Genome-Wide Characterization and Identification of the YABBY Gene Family in Mango (Mangifera indica)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Genome-Wide Identification of the MiYABBY Gene Family in Mango
2.3. Quantitative RT-PCR Data Analysis
3. Results
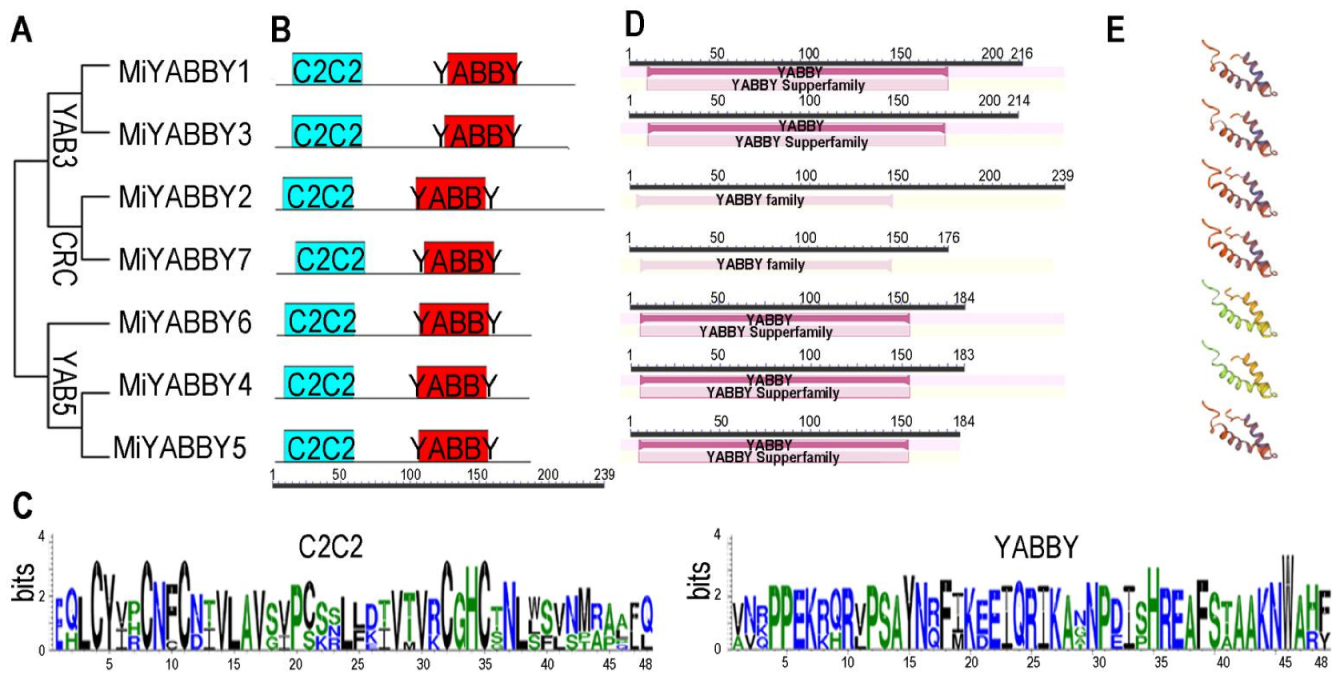
3.1. Identification and Characterization of MiYABBY
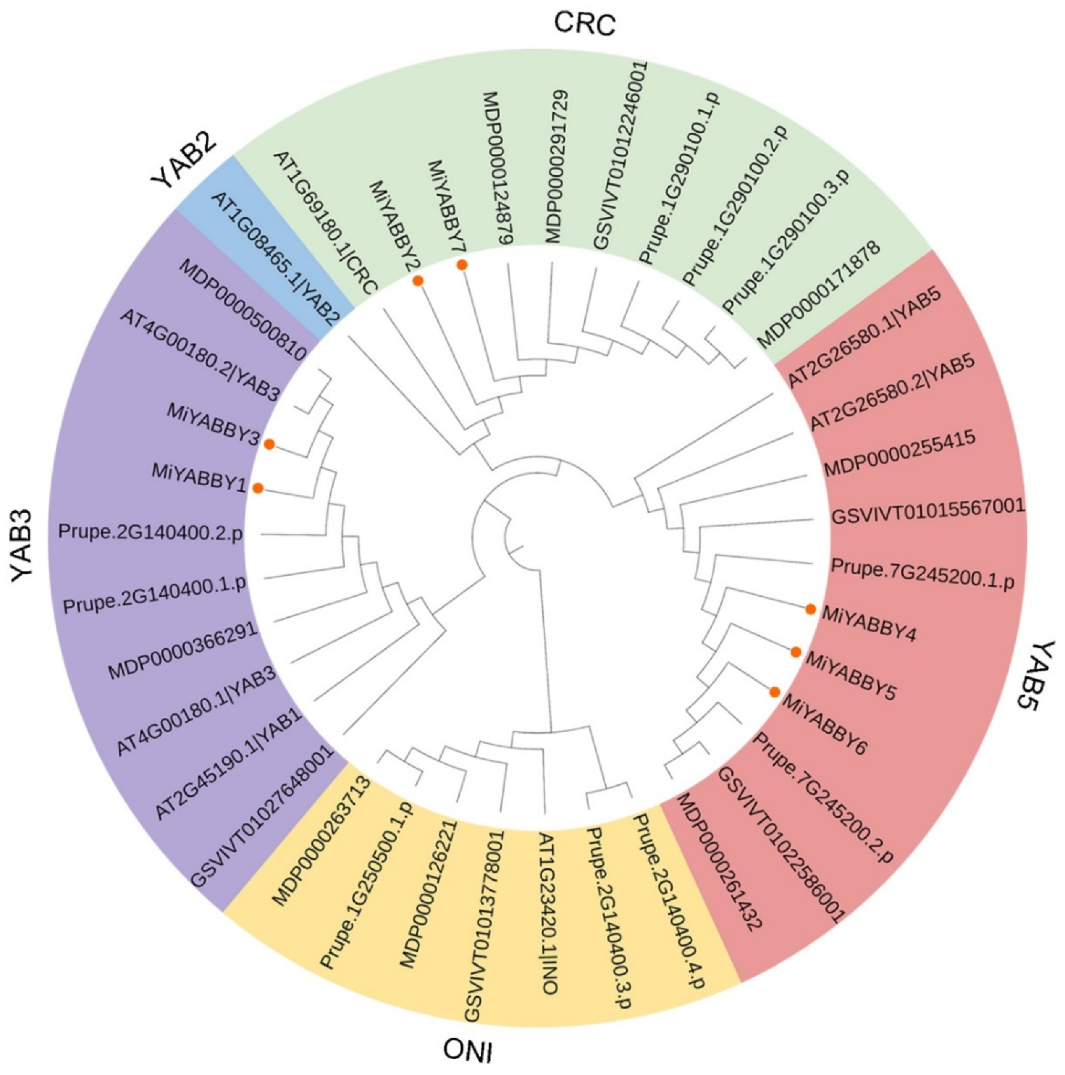
3.2. Phylogenetic Analysis and Classification of the MiYABBY Proteins
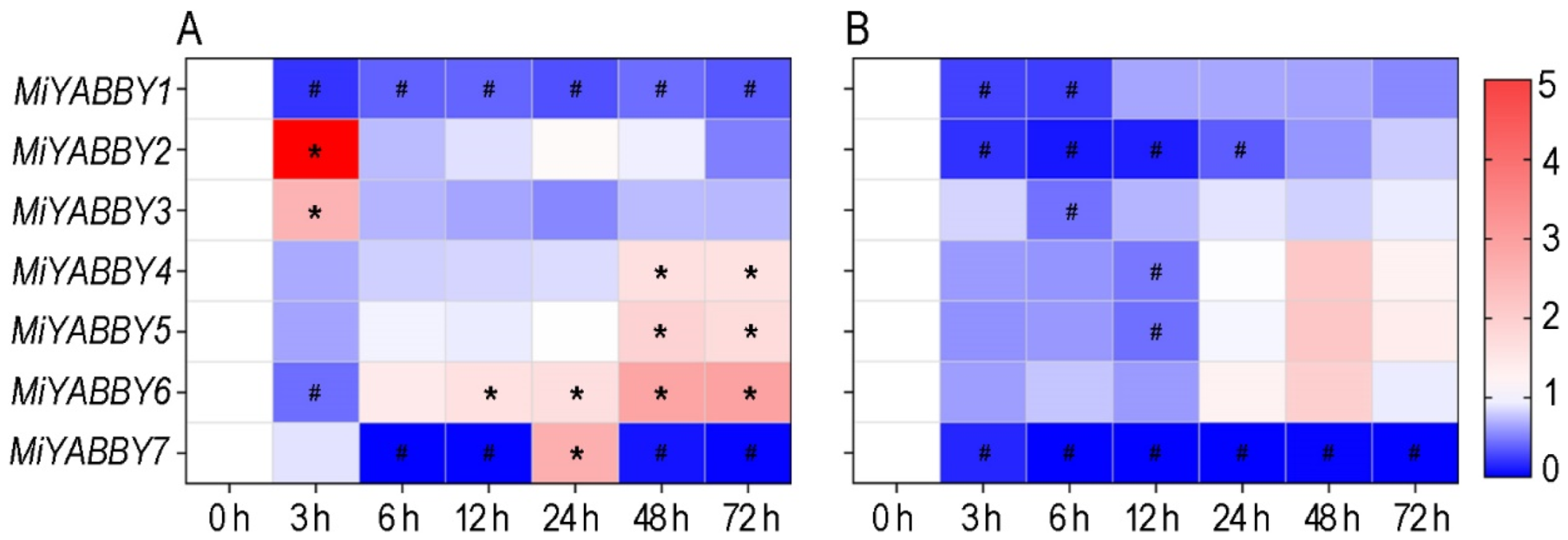
3.3. Expression Patterns of MiYABBY Genes in the Leaves and Fruits
3.4. MiYABBY Genes Expression Profiles in Response to C. gloeosporioides and X. campestris pv. Mangiferaeindicae Infection
3.5. MiYABBY Genes Expression Profiles upon Exposure to Drought- and Salt-Stresses
3.6. MiYABBY Genes Expression Profiles upon Exposure to MeJA and SA Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mirza, B.; Croley, C.R.; Ahmad, M.; Pumarol, J.; Das, N.; Sethi, G.; Bishayee, A. Mango (Mangifera indica L.): A magnificent plant with cancer preventive and anticancer therapeutic potential. Crit. Rev. Food Sci. Nutr. 2021, 61, 2125–2151. [Google Scholar] [CrossRef]
- Lebaka, V.R.; Wee, Y.J.; Ye, W.; Korivi, M. Nutritional composition and bioactive compounds in three different parts of mango fruit. Int. J. Environ. Res. Public Health 2021, 18, 741. [Google Scholar] [CrossRef]
- Li, W.; Zhu, X.; Zhang, Q.; Li, K.; Zhang, D.; Shi, C.; Gao, L. SMRT sequencing generates the chromosome-scale reference genome of tropical fruit mango, Mangifera indica. bioRxiv 2020, 25. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar] [CrossRef]
- Bowman, J.L. The YABBY gene family and abaxial cell fate. Curr. Opin. Plant Biol. 2000, 3, 17–22. [Google Scholar] [CrossRef]
