Population Genetic Differentiation on the Hydrothermal Vent Crabs Xenograpsus testudinatus along Depth and Geographical Gradients in the Western Pacific
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Collection
2.2. Molecular Analyses
2.3. Data Analyses
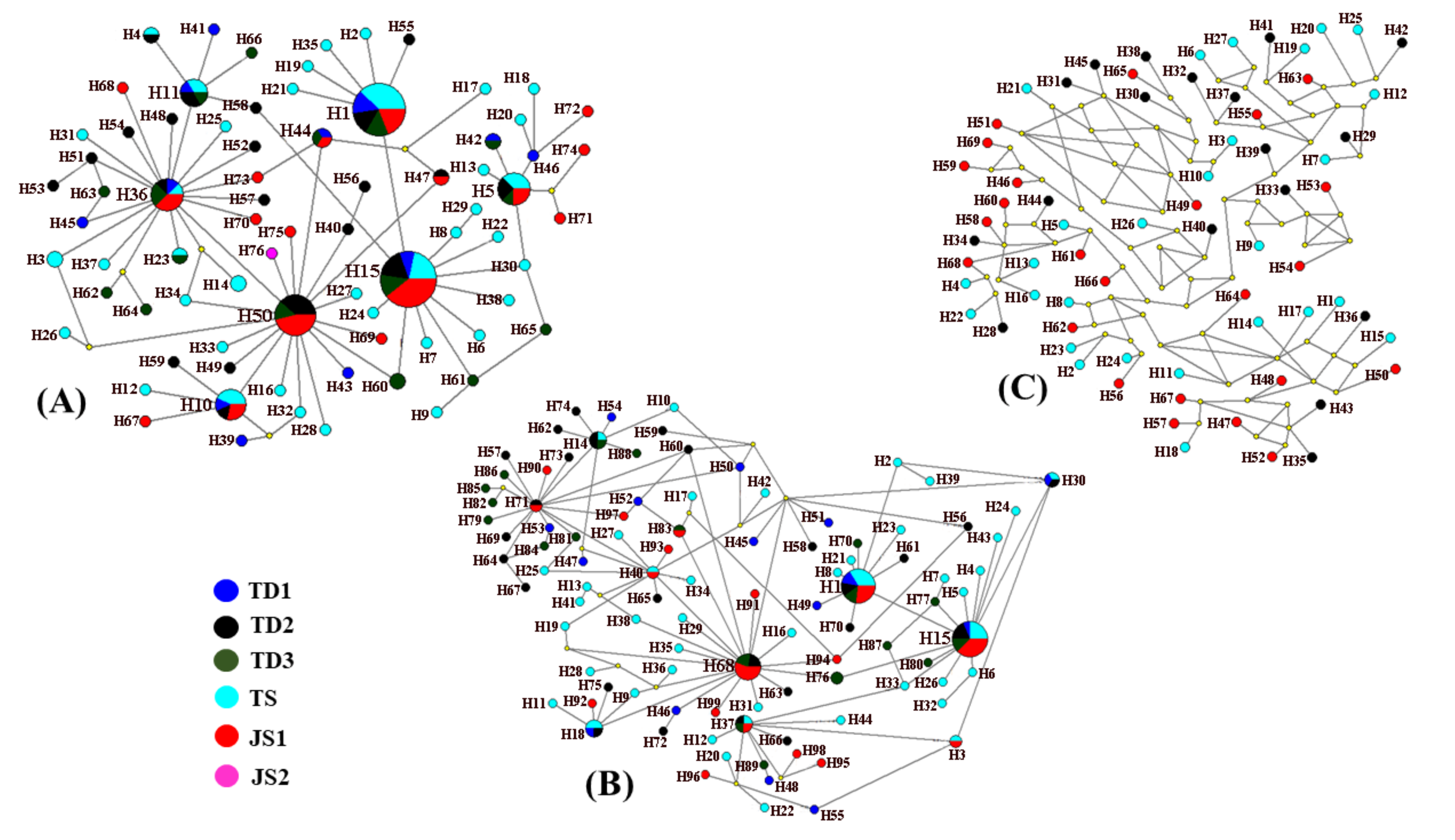
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, K.Y.K.; Grunbaum, D. Temperature and diet modified swimming behaviors of larval sand dollar. Mar. Ecol. Prog. Ser. 2010, 415, 49–59. [Google Scholar] [CrossRef]
- Wong, J.Y.; Chan, B.K.K.; Chan, K.Y.K. Evolution of feeding shapes swimming kinematics of barnacle naupliar larvae: A comparison between trophic modes. Integr. Organ. Biol. 2020, 2, obaa011. [Google Scholar] [CrossRef]
- Wong, J.Y.; Chan, B.K.K.; Chan, K.Y.K. Swimming kinematics and hydrodynamics of barnacle larvae throughout development. Proc. R. Soc. B 2020, 287, 20201360. [Google Scholar] [CrossRef]
- Young, C.M.; Sewell, M.A.; Tyler, P.A.; Metaxas, A. Biogeographic and bathymetric ranges of Atlantic deep-sea echinoderms and ascidians: The role of larval dispersal. Biodiver. Conser. 1997, 6, 1507–1522. [Google Scholar] [CrossRef]
- Arellano, S.M.; Van Gaest, A.L.; Johnson, S.B.; Vrijenhoek, R.C.; Young, C.M. Larvae from deep-sea methane seeps disperse in surface waters. Proc. R. Soc. B 2014, 281, 20133276. [Google Scholar] [CrossRef]
- Hilário, A.; Metaxas, A.; Gaudron, S.M.; Howell, K.L.; Mercier, A.; Mestre, N.C.; Ross, R.E.; Thurnherr, A.M.; Young, C. Estimating dispersal distance in the deep sea: Challenges and applications to marine reserves. Front. Mar. Sci. 2015, 13, 6. [Google Scholar] [CrossRef]
- Tsang, L.M.; Wu, T.H.; Shih, H.-T.; Williams, G.A.; Chu, K.H.; Chan, B.K.K. Genetic and morphological differentiation of the Indo-West Pacific intertidal barnacle Chthamalus malayensis. Integr. Comp. Biol. 2012, 52, 388–409. [Google Scholar] [CrossRef]
- Tsang, L.M.; Achituv, Y.; Chu, K.H.; Chan, B.K.K. Zoogeography of intertidal communities in the West Indian Ocean as determined by ocean circulation systems: Patterns from the Tetraclita barnacles. PLoS ONE 2012, 7, e45120. [Google Scholar] [CrossRef]
- Iacchei, M.; Gaither, M.R.; Bowen, B.W.; Toonen, R.J. Testing dispersal limits in the sea: Range-wide phylogeography of the pronghorn spiny lobster Panulirus penicillatus. J. Biogeogr. 2016, 43, 1032–1044. [Google Scholar] [CrossRef]
