Pacificimonas pallium sp. nov., an Isolated Bacterium from the Mantle of Pacific Oyster Crassostrea gigas in Germany, and Prediction of One-Carbon Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation
2.2. Morphological, Physiological, and Biochemical Studies
2.3. 16S rRNA Gene Analysis
2.4. Chemotaxonomy
2.5. Whole-Genome Analysis
2.6. Secondary Metabolite Production and Antimicrobial Activity
2.7. Chromatogram and Mass Analysis for Extraction of WHA3T
3. Result and Discussion
3.1. Morphological, Physiological, and Biochemical Results
3.2. 16S rRNA Gene Analysis
3.3. Chemotaxonomic Characterization
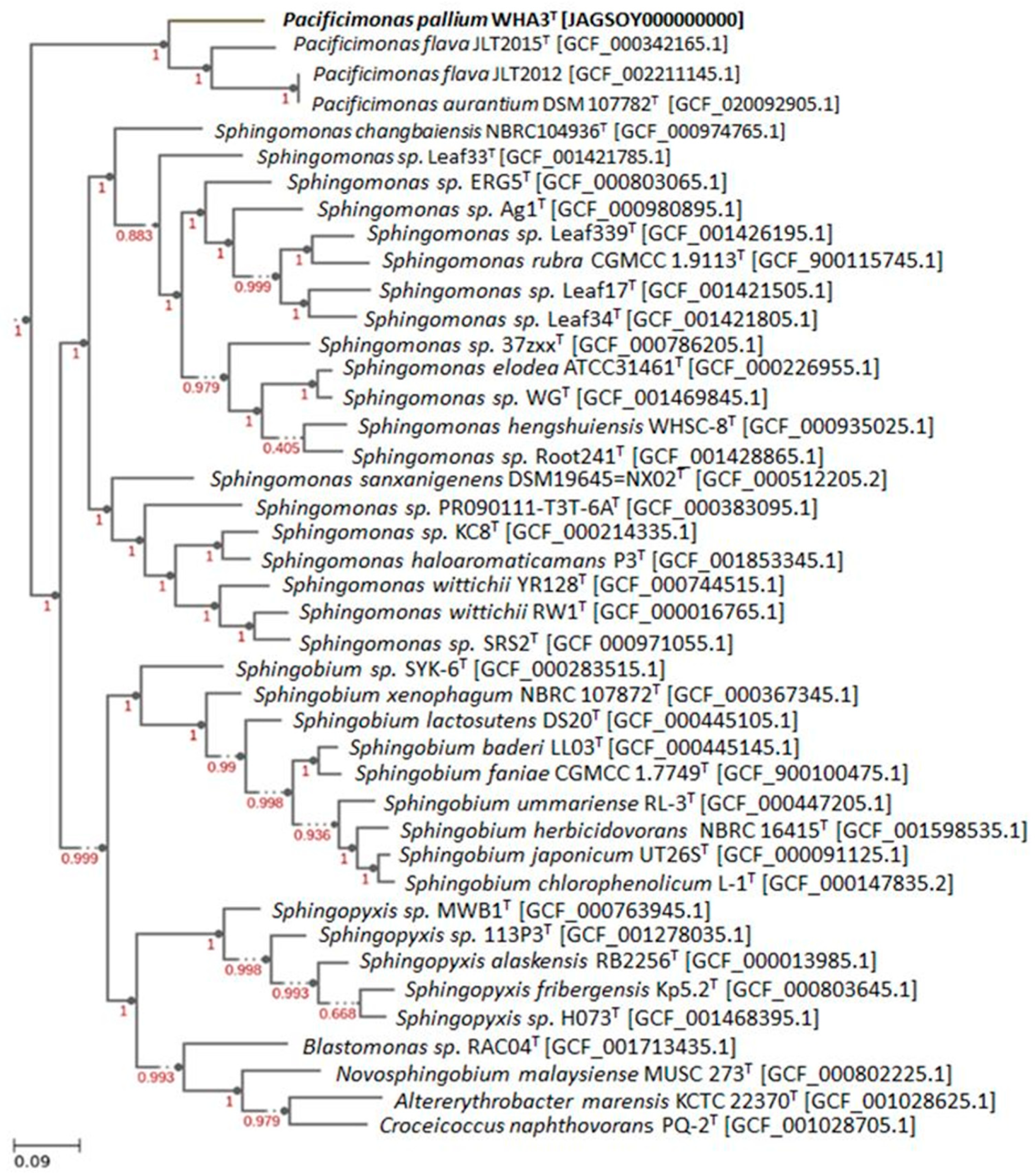
3.4. Genomic Characteristics and Phylogenomic Analysis
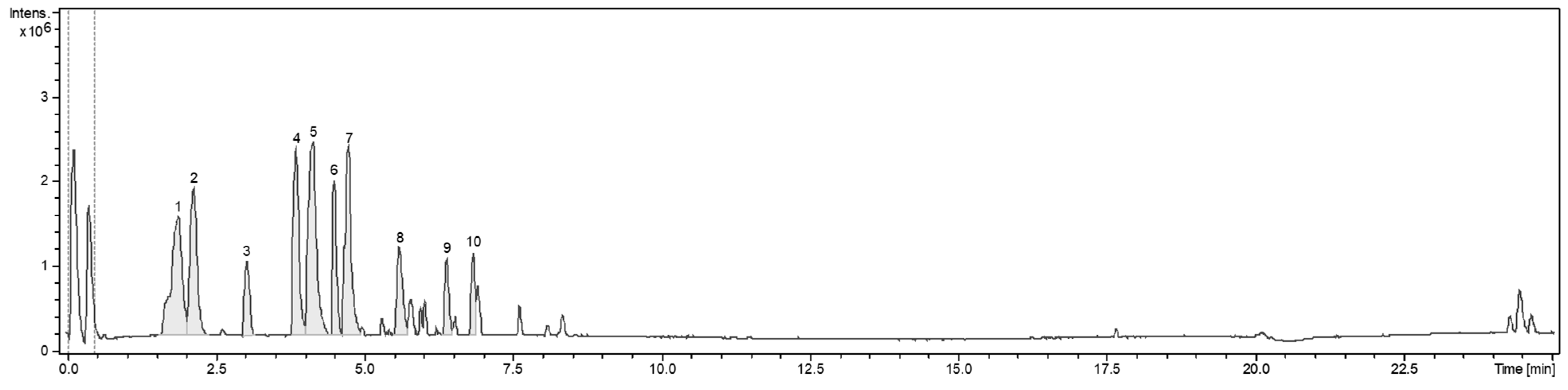
3.5. Chromatogram and Mass Analysis for Extraction of WHA3T
4. Conclusions
Description of Pacificimonas pallium sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kosako, Y.; Yabuuchi, E.; Naka, T.; Fujiwara, N.; Kobayashi, K. Proposal of Sphingomonadaceae fam. nov., consisting of Sphingomonas Yabuuchi et al. 1990, Erythrobacter Shiba and Shimidu 1982, Erythromicrobium Yurkov et al. 1994, Porphyrobacter Fuerst et al. 1993, Zymomonas Kluyver and van Niel 1936, and Sandaracinobacter Yurkov et al. 1997, with the type genus Sphingomonas Yabuuchi et al. 1990. Microbiol. Immunol. 2000, 44, 563–575. [Google Scholar] [CrossRef]
- Liu, K.; Li, S.; Jiao, N.; Tang, K. Pacificamonas flava gen. nov., sp. nov., a novel member of the family Sphingomonadaceae isolated from the Southeastern Pacific. Curr. Microbiol. 2014, 69, 96–101. [Google Scholar] [CrossRef]
- Li, S.; Zhou, W.; Lin, D.; Tang, K.; Jiao, N. Pacificimonas aurantium sp. nov., Isolated from the Seawater of the Pacific Ocean. Curr. Microbiol. 2016, 72, 752–757. [Google Scholar] [CrossRef]
- Lauro, F.M.; McDougald, D.; Thomas, T.; Williams, T.J.; Egan, S.; Rice, S.; DeMaere, M.Z.; Ting, L.; Ertan, H.; Johnson, J.; et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 15527–15533. [Google Scholar] [CrossRef] [Green Version]
- Sowell, S.M.; Wilhelm, L.J.; Norbeck, A.D.; Lipton, M.S.; Nicora, C.D.; Barofsky, D.F.; Carlson, C.A.; Smith, R.D.; Giovanonni, S.J. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 2009, 3, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, M.; Hamana, K.; Hiraishi, A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [Google Scholar] [CrossRef]
- Maruyama, T.; Park, H.D.; Ozawa, K.; Tanaka, Y.; Sumino, T.; Hamana, K.; Hiraishi, A.; Kato, K. Sphingosinicella microcystinivorans gen. nov., sp. nov., a microcystin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 85–89. [Google Scholar] [CrossRef]
