Seasonal Dynamics and Diversity of Haemosporidians in a Natural Woodland Bird Community in Slovakia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area, Field Methods and Study Species
2.2. DNA Extraction
2.3. PCR Analyses
2.4. Sequence and Statistical Analysis
3. Results
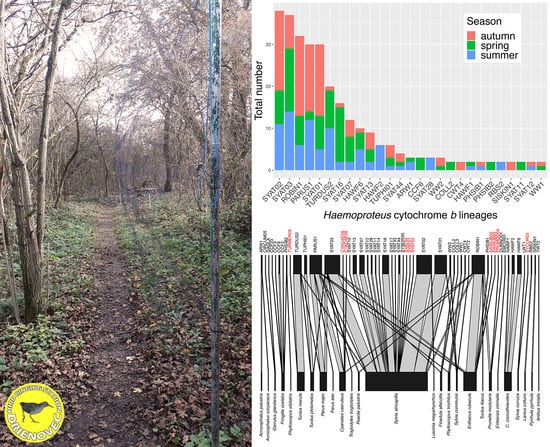
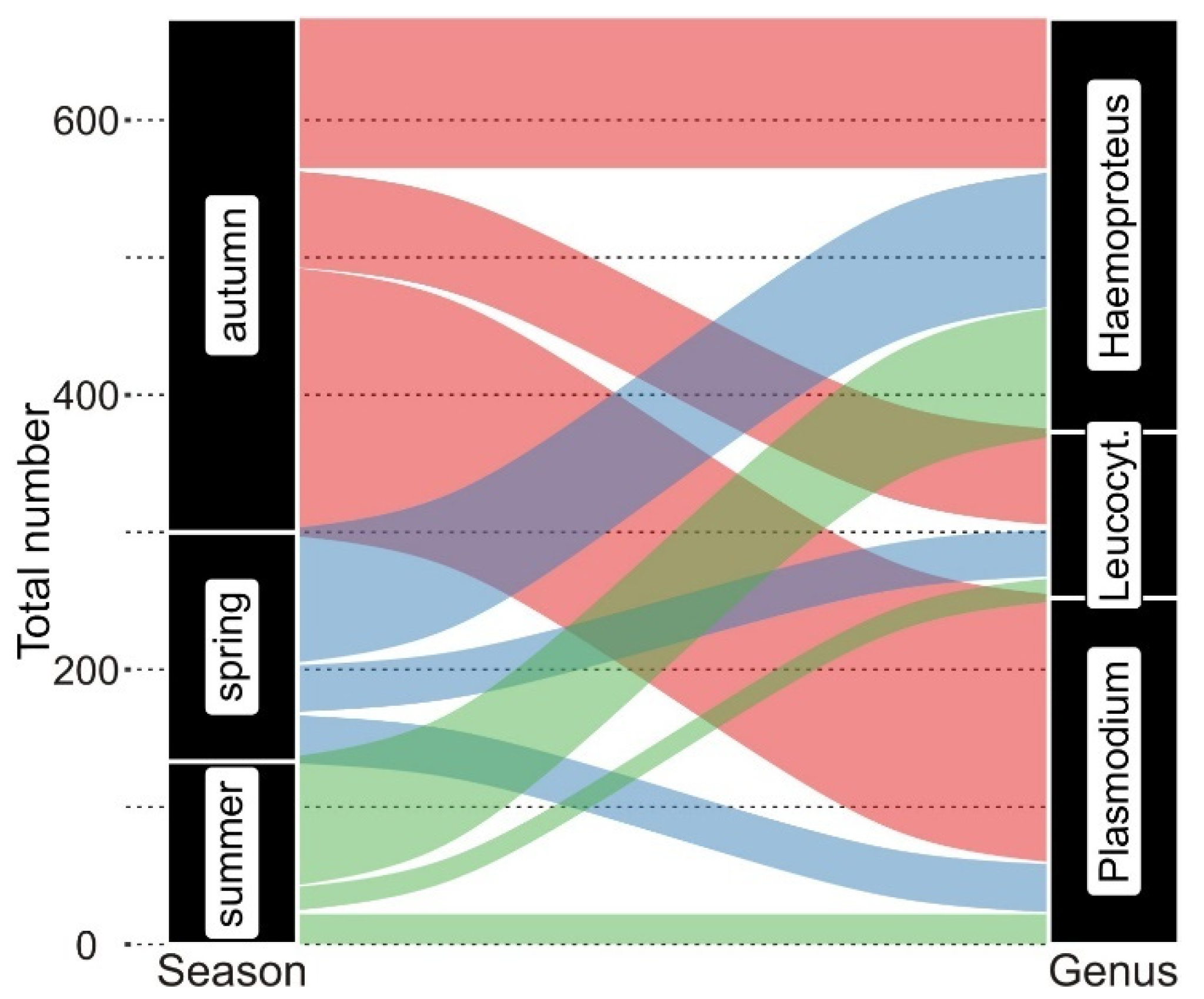
3.1. Avian and Haemosporidian Communities
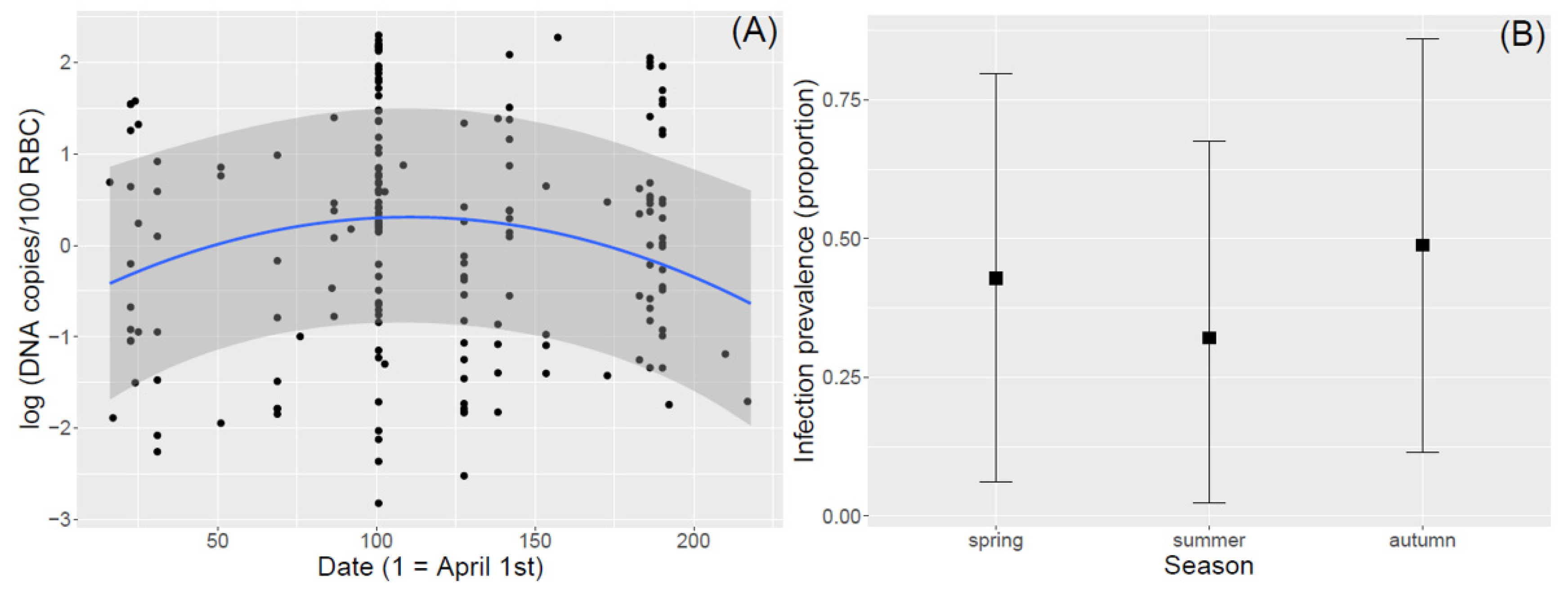
3.2. Seasonal Variation in Infection Intensity and Prevalence
3.3. Host-Parasitic Associations
4. Discussion
4.1. Haemosporidian Diversity
4.2. Haemosporidian Multiple Infections
4.3. Haemosporidian Infection Prevalence and Intensity
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dowell, S.F. Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg. Infect. Dis. 2001, 7, 369. [Google Scholar] [CrossRef]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef]
- Grassly, N.C.; Fraser, C. Seasonal infectious disease epidemiology. Proc. R. Soc. B Biol. Sci. 2006, 273, 2541–2550. [Google Scholar] [CrossRef]
- Ellis, V.A.; Medeiros, M.C.; Collins, M.D.; Sari, E.H.; Coffey, E.D.; Dickerson, R.C.; Lugarini, C.; Stratford, J.A.; Henry, D.R.; Merrill, L. Prevalence of avian haemosporidian parasites is positively related to the abundance of host species at multiple sites within a region. Parasitol. Res. 2017, 116, 73–80. [Google Scholar] [CrossRef]
- Fallon, S.M.; Bermingham, E.; Ricklefs, R.E. Island and taxon effects in parasitism revisited: Avian malaria in the lesser antilles. Evolution 2003, 57, 606–615. [Google Scholar] [CrossRef]
- Ishtiaq, F. Ecology and evolution of avian malaria: Implications of land use changes and climate change on disease dynamics. J. Indian Inst. Sci. 2021, 101, 213–225. [Google Scholar] [CrossRef]
- Danilewsky, V. La Parasitologie Comparée du Sang. 1. Nouvelles Recherches sur les Parasites du Sang. Des. Oiseaux; Darre: Kharkov, Ukraine, 1889. [Google Scholar]
- Beaudoin, R.L.; Applegate, J.E.; Davis, D.E.; McLean, R.G. A model for the ecology of avian malaria. J. Wildl. Dis. 1971, 7, 5–13. [Google Scholar] [CrossRef]
- Applegate, J.E. Spring relapse of plasmodium relictum infections in an experimental field population of english sparrows (passer domesticus). J. Wildl. Dis. 1971, 7, 37–42. [Google Scholar] [CrossRef]
- Cosgrove, C.L.; Wood, M.J.; Day, K.P.; Sheldon, B.C. Seasonal variation in plasmodium prevalence in a population of blue tits cyanistes caeruleus. J. Anim. Ecol. 2008, 77, 540–548. [Google Scholar] [CrossRef]
- Svensson-Coelho, M.; Loiselle, B.A.; Blake, J.G.; Ricklefs, R.E. Resource predictability and specialization in avian malaria parasites. Mol. Ecol. 2016, 25, 4377–4391. [Google Scholar] [CrossRef]
- Pigeault, R.; Cozzarolo, C.-S.; Choquet, R.; Strehler, M.; Jenkins, T.; Delhaye, J.; Bovet, L.; Wassef, J.; Glaizot, O.; Christe, P. Haemosporidian infection and co-infection affect host survival and reproduction in wild populations of great tits. Int. J. Parasitol. 2018, 48, 1079–1087. [Google Scholar] [CrossRef]