- Otwoma, L.M.; Kochzius, M. Genetic population structure of the coral reef sea star Linckia laevigata in the Western Indian Ocean and Indo-West Pacific. PLoS ONE 2016, 11, e0165552. [Google Scholar] [CrossRef]
- Zhang, B.-D.; Li, Y.-L.; Xue, D.-X.; Liu, J.-X. Population genomics reveals shallow genetic structure in a connected and ecologically important fish from the Northwestern Pacific Ocean. Front. Mar. Sci. 2020, 7, 374. [Google Scholar] [CrossRef]
- Taylor, M.L.; Roterman, C.N. Invertebrate population genetics across Earth’s largest habitat: The deep-sea floor. Mol. Ecol. 2017, 26, 4872–4896. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Baums, I.B.; Shank, T.M.; Morrison, C.L.; Cordes, E.E. Testing the depth-differentiation hypothesis in a deepwater octocoral. Proc. Biol. Sci. 2015, 282, 20150008. [Google Scholar] [CrossRef]
- Ng, N.K.; Huang, J.F.; Ho, P.H. Description of a new species of hydrothermal crab, Xenograpsus testudinatus (Crustacea: Decapoda: Brachyura: Grapsidae) from Taiwan. Nat. Taiwan Mus. Spec. Publ. Ser. 2000, 10, 191–199. [Google Scholar]
- Wang, T.W.; Chan, T.-Y.; Chan, B.K.K. Diversity and community structure of decapod crustaceans at hydrothermal vents and nearby deep-water fishing grounds off Kueishan Island, Taiwan: A high biodiversity deep-sea area in the NW Pacific. Bull. Mar. Sci. 2013, 89, 505–528. [Google Scholar] [CrossRef]
- Wang, T.W.; Chan, T.-Y.; Chan, B.K.K. Trophic relationships of hydrothermal vent and non-vent communities in the upper sublittoral and upper bathyal zones off Kueishan Island, Taiwan: A combined morphological, gut content analysis and stable isotope approach. Mar. Biol. 2014, 161, 2447–2463. [Google Scholar] [CrossRef]
- Yang, T.F.; Lan, T.F.; Lee, H.-F.; Fu, C.-C.; Chuang, P.-C.; Lo, C.-H.; Chen, C.-H.; Chen, C.A.; Lee, C.-S. Gas composition and helium isotopic ratios of fluid samples around Kueishantao, NE offshore Taiwan and its tectonic implications. Geochem. J. 2005, 39, 469–480. [Google Scholar] [CrossRef]
- Desbruyeres, D.; Segonzac, M.; Bright, M. Handbook of Deep-Sea Hydrothermal Vent Fauna; Biologiezentrum: Bonn, Germany, 2006; Volume 16, 544p. [Google Scholar]