- Stingl, U.; Desiderio, R.A.; Cho, J.-C.; Vergin, K.L.; Giovannoni, S.J. The SAR92 Clade: An abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Env. Microbiol. 2007, 73, 2290–2296. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Steindler, L.; Thrash, J.C.; Halsey, K.H.; Smith, D.P.; Carter, A.E.; Landry, Z.C.; Giovannoni, S.J. One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE 2011, 6, e23973. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Jiao, N.; Liu, K.; Zhang, Y.; Li, S. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: Implications for dissolved organic matter utilization. PLoS ONE 2012, 7, e41204. [Google Scholar] [CrossRef] [Green Version]
- Noinaj, N.; Guillier, M.; Barnard, T.J.; Buchanan, S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010, 64, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Landwehr, W.; Kampfer, P.; Glaeser, S.P.; Ruckert, C.; Kalinowski, J.; Blom, J.; Goesmann, A.; Mack, M.; Schumann, P.; Atasayar, E.; et al. Taxonomic analyses of members of the Streptomyces cinnabarinus cluster, description of Streptomyces cinnabarigriseus sp. nov. and Streptomyces davaonensis sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 382–393. [Google Scholar] [CrossRef]
- Kutzner, H.J. The Family Streptomycetaceae; Springer: Berlin/Heidelberg, Germany, 1981. [Google Scholar]
- Rüger, H.J.; Krambeck, H.J.J.S.; Microbiology, A. Evaluation of the BIOLOG substrate metabolism system for classification of marine bacteria. Syst. Appl. Microbiol. 1994, 17, 281–288. [Google Scholar] [CrossRef]
- Legat, A.; Gruber, C.; Zangger, K.; Wanner, G.; Stan-Lotter, H. Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species. Appl. Microbiol. Biotechnol. 2010, 87, 1119–1127. [Google Scholar] [CrossRef] [Green Version]
- Chaiya, L.; Matsumoto, A.; Wink, J.; Inahashi, Y.; Risdian, C.; Pathom-aree, W.; Lumyong, S. Amycolatopsis eburnea sp. nov., an actinomycete associated with arbuscular mycorrhizal fungal spores. Int. J. Syst. Evol. Microbiol. 2019, 69, 3603–3608. [Google Scholar] [CrossRef]
- Hall, T.A. BIOEDIT: A user- friendly biological sequence alignment editor and analysis for windows 95/98/ NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Goker, M.; Sproer, C.; Klenk, H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics-Int. J. Willi Hennig Soc. 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef]
