Microsatellite Genotyping of Two Bulgarian Sheep Breeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
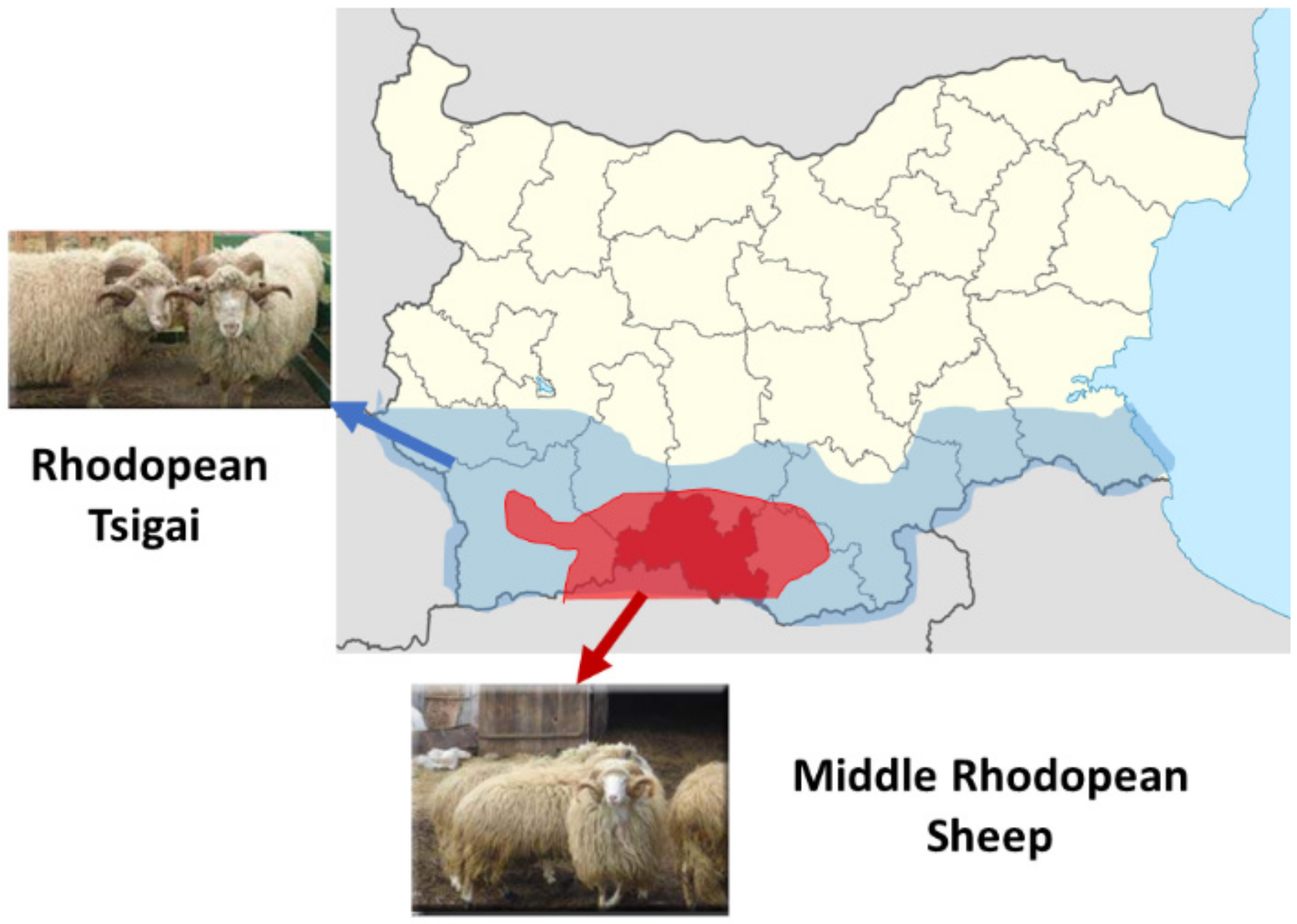
2.2. Animal Sampling and DNA Isolation
2.3. Microsatellite Markers
2.4. PCR Amplification and Fragment Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Population Genetic Diversity Based on Microsatellite Markers
3.2. Genetic Diversity between the Rhodopean Tsigai and the Middle Rhodopean Sheep
3.3. Genetic Population Structure
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaidarska, V.M.; Ignatova, M.M.; Lytskanov, P.I. Genetic resources of farm animals in bulgaria–conservation and management. Anim. Breed. Genet. 2017, 53, 35–43. [Google Scholar] [CrossRef]
- Tanchev, S. Conservation of genetic resources of autochthonous domestic livestock breeds in Bulgaria. A review. Bulg. J. Agric. Sci. 2015, 21, 1262–1271. [Google Scholar]
- Thornton, P.K.; Thornton, P.K. Livestock production: Recent trends, future prospects. Phil. Trans. R. Soc. B 2010, 365, 2853–2867. [Google Scholar] [CrossRef] [PubMed]
- Collard, R.-C. Putting animals back together, taking commodities apart. Ann. Am. Assoc. Geogr. 2014, 104, 151–165. [Google Scholar] [CrossRef]
- Dumroese, R.K.; Williams, M.I.; Stanturf, J.A.; Clair, J. Considerations for restoring temperate forests of tomorrow: Forest restoration, assisted migration, and bioengineering. New For. 2015, 46, 947–964. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Yang, J.; Lv, F.H.; Hu, X.J.; Xie, X.L.; Zhang, M.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef]
- Acatrinei, A.; Rusu, I.; Mircea, C.; Zagrean-Tuza, C.; Gál, E.; Păceșilă, D.; Gâza, O.; Urduzia, C.; Pinter, Z.K.; Dobrinescu, C.; et al. Shedding light on the dark ages: Sketching potential trade relationships in early Medieval Romania through mitochondrial DNA analysis of sheep remains. Diversity 2021, 13, 208. [Google Scholar] [CrossRef]
- Ilişiu, E.; Dărăban, S.; Radu, R.; Pădeanu, I.; Ilişiu, V.C.; Pascal, C.; Rahmann, G. The Romanian Tsigai sheep breed, their potential and the challenges for research. Appl. Agric. For. Res. 2013, 2, 161–170. [Google Scholar]
- Dimov, D. The autochthonous sheep breeds of Bulgaria. In Possible Way of Conservation the Multipurpose Tsigai and Other Indigenous Sheep Breeds in Central, Earstern European and Balkan countries; Kukovics, S., Kume, K., Eds.; Budapest Printing House: Budapest, Hungary, 2006; pp. 137–164. [Google Scholar]