- Chan, B.K.K.; Wang, T.-W.; Chen, P.-C.; Lin, C.-W.; Chan, T.-Y.; Tsang, L.M. Community structure of macrobiota and environmental parameters in shallow water hydrothermal vents off Kueishan Island, Taiwan. PLoS ONE 2016, 11, e0148675. [Google Scholar] [CrossRef]
- Ng, N.K.; Suzuki, H.; Shih, H.-T.; Dewa, S.; Ng, P.K.L. The hydrothermal crab, Xenograpsus testudinatus Ng, Huang & Ho, 2000 (Crustacea: Decapoda: Brachyura: Grapsidae) in southern Japan. Proc. Biol. Soc. Wash. 2014, 127, 391–399. [Google Scholar] [CrossRef]
- Suzuki, H.; Iwasaki, T.; Utsunomiya, Y.; Iwamoto, A. Biological study of the hydrothermal crab, Xenograpsus testudinatus Ng, Huang & Ho, 2000 (Crustacea: Decapoda: Brachyura: Grapsidae) inhabiting the adjacent water of Showa-Iwo-jima, southern Japan. Occasional papers/Kagoshima Univ. Res. Cent. South Pan. 2015, 56, 37–40. (In Japanese) [Google Scholar]
- Miyake, H.; Oda, A.; Wada, S.; Kodaka, T.; Kurosawa, S. First record of a shallow hydrothermal vent crab, Xenograpsus testudinatus, from Shikine-jima Island in the Izu archipelago. Biogeography 2019, 21, 31–36. [Google Scholar]
- Jeng, M.S.; Clark, P.F.; Ng, P.K.L. The first zoea, megalopa, and first crab stage of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda: Brachyura: Grapsoidea) and the systematic implications for the Varunidae. J. Crust. Biol. 2004, 24, 188–212. [Google Scholar] [CrossRef]
- Dahms, H.-U.; Tseng, L.-C.; Hwang, J.-S. Marine invertebrate larval distribution at the hydrothermal vent site of Gueishandao. J. Mar. Sci. Technol. 2014, 22, 67–74. [Google Scholar] [CrossRef]
- Yin, M.; Li, X.; Xiao, Z.; Li, C. Relationships between intensity of the Kuroshio current in the East China Sea and the East Asian winter monsoon. Acta Oceanol. Sin. 2018, 37, 8–19. [Google Scholar] [CrossRef]
- Ki, J.-S.; Dahms, H.-U.; Hwang, J.-S.; Lee, J.-S. The complete mitogenome of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda, Brachyura) and comparison with brachyuran crabs. Comp. Biochem. Physiol. Part D 2009, 4, 290–299. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Simons, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Crandall, K.A.; Fitzpatrick, J.F. Crayfish molecular systematics: Using a combination of procedures to estimate phylogeny. Syst. Biol. 1996, 45, 1–26. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA 7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many species in one: DNA barcoding over-estimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999, 41, 95–98. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Schubart, C.D.; Diesel, R.; Hedges, S.B. Rapid evolution to terrestrial life in Jamaican crabs. Nature 1998, 393, 363–365. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Jayne, S.R.; Hogg, N.G.; Waterman, S.N.; Rainville, L.; Donohue, K.A.D.; Watts, R.; Tracey, K.L.; McClean, J.L.; Maltrud, M.E.; Qiu, B.; et al. The Kuroshio Extension and its recirculation gyres. Deep-Sea Res. Part I Oceanogr. Res. Pap. 2009, 56, 2088–2099. [Google Scholar] [CrossRef]
- Kim, C.H.; Hwang, S.G. The complete larval development of the mitten crab Eriocheir sinensis H. Milne Edwards, 1853 (Decapoda, Brachyura, Grapsidae) reared in the laboratory and a key to the known zoeae of the Varuninae. Crustaceana 1995, 68, 793–812. [Google Scholar]
- Park, Y.-S.; Ko, H.-S. Complete larval development of Hemigrapsus longitarsis (Miers, 1879) (Crustacea, Decapoda, Grapsidae), with a key to the known grapsid zoeas of Korea. Korean J. Biol. Sci. 2002, 6, 107–123. [Google Scholar] [CrossRef][Green Version]
- Chan, B.K.K.; Tsang, L.M.; Chu, K.H. Morphological and genetic differentiation of the acorn barnacle Tetraclita squamosa (Crustacea, Cirripedia) in East Asia and description of a new species of Tetraclita. Zool. Scr. 2007, 36, 79–91. [Google Scholar] [CrossRef]
- Chang, Y.W.; Chan, J.S.M.; Hayashi, R.; Shuto, T.; Tsang, L.M.; Chu, K.H.; Chan, B.K.K. Genetic differentiation of the soft shore barnacle Fistulobalanus albicostatus (Cirripedia: Thoracica: Balanomorpha) in the West Pacific. Mar. Ecol. 2017, 38, e12422. [Google Scholar] [CrossRef]
- Ma, K.Y.; Chow, L.H.; Wong, K.J.H.; Chen, H.-N.; Ip, B.H.Y.; Schubart, C.D.; Tsang, L.M.; Chan, B.K.K.; Chu, K.H. Speciation pattern of the horned ghost crab Ocypode ceratophthalmus (Pallas, 1772): An evaluation of the drivers of Indo-Pacific marine biodiversity using a widely distributed species. J. Biogeogr. 2019, 45, 2658–2668. [Google Scholar] [CrossRef]