- Hasselquist, D.; Östman, Ö.; Waldenström, J.; Bensch, S. Temporal patterns of occurrence and transmission of the blood parasite haemoproteus payevskyi in the great reed warbler acrocephalus arundinaceus. J. Ornithol. 2007, 148, 401–409. [Google Scholar] [CrossRef]
- Møller, A.P.; Erritzøe, J. Host immune defence and migration in birds. Evol. Ecol. 1998, 12, 945–953. [Google Scholar] [CrossRef]
- Waldenström, J.; Bensch, S.; Kiboi, S.; Hasselquist, D.; Ottosson, U. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol. Ecol. 2002, 11, 1545–1554. [Google Scholar] [CrossRef] [PubMed]
- Ishak, H.D.; Loiseau, C.; Hull, A.C.; Sehgal, R.N. Prevalence of blood parasites in migrating and wintering california hawks. J. Raptor Res. 2010, 44, 215–223. [Google Scholar] [CrossRef]
- Walther, E.L.; Carlson, J.S.; Cornel, A.; Morris, B.K.; Sehgal, R.N. First molecular study of prevalence and diversity of avian haemosporidia in a central california songbird community. J. Ornithol. 2016, 157, 549–564. [Google Scholar] [CrossRef]
- Ciloglu, A.; Ergen, A.G.; Inci, A.; Dik, B.; Duzlu, O.; Onder, Z.; Yetismis, G.; Bensch, S.; Valkiūnas, G.; Yildirim, A. Prevalence and genetic diversity of avian haemosporidian parasites at an intersection point of bird migration routes: Sultan marshes national park, Turkey. Acta Trop. 2020, 210, 105465. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.R.; MacGillivray, H.L.; Sarquis-Adamson, Y.; Watson, M.J.; Hobson, K.A.; MacDougall-Shackleton, E.A. Seasonal migration distance varies with natal dispersal and predicts parasitic infection in song sparrows. Behav. Ecol. Sociobiol. 2016, 70, 1857–1866. [Google Scholar] [CrossRef]
- Sol, D.; Jovani, R.; Torres, J. Parasite mediated mortality and host immune response explain age-related differences in blood parasitism in birds. Oecologia 2003, 135, 542–547. [Google Scholar] [CrossRef]
- Huang, X.; Jönsson, J.; Bensch, S. Persistence of avian haemosporidians in the wild: A case study to illustrate seasonal infection patterns in relation to host life stages. Int. J. Parasitol. 2020, 50, 611–619. [Google Scholar] [CrossRef]
- Davidar, P.; Morton, E.S. Living with parasites: Prevalence of a blood parasite and its effect on survivorship in the purple martin. Auk 1993, 110, 109–116. [Google Scholar]
- Wilson, K.; Bjørnstad, O.N.; Dobson, A.P.; Merler, S.; Poglayen, G.; Randolph, S.E.; Read, A.F.; Skorping, A. Heterogeneities in macroparasite infections: Patterns and processes. Ecol. Wildl. Dis. 2002, 44, 6–44. [Google Scholar]
- Emmenegger, T.; Alves, J.A.; Rocha, A.D.; Costa, J.S.; Schmid, R.; Schulze, M.; Hahn, S. Population-and age-specific patterns of haemosporidian assemblages and infection levels in european bee-eaters (merops apiaster). Int. J. Parasitol. 2020, 50, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Parker, I.M.; Saunders, M.; Bontrager, M.; Weitz, A.P.; Hendricks, R.; Magarey, R.; Suiter, K.; Gilbert, G.S. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 2015, 520, 542–544. [Google Scholar] [CrossRef]