- Toriba, T.; Harada, K.; Takamura, A.; Nakamura, H.; Ichikawa, H.; Suzaki, T.; Hirano, H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet. Genom. 2007, 277, 457–468. [Google Scholar] [CrossRef]
- Hou, H.; Wu, P.; Gao, L.; Zhang, C.; Hou, X. Characterization and expression profile analysis of YABBY family genes in Pak-choi (Brassica rapa ssp. chinensis) under abiotic stresses and hormone treatments. Plant Growth Regul. 2019, 87, 421–432. [Google Scholar] [CrossRef]
- Jiu, S.; Zhang, Y.; Han, P.; Han, Y.; Xu, Y.; Liu, G.; Leng, X. Genome-wide identification and expression analysis of VviYABs family reveal its potential functions in the developmental switch and stresses response during grapevine development. Front. Genet. 2022, 12, 766221. [Google Scholar] [CrossRef]
- Ma, R.; Huang, B.; Huang, Z.; Zhang, Z. Genome-wide identification and analysis of the YABBY gene family in moso bamboo (Phyllostachys edulis (Carriere) J. Houz). PeerJ 2021, 9, e11780. [Google Scholar] [CrossRef]
- Yang, Z.; Gong, Q.; Wang, L.; Jin, Y.; Xi, J.; Li, Z.; Qin, W.; Yang, Z.; Lu, L.; Chen, Q.; et al. Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 2018, 9, 33. [Google Scholar] [CrossRef]
- Li, Z.; Li, G.; Cai, M.; Priyadarshani, S.V.G.N.; Aslam, M.; Zhou, Q.; Huang, X.; Wang, X.; Liu, Y.; Qin, Y. Genome-wide analysis of the YABBY transcription factor family in pineapple and functional identification of AcYABBY4 involvement in salt stress. Int. J. Mol. Sci. 2019, 20, 5863. [Google Scholar] [CrossRef]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef]
- Liu, H.; Ye, H.; Wang, J.; Chen, S.; Li, M.; Wang, G.; Hou, N.; Zhao, P. Genome-wide identification and characterization of YABBY gene family in Juglans regia and Juglans mandshurica. Agronomy 2022, 12, 1914. [Google Scholar] [CrossRef]