- Lin, H.C.; Cheang, C.C.; Corbari, L.; Chan, B.K.K. Trans-Pacific genetic differentiation in the deep-water stalked barnacle Scalpellum stearnsii (Cirripedia: Thoracica: Scalpellidae). Deep-Sea Res. Part I Oceanogr. Res. Pap. 2020, 164, 103359. [Google Scholar] [CrossRef]
- Kyuno, A.; Shintaku, M.; Fujita, Y.; Matsumoto, H.; Utsumi, M.; Watanabe, H.; Fujiwara, Y.; Miyazaki, J.-I. Dispersal and differentiation of deep-sea mussels of the genus Bathymodiolus (Mytilidae, Bathymodiolinae). J. Mar. Sci. 2009, 2009, 625672. [Google Scholar] [CrossRef]
- Herrera, S.; Watanabe, H.; Shank, T.M. Evolutionary and biogeographical patterns of barnacles from deep-sea hydrothermal vents. Mol. Ecol. 2015, 24, 673–689. [Google Scholar] [CrossRef]
- Yahagi, T.; Watanabe, H.; Ishibashi, J.-I.; Kojima, S. Genetic population structure of four hydrothermal vent shrimp species (Alvinocarididae) in the Okinawa Trough, Northwest Pacific. Mar. Ecol. Prog. Ser. 2015, 529, 159–169. [Google Scholar] [CrossRef]
- Yahagi, T.; Watanabe, H.; Kojima, S.; Kano, Y. Do larvae from deep-sea hydrothermal vents disperse in surface waters? Ecology 2017, 98, 1524–1534. [Google Scholar] [CrossRef]
- Mitarai, S.; Watanabe, H.; Nakajima, Y.; Schchepetkin, A.F.; McWilliams, J.C. Quantifying dispersal from hydrothermal vent fields in the western Pacific Ocean. Proc. Natl. Acad. Sci. USA 2016, 113, 2976–2981. [Google Scholar] [CrossRef]
- Voris, H.K. Maps of Pleistocene sea levels in Southeast Asia: Shorelines, river systems and time durations. J. Biogeogr. 2001, 27, 1153–1167. [Google Scholar] [CrossRef]
- Daigle, R.M.; Metaxas, A. Vertical distribution of marine invertebrate larvae in response to thermal stratification in the laboratory. J. Exp. Mar. Biol. Ecol. 2011, 409, 89–98. [Google Scholar] [CrossRef]
- Anger, K.; Queiroga, H.; Calado, R. Larval development and behaviour strategies in Brachyura. In Treatise on Zoology- Anatomy, Taxonomy, Biology; Castro, P., Davie, P., Guinot, D., Schram, F., von Vaupel Klein, C., Eds.; Brill: Leiden, The Netherlands, 2015; Volume 9, pp. 317–374. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, J.; Shi, W. Mapping the coastal upwelling east of Taiwan using geostationary satellite data. Remote Sens. 2021, 13, 170. [Google Scholar] [CrossRef]
- Watanabe, H.K.; Kado, R.; Kadia, M.; Tsuchida, S.; Kokima, S. Dispersal of vent-barnacle (genus Neoverruca) in the Western Pacific. Cah. Biol. Mar. 2006, 47, 353–357. [Google Scholar]