- Ellis, V.A.; Huang, X.; Westerdahl, H.; Jönsson, J.; Hasselquist, D.; Neto, J.M.; Nilsson, J.-A.; Nilsson, J.; Hegemann, A.; Hellgren, O. Explaining prevalence, diversity and host specificity in a community of avian haemosporidian parasites. Oikos 2020, 129, 1314–1329. [Google Scholar] [CrossRef]
- Vázquez, D.P.; Poulin, R.; Krasnov, B.R.; Shenbrot, G.I. Species abundance and the distribution of specialization in host–parasite interaction networks. J. Anim. Ecol. 2005, 74, 946–955. [Google Scholar] [CrossRef]
- Drovetski, S.V.; Aghayan, S.A.; Mata, V.A.; Lopes, R.J.; Mode, N.A.; Harvey, J.A.; Voelker, G. Does the niche breadth or trade-off hypothesis explain the abundance–occupancy relationship in avian haemosporidia? Mol. Ecol. 2014, 23, 3322–3329. [Google Scholar] [CrossRef]
- Neto, J.M.; Pérez-Rodríguez, A.; Haase, M.; Flade, M.; Bensch, S. Prevalence and diversity of plasmodium and haemoproteus parasites in the globally-threatened aquatic warbler acrocephalus paludicola. Parasitology 2015, 142, 1183–1189. [Google Scholar] [CrossRef]
- Slowinski, S.P.; Fudickar, A.M.; Hughes, A.M.; Mettler, R.D.; Gorbatenko, O.V.; Spellman, G.M.; Ketterson, E.D.; Atwell, J.W. Sedentary songbirds maintain higher prevalence of haemosporidian parasite infections than migratory conspecifics during seasonal sympatry. PLoS ONE 2018, 13, e0201563. [Google Scholar] [CrossRef]
- Lynton-Jenkins, J.G.; Bründl, A.C.; Cauchoix, M.; Lejeune, L.A.; Sallé, L.; Thiney, A.C.; Russell, A.F.; Chaine, A.S.; Bonneaud, C. Contrasting the seasonal and elevational prevalence of generalist avian haemosporidia in co-occurring host species. Ecol. Evol. 2020, 10, 6097–6111. [Google Scholar] [CrossRef] [PubMed]
- Neto, J.M.; Mellinger, S.; Halupka, L.; Marzal, A.; Zehtindjiev, P.; Westerdahl, H. Seasonal dynamics of haemosporidian (apicomplexa, haemosporida) parasites in house sparrows passer domesticus at four european sites: Comparison between lineages and the importance of screening methods. Int. J. Parasitol. 2020, 50, 523–532. [Google Scholar] [CrossRef]
- Santiago-Alarcon, D.; Bloch, R.; Rolshausen, G.; Schaefer, H.M.; Segelbacher, G. Prevalence, diversity, and interaction patterns of avian haemosporidians in a four-year study of blackcaps in a migratory divide. Parasitology 2011, 138, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, I.; Ilahiane, L.; Boano, G.; Cucco, M.; Pavia, M.; Prestridge, H.L.; Voelker, G. Avian haemosporidian diversity on sardinia: A first general assessment for the insular mediterranean. Diversity 2021, 13, 75. [Google Scholar] [CrossRef]
- Mtierová, Z.; Derdáková, M.; Chvostáč, M.; Didyk, Y.M.; Mangová, B.; Rusňáková Tarageľová, V.; Selyemová, D.; Šujanová, A.; Václav, R. Local population structure and seasonal variability of borrelia garinii genotypes in ixodes ricinus ticks, Slovakia. Int. J. Environ. Res. Public Health 2020, 17, 3607. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1989; ISBN 978-0-87969-309-1. [Google Scholar]