- Nikolov, V. Livestock Breeds in the Republic of Bulgaria. Catalog, 3rd ed.; Animal Genetic Resources/Resources Génétiques animales/Recursos Genéticos Animales: Sofia, Bulgaria, 2012; Volume 51, p. 150. [Google Scholar]
- Nikolov, V. Review of the specific measures for support of the autochthonous breeds in Bulgaria. J. Cent. Eur. Agric. 2015, 16, 38–46. [Google Scholar] [CrossRef]
- Genkovski, D. Comparative Characterization of Sheep of Different Breeds, Cultivated in the Conditions of Central Stara Planina, Dissertation; Bulgarian Academy of Sciences: Sofia, Bulgaria, 2002. [Google Scholar]
- Hlebarov, G. Study on Bulgarian Local Sheep and Possibilities for Their Improvement; Bulgarian Academy of Sciences: Sofia, Bulgaria, 1940; p. 187. [Google Scholar]
- Vasilev, D.; Dimov, G.; Odjacova, T.S. Dynamics of productive ability of srednorhoropska native sheep. Maced. Agric. Rev. 2000, 47, 65–69. [Google Scholar]
- Dimov, D.; Vuchkov, A. Sheep genetic resources in Bulgaria with focus on breeds with coloured wool. Genet. Resour. 2021, 2, 11–24. [Google Scholar] [CrossRef]
- Hristova, D.; Todorovska, E.; Vassilev, D.; Metodiev, S.; Popov, I.; Yablanski, T.; Zhelyazkov, E. Microsatellites based genetic diversity and population structure of seven Bulgarian indigenous sheep breeds. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 569–581. [Google Scholar]
- Hristova, D.; Metodiev, S.; Nikolov, V.; Vassilev, D.; Todorovska, E. Genetic variation of Bulgarian autochthonous sheep breeds using microsatellite markers. Genetika 2017, 49, 247–258. [Google Scholar] [CrossRef]
- Kusza, S.; Nagy, I.; Sasvári, Z.; Stágel, A.; Németh, T.; Molnár, A.; Kume, K.; Bosze, Z.; Jávor, A.; Kukovics, S. Genetic diversity and population structure of Tsigai and Zackel type of sheep breeds in the Central-, Eastern-and Southern-European regions. Small Rumin. Res. 2008, 78, 13–23. [Google Scholar] [CrossRef]
- Schandl, J. Juhteny´Eszt´Es (Sheep Breeding); Mez˝ogazdas´agi Kiad´o: Budapest, Hungary, 1960. [Google Scholar]
- Niżnikowski, R.; Strzelec, E.; Popielarczyk, D. Economics and profitability of sheep and goat production under new support regimes and market conditions in Central and Eastern Europe. Small Rumin. Res. 2006, 62, 159–165. [Google Scholar] [CrossRef]