- Yorisue, T.; Kado, R.; Watanabe, H.; Høeg, J.T.; Inoue, K.; Kojima, S.; Chan, B.K.K. Influence of water temperature on the larval development of Neoverruca sp. and Ashinkailepas seepiophila -implications for larval dispersal and settlement in the vent and seep environments. Deep-Sea Res. Part I Oceanogr. Res. Pap. 2013, 71, 33–37. [Google Scholar] [CrossRef]
- Hwang, J.S.; Dahms, H.-U.; Alekseev, V. Novel nursery habitat of hydrothermal vent crabs. Crustaceana 2008, 81, 375–380. [Google Scholar]
- Chan, B.K.K.; Chang, Y.-W. A new deep-sea scalpelliform barnacle, Vulcanolepas buckeridgei sp. nov. (Eolepadidae: Neolepadinae) from hydrothermal vents in the Lau Basin. Zootaxa 2018, 4407, 117–129. [Google Scholar] [CrossRef]
- Chan, B.K.K.; Ju, S.-J.; Kim, S.-J. A new species of hydrothermal vent stalked barnacle Vulcanolepas (Scalpelliforms: Eolepadidae) from the North Fiji Basin, Southwestern Pacific Ocean. Zootaxa 2019, 4563, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-K.; Kim, S.-J.; Hou, B.K.; Van Dover, C.L.; Ju, S.-J. Population genetic differentiation of the hydrothermal vent crab Austinograea alayseae (Crustacea: Bythograeidae) in the Southwest Pacific Ocean. PLoS ONE 2019, 14, e0215829. [Google Scholar] [CrossRef] [PubMed]
- Tsang, L.M.; Schubart, C.D.; Ahyong, S.T.; Lai, J.C.Y.; Au, E.Y.C.; Chan, T.-Y.; Ng, P.K.L.; Chu, K.H. Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origin of freshwater crabs. Mol. Biol. Evol. 2014, 31, 1173–1187. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Kurata, Y. Crabs of the Ogasawara Islands. IV. A collection made at the new volcanic island, Nishino-shima-shinto, in 1975. Bull. Natl. Sci. Mus. Ser. A 1977, 3, 91–111. [Google Scholar]
- Türkay, M.; Sakai, K. Decapod crustaceans from a volcanic hot spring in the Marianas. Senckenbergia Marit. 1995, 26, 25–35. [Google Scholar]
- McLay, C.L. New crabs from hydrothermal vents of the Kermadec Ridge submarine volcanoes, New Zealand: Gandalfus gen. nov. (Bythograeidae) and Xenograpsus (Varunidae) (Decapoda: Brachyura). Zootaxa 2007, 1524, 1–22. [Google Scholar] [CrossRef]
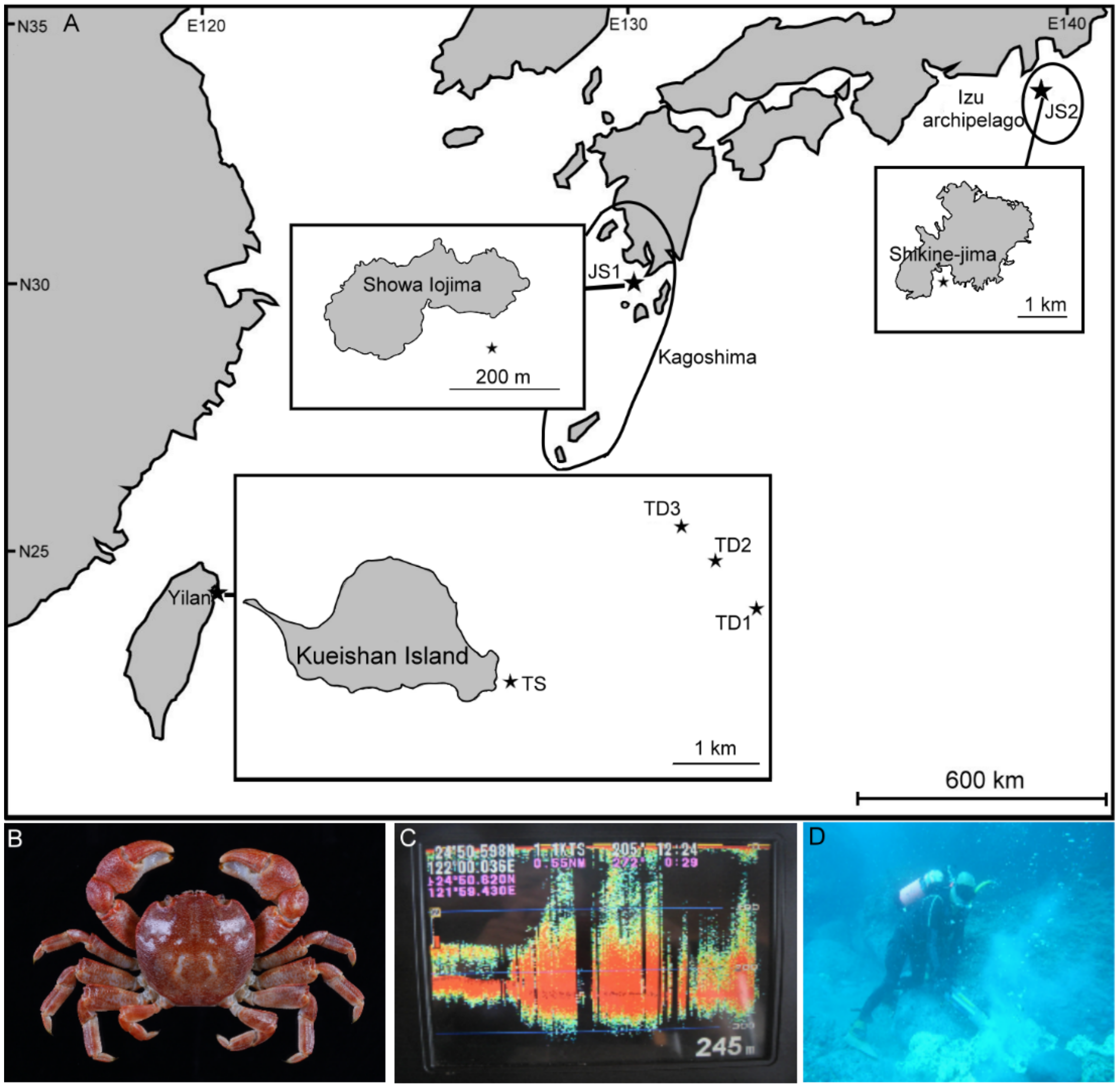

| Locality | Habitat & Population Name Abbreviation | Latitude & Longitude | Depth (m) | Sampling Date | Sample Sizes | Source |