- Bell, J.A.; Weckstein, J.D.; Fecchio, A.; Tkach, V.V. A new real-time pcr protocol for detection of avian haemosporidians. Parasites Vectors 2015, 8, 383. [Google Scholar] [CrossRef]
- Friedl, T.W.P.; Groscurth, E. A real-time PCR protocol for simple and fast quantification of blood parasite infections in evolutionary and ecological studies and some data on intensities of blood parasite infections in a subtropical weaverbird. J. Ornithol. 2012, 153, 239–247. [Google Scholar] [CrossRef]
- Bensch, S.; Stjernman, M.; Hasselquist, D.; Ostman, O.; Hansson, B.; Westerdahl, H.; Pinheiro, R.T. Host specificity in avian blood parasites: A study of plasmodium and haemoproteus mitochondrial DNA amplified from birds. Proc. Biol. Sci. 2000, 267, 1583–1589. [Google Scholar] [CrossRef]
- Hellgren, O.; Waldenström, J.; Bensch, S. A new PCR assay for simultaneous studies of leucocytozoon, plasmodium, and haemoproteus from avian blood. J. Parasitol. 2004, 90, 797–802. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. The UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Bensch, S.; Hellgren, O.; Pérez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Esser, H.J.; Herre, E.A.; Blüthgen, N.; Loaiza, J.R.; Bermúdez, S.E.; Jansen, P.A. Host specificity in a diverse neotropical tick community: An assessment using quantitative network analysis and host phylogeny. Parasites Vectors 2016, 9, 372. [Google Scholar] [CrossRef]
- Blüthgen, N.; Menzel, F.; Blüthgen, N. Measuring specialization in species interaction networks. BMC Ecol. 2006, 6, 9. [Google Scholar] [CrossRef][Green Version]
- Blüthgen, N.; Fründ, J.; Vázquez, D.P.; Menzel, F. What do interaction network metrics tell us about specialization and biological traits. Ecology 2008, 89, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- Poulin, R. Network analysis shining light on parasite ecology and diversity. Trends Parasitol. 2010, 26, 492–498. [Google Scholar] [CrossRef]
- Tiersch, T.R.; Wachtel, S.S. On the evolution of genome size of birds. J. Hered. 1991, 82, 363–368. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Hadfield, J.D.; Nakagawa, S. General quantitative genetic methods for comparative biology: Phylogenies, taxonomies and multi-trait models for continuous and categorical characters. J. Evol. Biol. 2010, 23, 494–508. [Google Scholar] [CrossRef]
- Jetz, W.; Thomas, G.H.; Joy, J.B.; Hartmann, K.; Mooers, A.O. The global diversity of birds in space and time. Nature 2012, 491, 444–448. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Bürkner, P.-C. Estimating Phylogenetic Multilevel Models with Brms. Available online: https://cran.r-project.org/web/packages/brms/vignettes/brms_phylogenetics.html (accessed on 9 August 2021).
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. Available online: https://CRAN.R-project.org/package=vegan (accessed on 9 August 2021).