- Yin, S.; Li, S.; Gao, Y.; Bartholomew, E.S.; Wang, R.; Yang, H.; Liu, C.; Chen, X.; Wang, Y.; Liu, X.; et al. Genome-wide identification of YABBYgene family in cucurbitaceae and expression analysis in cucumber (Cucumis sativus L.). Genes 2022, 13, 467. [Google Scholar] [CrossRef]
- Lu, Y.H.; Alam, I.; Yang, Y.Q.; Yu, Y.C.; Chi, W.C.; Chen, S.B.; Chalhoub, B.; Jiang, L.X. Evolutionary Analysis of the YABBY gene family in Brassicaceae. Plants 2021, 10, 2700. [Google Scholar] [CrossRef]
- Xia, J.; Wang, D.; Peng, Y.; Wang, W.; Wang, Q.; Xu, Y.; Li, T.; Zhang, K.; Li, J.; Xu, X. Genome-wide analysis of the YABBY transcription factor family in rapeseed (Brassica napus L.). Genes 2021, 12, 981. [Google Scholar] [CrossRef]
- Zhao, S.P.; Lu, D.; Yu, T.F.; Ji, Y.J.; Zheng, W.J.; Zhang, S.X.; Chai, S.C.; Chen, Z.Y.; Cui, X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017, 119, 32–146. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Li, D.; Liu, Y.; Yang, X. Roles of YABBY transcription factors in the modulation of morphogenesis, development, and phytohormone and stress responses in plants. J. Plant Res. 2020, 133, 751–763. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Marchler, B.A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Asma, A.; Saddam, S.; Huang, H.; Wajid, Z.; Lü, S.; Zhao, H. Genome-wide comparative analysis of long-chain acyl-CoA synthetases (LACSs) gene family: A focus on identification, evolution and expression profiling related to lipid synthesis. Plant Physiol. Biochem. 2021, 161, 1–11. [Google Scholar] [CrossRef]