- Swofford, D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2002; Volume 4. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Risdian, C.; Landwehr, W.; Rohde, M.; Schumann, P.; Hahnke, R.L.; Sproer, C.; Bunk, B.; Kampfer, P.; Schupp, P.J.; Wink, J. Streptomyces bathyalis sp. nov., an actinobacterium isolated from the sponge in a deep sea. Antonie Van Leeuwenhoek 2021, 114, 425–435. [Google Scholar] [CrossRef]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl. 1990, 20, 1–6. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic. Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic. Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic. Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Goker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic. Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-m.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [Green Version]
- Ateba, T.P.; Alayande, K.A.; Mwanza, M. Feces metagenomes and metagenome-assembled genome sequences from two separate dogs (Canis lupus familiaris) with multiple diarrheal episodes. Microbiol. Resour. Announc. 2020, 9, e01065-20. [Google Scholar] [CrossRef]
- Guse, A.; Stevenson, C.E.; Kuper, J.; Buchanan, G.; Schwarz, G.; Giordano, G.; Magalon, A.; Mendel, R.R.; Lawson, D.M.; Palmer, T. Biochemical and structural analysis of the molybdenum cofactor biosynthesis protein MobA. J. Biol. Chem. 2003, 278, 25302–25307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zhang, W.; Lu, J.; Yang, Y.; Chiao, J.; Zhao, G.; Jiang, W. MoeA, an enzyme in the molybdopterin synthesis pathway, is required for rifamycin SV production in Amycolatopsis mediterranei U32. Appl. Microbiol. Biotechnol. 2002, 60, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.A.; Hickerson, S.M.; Vickers, T.J.; Beverley, S.M. The role of the mitochondrial glycine cleavage complex in the metabolism and virulence of the protozoan parasite Leishmania major. J. Biol. Chem. 2008, 283, 155–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dworkin, M. The Prokaryotes: Vol. 1: Symbiotic Associations, Biotechnology, Applied Microbiology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Toulza, E.; Tagliabue, A.; Blain, S.; Piganeau, G. Analysis of the global ocean sampling (GOS) project for trends in iron uptake by surface ocean microbes. PLoS ONE 2012, 7, e30931. [Google Scholar] [CrossRef] [PubMed]
- Bibby, T.S.; Zhang, Y.; Chen, M. Biogeography of photosynthetic light-harvesting genes in marine phytoplankton. PLoS ONE 2009, 4, e4601. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, M.; Hedges, J.; Benner, R.J.M.C. Major biochemical composition of dissolved high molecular weight organic matter in seawater. Mar. Chem. 1996, 55, 281–297. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Pira, H.; Risdian, C.; Kämpfer, P.; Müsken, M.; Schupp, P.J.; Wink, J. Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov. Diversity 2021, 13, 641. [Google Scholar] [CrossRef]