|---|---|---|---|---|---|---|
| Kueishan Is., Yilan, Taiwan | Shallow waters (TS) ★ | 24°50.112′ N 121°57.741′ E | 10–15 | 9 August 2013 | 51 | This study |
| n/a | n/a | n/a | 1 | EU727203 [26] | ||
| n/a | 15 | 11 August 2008 | 4 # | AB933570-AB933573 [20] | ||
| Deep sea (TD1) | 24°50.662′ N 121°59.697′ E | 200 | 13 July 2005 | 16 | This study | |
| Deep sea (TD2) ★ | 24°50.994′ N 121°59.431′ E | 209–224 | 12 August 2010 | 31 | This study | |
| Deep sea (TD3) | 24°51.231′ N 121°59.204′ E | 252–275 | 4 September 2008 | 23 | This study | |
| Showa Iojima Is., Kagoshima, Japan | Shallow waters (JS1) ★ | 30°48.16′ N 130°20.5′ E | 3–5 | 14 May 2011 | 30 | This study |
| 30°48.16′ N 130°20.5′ E | 3–5 | 14 May 2011 | 5 # | AB933569, AB933574-AB933577 [20] | ||
| Shikine-Jima Is., Izu archipelago, Japan | Shallow waters (JS2) | n/a | 10–14 | 17 July 2018 | 3 # | LC490686-LC490688 [22] |
| Population | N | H | S | h | π | T’s D |
|---|---|---|---|---|---|---|
| COI gene | ||||||
| Kueishan Is., Shallow waters (TS) | 56 | 38 | 40 | 0.969 (0.013) | 0.006 (0.0004) | −1.95 * |
| Kueishan Is., Deep-sea (TD1) | 16 | 13 | 18 | 0.967 (0.036) | 0.006 (0.0007) | −1.16 |
| Kueishan Is., Deep-sea (TD2) | 31 | 20 | 20 | 0.955 (0.022) | 0.005 (0.0004) | −1.39 |
| Kueishan Is., Deep-sea (TD3) | 23 | 16 | 16 | 0.964 (0.022) | 0.005 (0.0005) | −0.95 |
| Showa Iojima Is., Shallow waters (JS1) | 35 | 17 | 20 | 0.921 (0.026) | 0.005 (0.0005) | −1.28 |
| Shikine-Jima Is., Shallow waters (JS2) | 3 | 2 | 2 | 0.667 (0.314) | 0.004 (0.0019) | None |
| Total | 164 | 78 | 54 | 0.898 (0.018) | 0.006 (0.0003) | −2.19 ** |
| 16S rRNA gene | ||||||
| Kueishan Is., Shallow waters (TS) | 52 | 5 | 5 | 0.370 (0.081) | 0.0009 (0.0002) | −1.45 |
| Kueishan Is., Deep-sea (TD1) | 16 | 4 | 3 | 0.592 (0.122) | 0.001 (0.0004) | −0.47 |
| Kueishan Is., Deep-sea (TD2) | 31 | 4 | 3 | 0.553 (0.060) | 0.001 (0.001) | −0.41 |
| Kueishan Is., Deep-sea (TD3) | 23 | 6 | 4 | 0.632 (0.089) | 0.002 (0.0003) | −0.69 |
| Showa Iojima Is., Shallow waters (JS1) | 30 | 4 | 3 | 0.303 (0.104) | 0.0006 (0.0002) | −1.36 |
| Total | 152 | 14 | 12 | 0.435 (0.0001) | 0.001 (0.0001) | −1.92 * |
| D-loop gene | ||||||
| Kueishan Is., Shallow waters (TS) | 27 | 27 | 81 | 1 (0.011) | 0.037 (0.002) | −1.28 |