- Staikova, G.; Stancheva, N. Effect of some factors on the wool yield and staple length at different ages in sheep from the Northeast Bulgarian Fine Fleece Breed-Shumen type. Bulg. J. Agric. Sci. 2009, 15, 463–470. [Google Scholar]
- Kukovics, S.; Kume, K. Cooperation in the preservation of sheep breeds. In Possible Way of Conservation the Multipurpose Tsigai and other Indigenous Sheep Breeds in Central, Earstern European and Balkan Countries; Kukovics, S., Kume, K., Eds.; Budapest Printing House: Budapest, Hungary, 2006; pp. 117–122. [Google Scholar]
- The Ministry of Agriculture, Food and Forestry of Bulgaria. 2017. Available online: https://www.mzh.government.bg/bg/statistika-i-analizi/izsledvane-zhivotnovdstvo/normativna-uredba (accessed on 16 March 2021).
- Fischer, M.C.; Rellstab, C.; Leuzinger, M.; Roumet, M.; Gugerli, F.; Shimizu, K.K.; Holderegger, R.; Widmer, A. Estimating Genomic Diversity and Population Differentiation—An Empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genom. 2017, 18, 69. [Google Scholar] [CrossRef]
- Xia, Q.; Wang, X.; Pan, Z.; Zhang, R.; Wei, C.; Chu, M.; Di, R. Genetic diversity and phylogenetic relationship of nine sheep populations based on microsatellite markers. Arch. Anim. Breed. 2021, 64, 7–16. [Google Scholar] [CrossRef]
- Olschewsky, A.; Hinrichs, D. An overview of the use of genotyping techniques for assessing genetic diversity in local farm animal breeds. Animals 2021, 11, 2016. [Google Scholar] [CrossRef]
- Abebe, A.S.; Mikko, S.; Johansson, A.M. Genetic diversity of five local Swedish chicken breeds detected by microsatellite markers. PLoS ONE 2015, 10, e0120580. [Google Scholar] [CrossRef]
- Abdelkader, A.A.; Ata, N.; Benyoucef, M.T.; Djaout, A.; Azzi, N.; Yilmaz, O.; Cemal, İ.; Gaouar, S.B.S. New genetic identification and characterisation of 12 Algerian sheep breeds by microsatellite markers. Ital. J. Anim. Sci. 2018, 17, 38–48. [Google Scholar] [CrossRef]
- Srivastava, S.; Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019, 20, 153. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; de Freitas Munhoz, C. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef] [PubMed]
- FAO. Molecular Genetic Characterization of Animal Genetic Resource. Available online: http://www.fao.org/docrep/ (accessed on 12 September 2021).