- Khosravi Babadi, Z.; Ebrahimipour, G.; Wink, J.; Narmani, A.; Risdian, C. Isolation and identification of Streptomyces sp. Act4Zk, a good producer of Staurosporine and some derivatives. SfAM 2021, 72, 206–218. [Google Scholar] [CrossRef]
- Rodriguez, R.L.M.; Konstantinidis, K.J.M.M. Bypassing cultivation to identify bacterial species: Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]



| Culture Media | Pacificimonas pallium WHA3T | Pacificimonas flava DSM 107612T | Pacificimonas aurantium DSM 107782T |
|---|---|---|---|
| MB 514f | RAL2003 Pastel orange | RAL1037 Sun yellow | RAL1032 Broom yellow |
| CSY-3 | RAL2008 Bright red orange | RAL1002 Sand yellow | RAL1002 Sand yellow |
| 1.5 LBM | RAL2003 Pastel orange | RAL1033 Dahlia yellow | RAL1037 Sun yellow |
| YED | RAL2000 Yellow orange | RAL1002 Sand yellow | RAL1005 Honey yellow |
| TSA | NG | RAL2000 Yellow orange | NG |
| SSM + T | NG | RAL1011 Brown beige | NG |
| SSM-T | NG | RAL1002 Sand yellow | NG |
| YTSS | RAL1017 Saffron yellow | RAL1003 Signal yellow | RAL1003 Signal yellow |
| ASG + Cycloheximide | NG | NG | RAL1015 Light ivory |
| YEA | NG | RAL1023 Traffic yellow | NG |
| Enzyme | Observation | Enzyme | Observation |
|---|---|---|---|
| Phosphatase alkaline | ++ | Naphtol-AS-BI-phosphohydrolase | (+) |
| Esterase (C4) | + | α-galactosidase | - |
| Esterase lipase (C8) | + | β-galactosidase | - |
| Lipase (C14) | (+) | β-glucurunidase | - |
| Leucin arylamidase | ++ | α-glucosidase | - |
| Valine arylamidase | (+) | β-glucosidase | - |
| Cystine arylamidase | (+) | N-acetyl-beta- glucosaminidase | - |
| Trypsin | + | α-mannosidase | - |
| Chymotrypsin | ++ | α-fucosidase | - |
| Phosphatase acid | (+) |
| Characteristics | 1 | 2 | 3 |
|---|---|---|---|
| Color of colony | orange | orange | orange |
| Temperature range for growth (°C) | 15–40 | 20–30 * | 10–40 † |
| pH spectrum for growth | 5–11 | 5–12 * | 5–10 † |
| NaCl optimum for growth (%) | 2.5–5 | 0.5–7 * | 0.5–4 † |
| Trypsin | + | + | (+) |
| α-Chymotrypsin | ++ | (+) | - |
| Acid phosphatase | (+) | ++ | (+) |
| Naphthol-AS-BI-phosphohydrolase | (+) | + | (+) |
| N-acetyl-β-glucosaminidase | - | (+) | (+) |
| gelatine (hydrolysis) | - | + | + |
| Oxidation of (Biolog GN2) | |||
| Acetic acid | + | + * | - † |
| Methyl pyruvate | - | + * | - † |
| D-galactose | - | ND * | + † |
| D-raffinose | - | - * | + † |
| D-trehalose | - | - * | + † |
| D-Turanose | - | - * | + † |
| α -keto glutaric acid | - | - * | + † |
| L-alanine | + | - * | + † |
| L-glutamic acid | - | - * | + † |
| L-aspartic acid | - | - * | + † |
| γ-amino butyric acid | - | ND * | + † |
| Dextrin | - | + * | + † |
| D-fructose | - | + * | + † |
| α-D-glucose | (+) | + * | + † |