- Dormann, C.F.; Fründ, J.; Blüthgen, N.; Gruber, B. Indices, graphs and null models: Analyzing bipartite ecological networks. Open Ecol. J. 2009, 2, 7–24. [Google Scholar] [CrossRef]
- Dormann, C.F. How to be a specialist? Quantifying specialisation in pollination networks. Netw. Biol. 2011, 1, 1–20. [Google Scholar]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Bürkner, P.-C. Brms: An R package for bayesian multilevel models using stan. J. Stat. Softw. 2017, 80, 1–28. [Google Scholar] [CrossRef]
- Bürkner, P.-C. Advanced bayesian multilevel modeling with the R package brms. R J. 2018, 10, 395–411. [Google Scholar] [CrossRef]
- Morand, S.; Krasnov, B.R. The Biogeography of Host-Parasite Interactions; Oxford University Press: Oxford, UK, 2010; ISBN 978-0-19-956134-6. [Google Scholar]
- Santiago-Alarcon, D.; MacGregor-Fors, I.; Kühnert, K.; Segelbacher, G.; Schaefer, H.M. Avian haemosporidian parasites in an urban forest and their relationship to bird size and abundance. Urb. Ecosyst. 2016, 19, 331–346. [Google Scholar] [CrossRef]
- Bensch, S.; Jönsson, J.; Copete, J.L. Low prevalence of haemoproteus infections in chiffchaffs. Parasitology 2012, 139, 302–309. [Google Scholar] [CrossRef]
- Santiago-Alarcon, D.; MacGregor-Fors, I.; Falfán, I.; Lüdtke, B.; Segelbacher, G.; Schaefer, H.M.; Renner, S. Parasites in space and time: A case study of haemosporidian spatiotemporal prevalence in urban birds. Int. J. Parasitol. 2019, 49, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G. The role of seasonal migrations in the distribution of haemosporidia of birds in north palaearctic. Ekologija 1993, 2, 57–67. [Google Scholar]
- Cepák Jaroslav, J.; Klvaňa, P.; Škopek, J.; Schröpfer, L.; Jelínek, M.; Hořák, D.; Formánek, J.; Zárybnický, J. Czech and Slovak Bird Migration Atlas; Aventinum: Prague, Czech Republic, 2008. [Google Scholar]
- Clement, P.; Christie, D.A. Hawfinch (coccothraustes coccothraustes). In Birds of the World, 1st ed.; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2000. [Google Scholar]