- ISAG. Applied Genetics in sheep and goats workshop. In Proceedings of the 32nd International Conference on Animal Genetics, Edinburgh, UK, 23 November 2021. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358. [Google Scholar] [PubMed]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.; Ye, Z.H.; Mao, J.X. PopGene, the User-Friendly Shareware for Population Genetic Analysis, Molecular Biology and Biotechnology Center; University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Belkhir, K. GENETIX 4.04, Logiciel Sous Windows TM Pour la Genetique des Populations; Laboratories Genome, Populations, Interactions, Universite de Montpellier II, CNRS UMR 5000: Montpellier, France, 2004. [Google Scholar]
- Agung, P.P.; Saputra, F.; Zein, M.; Wulandari, A.S.; Putra, W.; Said, S.; Jakaria, J. Genetic diversity of Indonesian cattle breeds based on microsatellite markers. Asian-Australas. J. Anim. Sci. 2019, 32, 467–476. [Google Scholar] [CrossRef]
- Ren, A.; Du, K.; Jia, X.; Yang, R.; Wang, J.; Chen, S.-Y.; Lai, S.-J. Genetic diversity and population structure of four Chinese rabbit breeds. PLoS ONE 2019, 14, e0222503. [Google Scholar] [CrossRef]
- Mihailova, Y. Genetic diversity and structure of 2 indigenous sheep breeds (Kotel and Teteven) in Bulgaria using microsatellite markers. Biotechnol. Biotechnol. Equip. 2021, 35, 576–585. [Google Scholar] [CrossRef]
- Kusza, S.; Dimov, D.; Nagy, I.; Bõsze, Z.; Jávor, A.; Kukovics, S. Microsatellite analysis to estimate genetic relationships among five bulgarian sheep breeds. Genet. Mol. Biol. 2010, 33, 51–56. [Google Scholar] [CrossRef]
- Kdidi, S.; Calvo, J.H.; González-Calvo, L.; Ben Sassi, M.; Khorchani, T.; Yahyaoui, M.H. Genetic relationship and admixture in four Tunisian sheep breeds revealed by microsatellite markers. Small Rumin. Res. 2015, 131, 64–69. [Google Scholar] [CrossRef]
- Gaouar, S.B.S.; Kdidi, S.; Ouragh, L. Estimating population structure and genetic diversity of five Moroccan sheep breeds by microsatellite markers. Small Rumin. Res. 2016, 144, 23–27. [Google Scholar] [CrossRef]
- Yilmaz, O.; Cemal, I.; Karaca, O.; Ata, N. Molecular genetic characterization of Kivircik sheep breed raised in Western Anatolia. Livest. Res. Rural Dev. 2016, 28, 40. [Google Scholar]
- Álvarez, I.; Capote, J.; Traoré, A.; Fonseca, N.; Pérez, K.; Cuervo, M.; Fernández, I.; Goyache, F. Genetic relationships of the Cuban hair sheep inferred from microsatellite polymorphism. Small Rumin. Res. 2012, 104, 89–93. [Google Scholar] [CrossRef][Green Version]
- De Meeûs, T. Revisiting FIS, FST, Wahlund Effects, and Null Alleles. J. Hered. 2018, 109, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Manangwa, O.; De Meeûs, T.; Grébaut, P.; Ségard, A.; Byamungu, M.; Ravel, S. Detecting Wahlund effects together with amplification problems: Cryptic species, null alleles and short allele dominance in Glossina pallidipes populations from Tanzania. Mol. Ecol. Resour. 2019, 19, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Oner, Y.; Ustüner, H.; Orman, A.; Yilmaz, O.; Yılmaz, A. Genetic Diversity of Kıvırcık Sheep Breed Reared in Different Regions and its Relationship with Other Sheep Breeds in Turkey. Ital. J. Anim. Sci. 2014, 13, 3. [Google Scholar] [CrossRef]