| Tween 40 | - | + * | + † |
| Susceptibility to | |||
| Ampicillin | - | - * | + † |
| Gentamycin | + | - * | + † |
| Kanamycin | + | - * | + † |
| Polar lipids | DPG-PE-PG-SGL-PL-GL-L | DPG-PE-PG-SGL-L | DPG-PE-PG-SGL † |
| Major fatty acid | C18:1ω7c C16:1ω7c | C18:1ω7c C14:02-OH | C18:1ω7c C14:02-OH Unknown fatty acid |
| Contigs | 11 | 32 | 12 |
| No. of proteins | 2794 | 2908 | 3104 |
| rRNA | 3 | 4 | 3 |
| tRNA | 44 | 45 | 45 |
| No. of Genes | 2859 | 3038 | 3185 |
| Other RNA | 3 | 4 | 4 |
| Pseudogenes | 15 | 77 | 29 |
| G + C content (%) | 61.69 | 64.2 * | 65.5 † |
| Total sequence length (Mbp) | 3.01 | 3.25 | 3.34 |
| Pacificimonas pallium WHA3T | Pacificimonas flava DSM 107612T | Pacificimonas aurantium DSM 107782T | |
|---|---|---|---|
| Fatty acid | |||
| C14:0 | -- | 5.0 | -- |
| C15:0 | -- | 2.2 | -- |
| C16:0 | 4.2 | 9.1 | 9.8 |
| C17:0 | -- | -- | 5.2 |
| C19:0 | -- | 2.4 | 8.3 |
| anteiso-C15:0 | -- | 5.7 | -- |
| anteiso-C17:0 | -- | 1.9 | -- |
| iso-C16:0 | -- | 4.1 | -- |
| C16:1ω7c | 20.5 | 5.8 | -- |
| C17:1ω6c | 5.1 | 4.0 | 5.3 |
| C18:1ω7c | 50.6 | 26.9 | 22.0 |
| C14:02-OH | 8.4 | 22.8 | 27.6 |
| C16:02-OH | 3.6 | -- | -- |
| cyclo-C19:0d8,9 | 1.3 | 5.7 | 4.0 |
| Unknown fatty acid | 6.3 | 4.4 | 17.8 |
| Strain | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| POCP (%) | AAI (%) | POCP (%) | AAI (%) | POCP (%) | AAI (%) | POCP (%) | AAI (%) | |
| Pacificimonas pallium WHA3T (JAGSPA000000000) | 60.36 | 66.00 | 60.55 | 64.48 | 49.96 | 54.47 | 33.01 | 46.38 |
| Pacificimonas flava DSM 107612T (JACHGC010000001) | 100 | 100 | 63.39 | 69.22 | 46.92 | 55.02 | 32.12 | 46.42 |
| Pacificimonas aurantium DSM 107782T (JAGSGB000000000) | 63.39 | 69.22 | 100 | 100 | 47.20 | 54.58 | 33.63 | 46.85 |
| Strain | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Ortho ANIu (%) | dDDH (%) | Ortho ANIu (%) | dDDH (%) | Ortho ANIu (%) | dDDH (%) | Ortho ANIu (%) | dDDH (%) | |
| Pacificimonas pallium WHA3T (JAGSPA000000000) | 71.95 | 19.20 | 72.25 | 18.40 | 68.84 | 19.00 | 66.73 | 17.90 |
| Pacificimonas flava DSM 107612T (JACHGC010000001) | 74.36 | 20.30 | 100 | 100 | 69.17 | 19.20 | 67.15 | 16.30 |
| Pacificimonas aurantium DSM 107782T (JAGSGB000000000) | 100 | 100 | 74.36 | 20.30 | 69.13 | 18.90 | 67.05 | 18.30 |
| Genes for C1 Oxidation and Methylotrophy | 1 | 2 | 3 | |
|---|---|---|---|---|
| THF-linked oxidation | ||||
| Methylene -THF methylenetetrahydrofolate reductase metF | + | - | - | |
| Bifunctional methylene-THF dehydrogenase-methenyl-THF cyclohydrolase folD | + | - | - | |
| AMTs | ||||
| Glycine system cleavage T-protein gcvT | + | - | - | |
| Glycine cleavage system H protein gcvH | + | - | - | |