- Collar, N. European robin (erithacus rubecula). In Birds of the World, 1st ed.; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2000. [Google Scholar]
- Valkiūnas, G. Avian Malaria Parasites and Other Haemosporidia; CRC press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Deviche, P.; Parris, J. Testosterone treatment to free-ranging male dark-eyed juncos (junco hyemalis) exacerbates hemoparasitic infection. Auk 2006, 123, 548–562. [Google Scholar] [CrossRef]
- Cornelius, J.; Zylberberg, M.; Breuner, C.; Gleiss, A.C.; Hahn, T. Assessing the role of reproduction and stress in the spring emergence of haematozoan parasites in birds. J. Exp. Biol. 2013, 217, 841–849. [Google Scholar] [CrossRef]
- Bernotienė, R.; Palinauskas, V.; Iezhova, T.; Murauskaitė, D.; Valkiūnas, G. Avian haemosporidian parasites (haemosporida): A comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp. Parasitol. 2016, 163, 31–37. [Google Scholar] [PubMed]
- Evans, K.L.; Gaston, K.J.; Sharp, S.P.; McGowan, A.; Simeoni, M.; Hatchwell, B.J. Effects of urbanisation on disease prevalence and age structure in blackbird turdus merula populations. Oikos 2009, 118, 774–782. [Google Scholar] [CrossRef]
- Bentz, S.; Rigaud, T.; Barroca, M.; Martin-Laurent, F.; Bru, D.; Moreau, J.; Faivre, B. Sensitive measure of prevalence and parasitaemia of haemosporidia from european blackbird (turdus merula) populations: Value of PCR-RFLP and quantitative PCR. Parasitology 2006, 133, 685–692. [Google Scholar] [CrossRef]


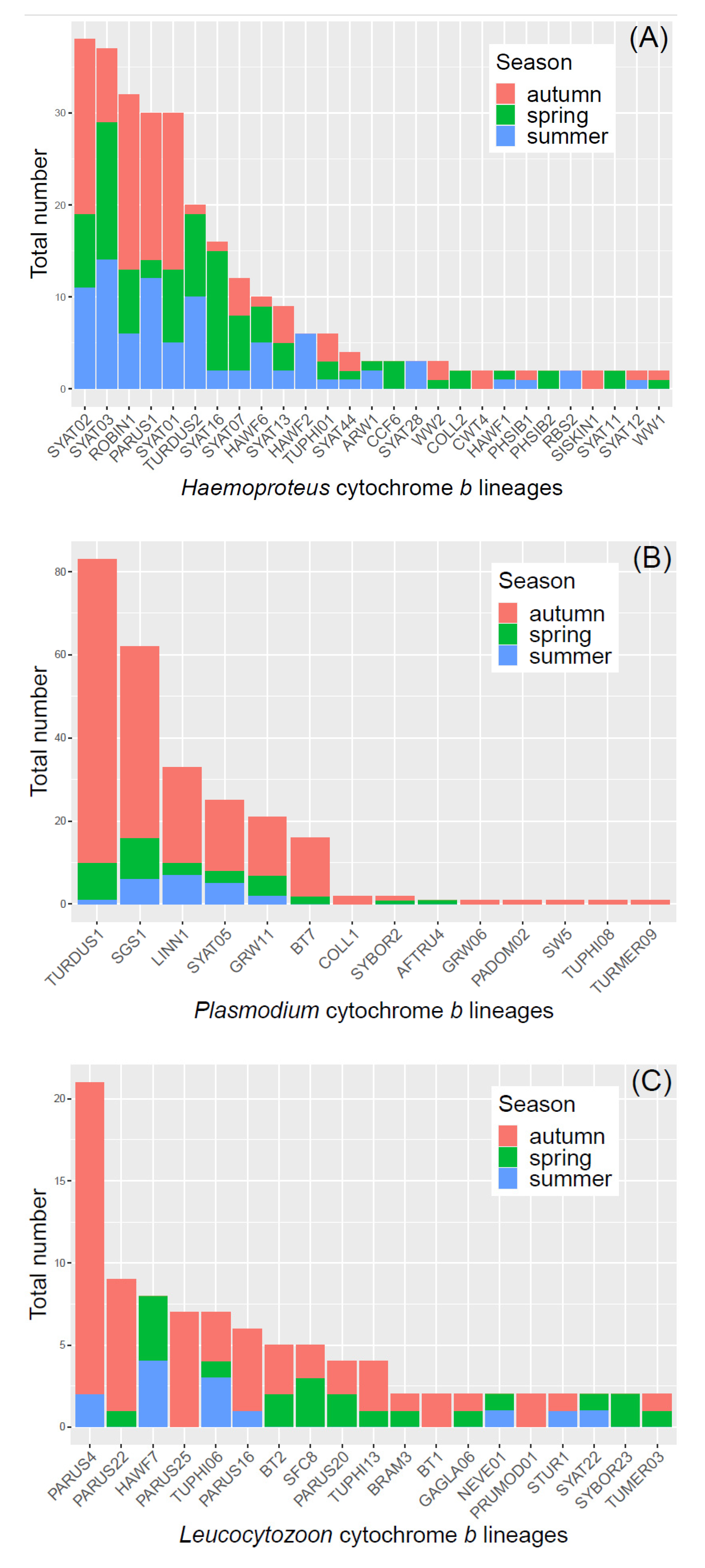
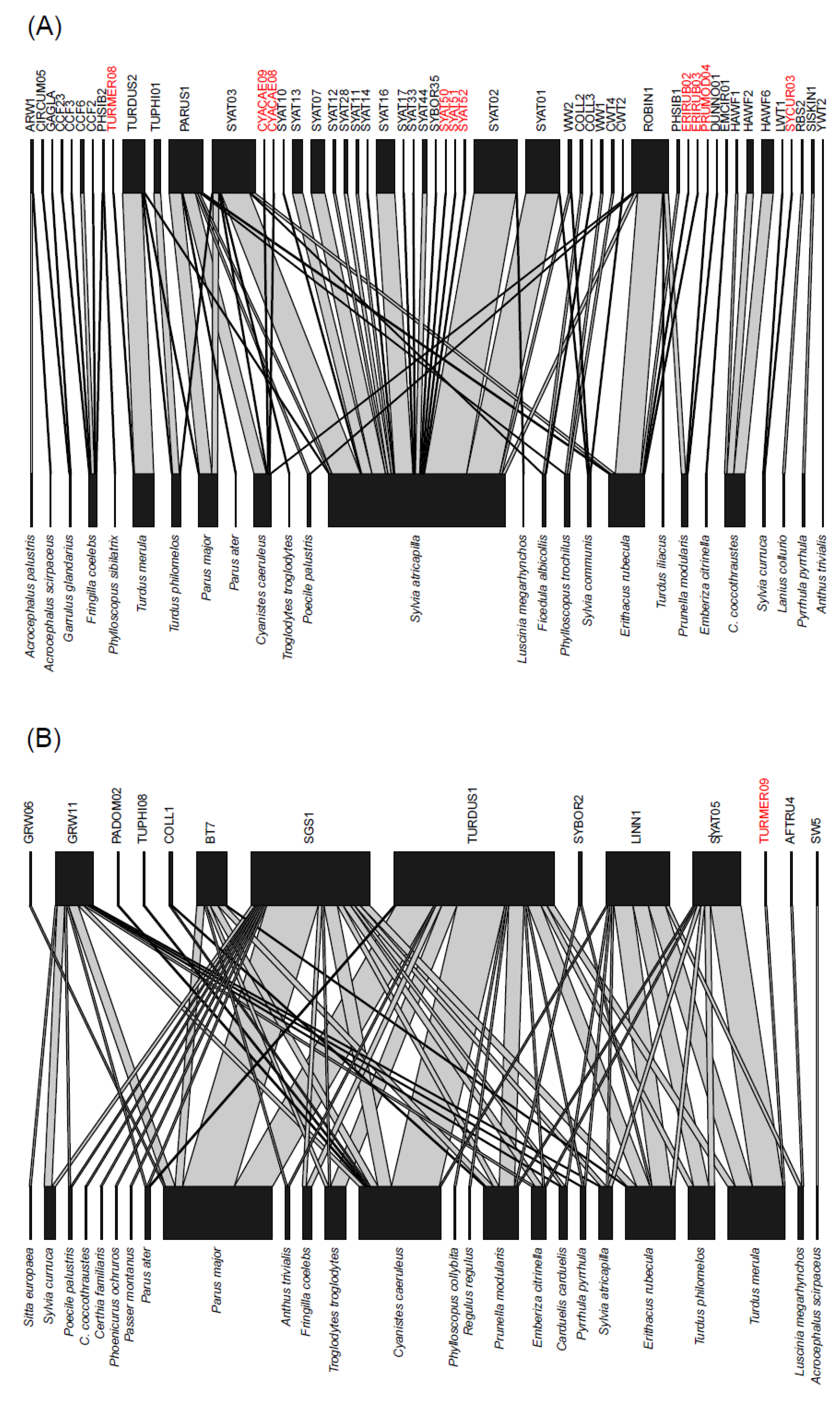
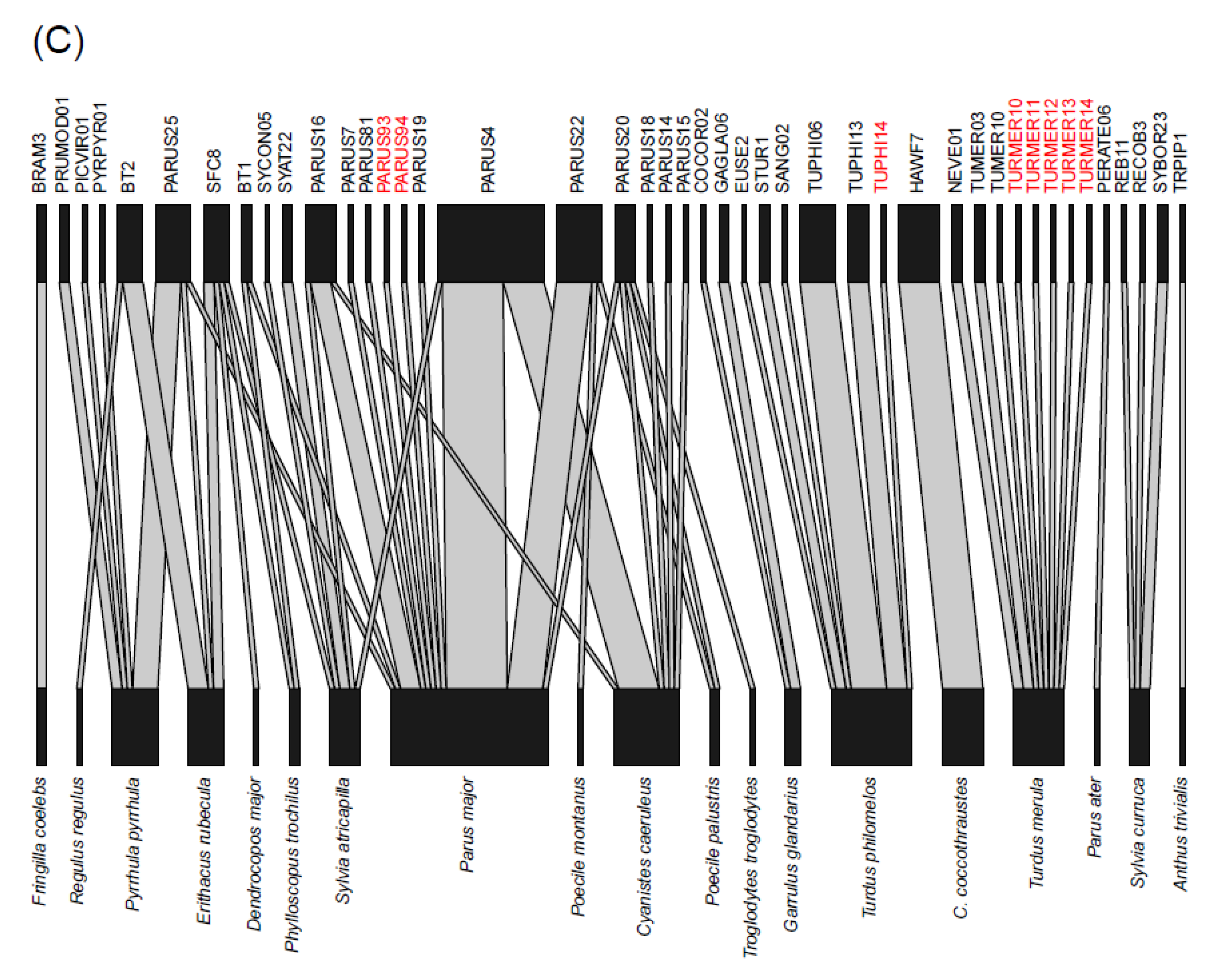
| Bird Species | MS | n (’17/’18/’19) | HY | AHY | Prevalence | d′ H | d′ P | d′ L | Haemosporidian Lineage (n) |
|---|---|---|---|---|---|---|---|---|---|
| Caprimulgiformes | |||||||||
| Caprimulgus europaeus | LD | 1 (0/1/0) | 1 | 0 | 0 (0/1) | - | - | - | |
| Gruiformes | |||||||||
| Crex crex | LD | 1 (0/1/0) | 1 | 0 | 0 (0/1) | - | - | - | |
| Rallus aquaticus | SD | 1 (0/1/0) | 0 | 1 | 100 (1/1) | - | - | - | |
