WA, USA, 2010. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).