- Kusza, S.; Nagy, I.; Németh, T.; Molnár, A.; Jávor, A.; Kukovics, S. The genetic variability of Hungarian Tsigai sheep. Arch. Anim. Breed. 2010, 53, 309–317. [Google Scholar] [CrossRef]
- Ćinkulov, M.; Tapio, M.; Ozerov, M.; Kiselyova, T.; Marzanov, N.; Pihler, I.; Olsaker, I.; Vegara, M.; Kantane, J. Genetic differentiation between the Old and New types of Serbian Tsigai sheep. Genet. Sel. Evol. 2008, 40, 321–331. [Google Scholar] [CrossRef]
- Kusza, S.; Gyarmathy, E.; Dubravska, J.; Nagy, I.; Jávor, A.; Kukovics, S. Study of genetic differences among Slovak Tsigai populations using microsatellite markers. Czech J. Anim. Sci. 2009, 54, 468–474. [Google Scholar] [CrossRef]
- Zãhan, M.; Miclea, V.; Raica, P.; Miclea, I.; Ilisiu, E. Analysis of Genetic Variation within Tsigai Population from Romania Using Microsatellite Markers. Anim. Sci. Biotechnol. 2011, 68, 396–400. [Google Scholar]
- Herold, P.; Roessler, R.; Willam, A.; Momm, H.; Zárate, A.V. Breeding and supply chain systems incorporating local pig breeds for small–scale pig producers in Northwest Vietnam. Livest. Prod. Sci. 2012, 129, 63–72. [Google Scholar] [CrossRef]
- Rege, J.E.O.; Gibson, J.P. Animal genetic resources and economic development: Issues in relation to economic evaluation. Ecol. Econ. 2003, 45, 319–330. [Google Scholar] [CrossRef]

| Microsatellite Marker | Chr. No. | Motif | Primer Sequence 5′–3′ | Product Size (bp) | AnnealingT (°C) |
|---|---|---|---|---|---|
| McM042 | 9 | (AC)n | F: GTTCGTACTTCTGGGTACTGGTCTC R: GTCCATGGATTTGCAGAGTCAG | 81–107 | 60 |
| McM527 | 5 | (TG)n | F: GTCCATTGCCTCAAATCAATTC R: AAACCACTTGACTACTCCCCAA | 158–182 | 56 |
| INRA005 | 12 | (GT)n | F: TCAGGCATACCCTACACCACATG R: AATATTAGCCAACTGAAAACTGGG | 125–147 | 54 |
| INRA006 | 1 | (CA)n | F: AGGAATATCTGTATCAACCGCAGTC R: CTGAGCTGGGGTGGGAGCTATAAATA | 110–132 | 58 |
| INRA023 | 3 | (AC)n | F: GAGTAGAGCTACAAGATAAACTTC R: TAACTACAGGGTGTTAGATGAACTC | 194–216 | 58 |
| INRA063 | 14 | (AC)n | F: GACCACAAAGGGATTTGCACAAGC R: AAACCACAGAAATGCTTGGAAG | 169–201 | 56 |
| INRA172 | 22 | (TG)n | F: CCAGGGCAGTAAAATGCATAACTG R: GGCCTTGCTAGCCTCTGCAAAC | 126–160 | 58 |
| ETH152 | 5 | (AC)n | F: TACTCGTAGGGCAGGCTGCCTG R: GAGACCTCAGGGTTGGTGATCAG | 186–200 | 60 |
| CSRD247 | 14 | (CA)n | F: GGACTTGCCAGAACTCTGCAAT R: CACTGTGGTTTGTATTAGTCAGG | 209–255 | 58 |
| OarFSB20 | 2 | (TG)n | F: GGAAAACCCCCATATATACCTATAC R: AAATGTGTTTAAGATTCCATACATGTG | 87–113 | 58 |
| MAF065 | 15 | (CA)n | F: AAAGGCCAGAGTATGCAATTAGGAG R: CCACTCCTCCTGAGAATATAACATG | 125–137 | 56 |
| MAF214 | 16 | (GT)n | F: AATGCAGGAGATCTGAGGCAGGGACG R: GGGTGATCTTAGGGAGGTTTTGGAGG | 189–265 | 58 |
| Locus | Na | Ne | PIC | Ho | He | I | FITa | FIS | FSTa | DST | HT | GST |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| McM042 | 8 | 2.65 | 0.44 | 0.60 | 0.62 | 1.35 | 0.048 | 0.026 | 0.023 | 0.056 | 0.633 | 0.017 |
| INRA006 | 10.5 | 3.52 | 0.67 | 0.73 | 0.72 | 1.63 | −0.006 | −0.018 | 0.011 | 0.030 | 0.724 | 0.006 |