| Accipitriformes | |||||||||
| Buteo buteo | SD | 2 (1/0/1) | 0 | 2 | 0 (0/2) | - | - | - | |
| Coraciiformes | |||||||||
| Alcedo atthis | SD | 1 (0/1/0) | 1 | 0 | 0 (0/1) | - | - | - | |
| Piciformes | |||||||||
| Dendrocopus major | SD | 7 (1/1/5) | 3 | 4 | 17 (1/6) | - | - | 0.47 | L: SFC8 (1) |
| Dryobates minor | SD | 1 (1/0/0) | 1 | 0 | - (0/0) | - | - | - | |
| Jynx torquilla | LD | 2 (1/0/1) | 1 | 1 | 0 (0/2) | - | - | - | |
| Passeriformes | |||||||||
| Laniidae | |||||||||
| Lanius collurio | LD | 5 (4/0/1) | 2 | 3 | 40 (2/5) | 1.00 | - | - | H: RBS2 (2) |
| Corvidae | |||||||||
| Garrulus glandarius | SD | 11 (3/4/4) | 4 | 7 | 30 (3/10) | 1.00 | - | 1.00 | H: CIRCUM05 (1), GAGLA02 (1), L: COCOR02 (1), GAGLA06 (2) |
| Remizidae | |||||||||
| Remiz pendulinus | SD | 6 (3/0/3) | 5 | 1 | 0 (0/5) | - | - | - | |
| Paridae | |||||||||
| Cyanistes caeruleus | SD | 121 (42/48/31) | 94 | 28 | 64 (76/118) | 0.56 | 0.17 | 0.52 | H: CYACAE08 (1), CYACAE09 (1), PARUS1 (11), ROBIN1 (1), SYAT03 (1), P: BT7 (4), COLL1 (1), GRW11 (3), LINN1 (1), PADOM02 (1), SGS1 (7), TUPHI08 (1), TURDUS1 (25), L: PARUS4 (8), PARUS14 (1), PARUS15 (1), PARUS16 (1), PARUS18 (1), PARUS20 (1) |
| Parus ater | SD | 10 (0/7/3) | 9 | 1 | 70 (7/10) | 0.06 | 0.04 | 1.00 | H: PARUS1 (1), P: GRW11 (2), SGS1 (1), TURDUS1 (1), L: PERATE06 (1) |
| Parus major | SD | 175 (35/86/54) | 126 | 49 | 53 (92/172) | 0.46 | 0.23 | 0.58 | H: PARUS1 (11), SYAT03 (5), TURDUS2 (1), P: BT7 (4), GRW06 (1), GRW11 (5), SGS1 (27), TURDUS1 (20), L: BT1 (1), PARUS4 (12), PARUS7 (1), PARUS16 (4), PARUS19 (1), PARUS20 (1), PARUS22 (7), PARUS25 (1), PARUS81 (1), PARUS93 (1), PARUS94 (1) |
| Lophophanes cristatus | SD | 1 (0/1/0) | 1 | 0 | 0 (0/1) | - | - | - | |
| Poecile montanus | SD | 1 (1/0/0) | 0 | 1 | 100 (1/1) | - | - | 0.28 | L: PARUS22 (1) |
| Poecile palustris | SD | 23 (4/8/11) | 14 | 9 | 30 (7/23) | 0.17 | 0.17 | 0.32 | H: PARUS1 (2), ROBIN1 (1), P: GRW11 (1), SGS1 (1), L: PARUS20 (1), PARUS22 (1) |
| Acrocephalidae | |||||||||
| Hippolais icterina | LD | 2 (0/1/1) | 1 | 1 | 0 (0/1) | - | - | - | |