- Muhammad, N.; Rafia, M.; Muhammad, W. The Solanum melongena COP1 delays fruit ripening and influences ethylene signaling in tomato. J. Plant Physiol. 2019, 240, 152997. [Google Scholar] [CrossRef]
- Combet, C.; Blanchet, C.; Geourjon, C.; Deléage, G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Li, C.; Dong, N.; Shen, L.; Lu, M.; Zhai, J.; Zhao, Y.; Chen, L.; Wan, Z.; Liu, Z.; Ren, H.; et al. Genome-wide identification and expression profile of YABBY genes in Averrhoa carambola. PeerJ 2022, 9, e12558. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, C.; Ge, D.; Yan, M.; Ren, Y.; Huang, X.; Yuan, Z. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 2020, 752, 144784. [Google Scholar] [CrossRef]
- She, Z.Y.; Huang, X.Y.; Aslam, M.; Wang, L.L.; Yan, M.K.; Qin, R.J.; Chen, Y.Z.; Qin, Y.; Niu, X.P. Expression characterization and cross-species complementation uncover the functional conservation of YABBY genes for leaf abaxial polarity and carpel polarity establishment in Saccharum spontaneum. BMC Plant Biol. 2022, 22, 124. [Google Scholar] [CrossRef]
- Orashakova, S.; Lange, M.; Lange, S.; Wege, S.; Becker, A. The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 2009, 58, 682–693. [Google Scholar] [CrossRef]
- Lora, J.; Hormaza, J.I.; Herrero, M.; Gasser, C.S. Seedless fruits and the disruption of a conserved genetic pathway in angiosperm ovule development. Proc. Natl. Acad. Sci. USA 2011, 108, 5461–5465. [Google Scholar] [CrossRef]
- Lee, J.Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef]
- Bartholmes, C.; Hidalgo, O.; Gleissberg, S. Evolution of the YABBY gene family with emphasis on the basal eudicot Eschscholzia californica (Papaveraceae). Plant Biol. 2012, 14, 11–23. [Google Scholar] [CrossRef]