| McM527 | 8.0 | 4.51 | 0.76 | 0.69 | 0.78 | 1.71 | 0.125 | 0.110 | 0.018 | 0.084 | 0.791 | 0.011 |
| ETH152 | 5.5 | 3.48 | 0.66 | 0.73 | 0.71 | 1.36 | −0.010 | −0.017 | 0.007 | 0.006 | 0.717 | 0.001 |
| CSRD247 | 12.0 | 6.09 | 0.73 | 0.83 | 0.82 | 2.01 | −0.022 | −0.033 | 0.010 | 0.043 | 0.829 | 0.004 |
| OarFSB20 | 10.5 | 6.85 | 0.81 | 0.85 | 0.85 | 2.06 | 0.015 | 0.000 | 0.015 | 0.116 | 0.865 | 0.009 |
| INRA172 | 8.0 | 3.84 | 0.72 | 0.74 | 0.74 | 1.61 | 0.017 | 0.003 | 0.014 | 0.046 | 0.749 | 0.008 |
| INRA063 | 12.5 | 5.92 | 0.80 | 0.77 | 0.83 | 2.07 | 0.090 | 0.071 | 0.020 | 0.147 | 0.847 | 0.014 |
| INRA005 | 14.5 | 7.83 | 0.88 | 0.86 | 0.87 | 2.32 | 0.035 | 0.017 | 0.019 | 0.202 | 0.889 | 0.013 |
| INRA023 | 10.5 | 6.82 | 0.85 | 0.82 | 0.85 | 2.08 | 0.054 | 0.041 | 0.014 | 0.099 | 0.865 | 0.008 |
| MAF065 | 9.0 | 4.06 | 0.70 | 0.69 | 0.75 | 1.64 | 0.096 | 0.085 | 0.013 | 0.042 | 0.763 | 0.006 |
| MAF214 | 12.0 | 4.00 | 0.73 | 0.67 | 0.75 | 1.76 | 0.114 | 0.109 | 0.005 | −0.011 | 0.753 | −0.002 |
| Mean | 10.08 | 4.96 | 0.73 | 0.75 | 0.77 | 1.79 | 0.046 | 0.033 | 0.014 | 0.059 | 0.786 | 0.008 |
| Breed | Na | Ne | Ho | He | FIS | NPA | No. Different Alleles (Freq ≥ 5%) |
|---|---|---|---|---|---|---|---|
| RT | 9.58 | 4.84 | 0.76 | 0.77 | 0.01 | 1.58 | 4.92 |
| MRS | 10.58 | 5.09 | 0.73 | 0.77 | 0.06 | 2.58 | 5.17 |
| Mean | 10.08 | 5.02 | 0.74 | 0.77 | 0.035 | 2.08 | 5.04 |
| Locus | Allele | Frequency (%) | Breed |
|---|---|---|---|
| McM527 | 176 | 8.7 | Middle Rhodopean Sheep |
| CSRD247 | 255 | 4.4 | Middle Rhodopean Sheep |
| OarFSB20 | 103 109 | 4.4 5.4 | Middle Rhodopean Sheep |
| INRA172 | 156 162 | 4.4 11.0 | Middle Rhodopean Sheep |
| INRA063 | 193 | 3.7 | Rhodopean Tsigai |
| INRA023 | 220 | 3.7 | Rhodopean Tsigai |
| MAF065 | 137 | 3.7 | Rhodopean Tsigai |
| MAF214 | 261 | 2.4 | Rhodopean Tsigai |
| Breed | Locus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| McM042 | INRA006 | McM527 | ETH152 | CSRD247 | OarFSB20 | INRA172 | INRA063 | INRA005 | INRA023 | MAF065 | MAF214 | |
| RT | 0.271 | 0.925 | 0.869 | 0.000 *** | 0.966 | 0.463 | 0.802 | 0.000 *** | 0.176 | 0.497 | 0.051 | 0.111 |
| MRS | 0.059 | 0.978 | 0.215 | 0.043 * | 0.179 | 0.226 | 0.341 | 0.368 | 0.676 | 0.855 | 0.086 | 0.003 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odjakova, T.; Todorov, P.; Radoslavov, G.; Hristov, P. Microsatellite Genotyping of Two Bulgarian Sheep Breeds. Diversity 2022, 14, 210. https://doi.org/10.3390/d14030210
Odjakova T, Todorov P, Radoslavov G, Hristov P. Microsatellite Genotyping of Two Bulgarian Sheep Breeds. Diversity. 2022; 14(3):210. https://doi.org/10.3390/d14030210
Chicago/Turabian StyleOdjakova, Tsonka, Pavel Todorov, Georgi Radoslavov, and Peter Hristov. 2022. "Microsatellite Genotyping of Two Bulgarian Sheep Breeds" Diversity 14, no. 3: 210. https://doi.org/10.3390/d14030210
APA StyleOdjakova, T., Todorov, P., Radoslavov, G., & Hristov, P. (2022). Microsatellite Genotyping of Two Bulgarian Sheep Breeds. Diversity, 14(3), 210. https://doi.org/10.3390/d14030210