| Kueishan Is., Deep-sea (TD2) | 18 | 18 | 73 | 1 (0.019) | 0.041 (0.002) | −1.00 |
| Showa Iojima Is., Shallow waters (JS1) | 24 | 24 | 68 | 1 (0.012) | 0.032 (0.002) | −1.38 |
| Total | 69 | 60 | 111 | 1 (0.003) | 0.034 (0.001) | −1.68 |
| Populations | TD1 | TD2 | TD3 | TS | JS1 | JS2 |
|---|---|---|---|---|---|---|
| COI gene | ||||||
| TD1 | -- | |||||
| TD2 | −0.0193 | -- | ||||
| TD3 | −0.0293 | −0.0129 | -- | |||
| TS | −0.0188 | 0.0143 | −0.0054 | -- | ||
| JS1 | −0.0189 | 0.0029 | −0.0031 | 0.0100 | -- | |
| JS2 | −0.2990 | −0.1308 | −0.1720 | −0.2825 | −0.1709 | -- |
| COI + 16S rRNA genes | ||||||
| TD1 | -- | n/s | ||||
| TD2 | −0.0138 | -- | n/s | |||
| TD3 | −0.0241 | 0.0136 | -- | n/s | ||
| TS | −0.0105 | 0.0328 * | 0.0108 | -- | n/s | |
| JS1 | −0.0123 | 0.0249 | 0.0076 | −0.0023 | -- | n/s |
| COI + 16S rRNA + D-loop genes | ||||||
| TD2 | n/s | -- | n/s | n/s | ||
| TS | n/s | −0.0048 | n/s | -- | n/s | |
| JS1 | n/s | 0.0109 | n/s | −0.0039 | -- | n/s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-H.; Wang, T.-W.; Ng, P.K.L.; Chan, T.-Y.; Lin, Y.-Y.; Chan, B.K.K. Population Genetic Differentiation on the Hydrothermal Vent Crabs Xenograpsus testudinatus along Depth and Geographical Gradients in the Western Pacific. Diversity 2022, 14, 162. https://doi.org/10.3390/d14030162
Yang C-H, Wang T-W, Ng PKL, Chan T-Y, Lin Y-Y, Chan BKK. Population Genetic Differentiation on the Hydrothermal Vent Crabs Xenograpsus testudinatus along Depth and Geographical Gradients in the Western Pacific. Diversity. 2022; 14(3):162. https://doi.org/10.3390/d14030162
Chicago/Turabian StyleYang, Chien-Hui, Teng-Wei Wang, Peter Kee Lin Ng, Tin-Yam Chan, Yi-Yang Lin, and Benny Kwok Kan Chan. 2022. "Population Genetic Differentiation on the Hydrothermal Vent Crabs Xenograpsus testudinatus along Depth and Geographical Gradients in the Western Pacific" Diversity 14, no. 3: 162. https://doi.org/10.3390/d14030162
APA StyleYang, C.-H., Wang, T.-W., Ng, P. K. L., Chan, T.-Y., Lin, Y.-Y., & Chan, B. K. K. (2022). Population Genetic Differentiation on the Hydrothermal Vent Crabs Xenograpsus testudinatus along Depth and Geographical Gradients in the Western Pacific. Diversity, 14(3), 162. https://doi.org/10.3390/d14030162