| GSH-dependent pathway | ||||
| S-formyl-glutathione hydrolase (FGH) fghA | + | - | - | |
| methylamine oxidation | ||||
| Sarcosine oxidase subunit beta soxB | + | - | - | |
| Genes TonB-Dependent Transporters (TBDTs) | 1 | 2 | 3 |
|---|---|---|---|
| btuB_1 btuB_3 btuB_4 btuB_6 btuB_8 btuB_9 btuB_10 btuB_12 btuB_13 btuB_14 btuB_15 btuB_17 btuB_19 btuB_20 btuB_21 btuB_23 | btuB | btuB | |
| fyuA_1 fyuA_2 | TonB | TonB | |
| putative TonB-dependent receptor BfrD | + | - | - |
| TonB-dependent hemin and ferrichrome receptor hemR | + | - | - |
| TonB-dependent receptor and iron siderophore receptor protein btuB_7 | + | - | - |
| 1 | 2 | 3 | |
| Vitamin B12 transporter btuB | + | - | - |
| Outer membrane vitamin B12 receptor btuB | + | + | + |
| Biopolymer transport protein | exbD_1 exbD_2 exbD_3 | exbD | exbD |
| Fe(3+) ions import ATP-binding protein FbpC | + | - | - |
| Ferric uptake regulation protein fur | + | + | + |
| Fe(2+) transporter FeoB | + | + | + |
| Superoxide dismutase [Fe] sodB | + | - | - |
| Lipopolysaccharide ABC transporter, ATP-binding protein lptB | + | + | + |
| Capsular polysaccharide ABC transporter, ATP-binding protein kpsT | + | + | - |
| Lipopolysaccharide export system protein lptA | + | - | + |
| Lipopolysaccharide export system permease protein lptF-lptG | + | + | + |
| Capsular polysaccharide export system protein kpsS | + | + | - |
| Capsular polysaccharide ABC transporter, permease protein kpsM | + | + | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pira, H.; Risdian, C.; Müsken, M.; Schupp, P.J.; Wink, J. Pacificimonas pallium sp. nov., an Isolated Bacterium from the Mantle of Pacific Oyster Crassostrea gigas in Germany, and Prediction of One-Carbon Metabolism. Diversity 2022, 14, 181. https://doi.org/10.3390/d14030181
Pira H, Risdian C, Müsken M, Schupp PJ, Wink J. Pacificimonas pallium sp. nov., an Isolated Bacterium from the Mantle of Pacific Oyster Crassostrea gigas in Germany, and Prediction of One-Carbon Metabolism. Diversity. 2022; 14(3):181. https://doi.org/10.3390/d14030181
Chicago/Turabian StylePira, Hani, Chandra Risdian, Mathias Müsken, Peter J. Schupp, and Joachim Wink. 2022. "Pacificimonas pallium sp. nov., an Isolated Bacterium from the Mantle of Pacific Oyster Crassostrea gigas in Germany, and Prediction of One-Carbon Metabolism" Diversity 14, no. 3: 181. https://doi.org/10.3390/d14030181
APA StylePira, H., Risdian, C., Müsken, M., Schupp, P. J., & Wink, J. (2022). Pacificimonas pallium sp. nov., an Isolated Bacterium from the Mantle of Pacific Oyster Crassostrea gigas in Germany, and Prediction of One-Carbon Metabolism. Diversity, 14(3), 181. https://doi.org/10.3390/d14030181