- Villanueva, J.M.; Broadhvest, J.; Hauser, B.A.; Meister, R.J.; Schneitz, K.; Gasser, C.S. INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 1999, 13, 3160–3169. [Google Scholar] [CrossRef]
- Rachappanavar, V.; Padiyal, A.; Sharma, J.K.; Gupta, S.K. Plant hormone-mediated stress regulation responses in fruit crops- a review. Sci. Hortic. 2022, 304, 111302. [Google Scholar] [CrossRef]
- Kondo, S. Usage and action mechanism of oxylipins including jasmonic acid on physiological aspects of fruit production. Sci. Hortic. 2022, 295, 110893. [Google Scholar] [CrossRef]
- Khan, M.S.S.; Islam, F.; Chen, H.; Chang, M.; Wang, D.W.; Liu, F.Q.; Fu, Z.Q.; Chen, J. Transcriptional coactivators: Driving force of plant immunity. Front. Plant Sci. 2022, 13, 823937. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, Y.; Luo, R.; Sun, R.; Yang, N.; Pu, J.; Gao, A.; Zhang, H. Genome-Wide Characterization and Identification of the YABBY Gene Family in Mango (Mangifera indica). Diversity 2022, 14, 861. https://doi.org/10.3390/d14100861
Xia Y, Luo R, Sun R, Yang N, Pu J, Gao A, Zhang H. Genome-Wide Characterization and Identification of the YABBY Gene Family in Mango (Mangifera indica). Diversity. 2022; 14(10):861. https://doi.org/10.3390/d14100861
Chicago/Turabian StyleXia, Yuqi, Ruixiong Luo, Ruiqing Sun, Nan Yang, Jinji Pu, Aiping Gao, and He Zhang. 2022. "Genome-Wide Characterization and Identification of the YABBY Gene Family in Mango (Mangifera indica)" Diversity 14, no. 10: 861. https://doi.org/10.3390/d14100861
APA StyleXia, Y., Luo, R., Sun, R., Yang, N., Pu, J., Gao, A., & Zhang, H. (2022). Genome-Wide Characterization and Identification of the YABBY Gene Family in Mango (Mangifera indica). Diversity, 14(10), 861. https://doi.org/10.3390/d14100861