| Acrocephalus arundinaceus | LD | 1 (0/1/0) | 1 | 0 | 0 (0/1) | - | - | - | |
| A. palustris | LD | 13 (2/5/6) | 3 | 10 | 31 (4/13) | 0.89 | - | - | H: ARW1 (2), |
| A. scirpaceus | LD | 5 (0/5/0) | 3 | 2 | 40 (2/5) | 0.70 | 1.00 | - | H: ARW1 (1), P: SW5 (1) |
| A. schoenobaenus | LD | 5 (0/5/0) | 5 | 0 | 0 (0/5) | - | - | - | |
| Locustellidae | |||||||||
| Locustella naevia | LD | 4 (0/4/0) | 4 | 0 | 0 (0/4) | - | - | - | |
| Hirundinidae | |||||||||
| Hirundo rustica | LD | 7 (4/3/0) | 5 | 2 | 0 (0/7) | - | - | - | |
| Sylvidae | |||||||||
| Sylvia atricapilla | SD | 372 (109/158/85) | 158 | 214 | 60 (213/355) | 0.71 | 0.14 | 0.50 | H: PARUS1 (2), ROBIN1 (3), SYAT01 (29), SYAT02 (37), SYAT03 (25), SYAT07 (12), SYAT10 (1), SYAT11 (2), SYAT12 (2), SYAT13 (9), SYAT14 (1), SYAT16 (16), SYAT17 (1), SYAT28 (3), SYAT33 (1), SYAT44 (4), SYAT50 (1), SYAT51 (1), SYAT52 (1), SYBOR35 (1), TURDUS2 (1), WW2 (2), P: GRW11 (1), LINN1 (1), SGS1 (3), SYAT05 (2), L: PARUS4 (1), PARUS16 (1), SYAT22 (2), SYCON05 (1), SFC8 (1) |
| S. borin | LD | 7 (1/6/0) | 4 | 3 | 14 (1/7) | - | - | - | |
| S. communis | LD | 12 (3/9/0) | 4 | 8 | 45 (5/11) | 0.58 | - | - | H: CWT2 (1), SYAT01 (1), WW2 (1) |
| S. curruca | LD | 35 (14/10/11) | 2 | 33 | 41 (13/32) | 1.00 | 0.38 | 1.00 | LWT1 (1), SYCUR03 (1), P: GRW11 (4), SGS1 (2), L: REB11 (1), RECOB3 (1), SYBOR23 (2) |
| Phylloscopidae | |||||||||
| Phylloscopus collybita | SD | 73 (14/16/43) | 9 | 53 | 9 (6/65) | - | 0.00 | - | P: TURDUS1 (1) |
| P. sibilatrix | LD | 4 (1/1/2) | 1 | 3 | 50 (2/4) | 0.81 | - | - | H: PHSIB2 (1) |
| P. trochilus | LD | 28 (7/16/5) | 1 | 27 | 32 (9/28) | 0.81 | - | 0.56 | CWT4 (2), PARUS1 (1), WW1 (2), L: BT1 (1), SFC8 (1) |
| Aegithalidae | |||||||||
| Aegithalos caudatus | SD | 8 (2/3/3) | 0 | 8 | 13 (1/8) | - | - | - | |
| Regulidae | |||||||||
| Regulus ignicapilla | SD | 4 (0/1/3) | 0 | 4 | 0 (0/3) | - | - | - | |
| Regulus regulus | SD | 20 (3/6/11) | 10 | 10 | 20 (4/20) | - | 0.00 | 0.47 | P: TURDUS1 (1), L: BT2 (1) |
| Sittidae | |||||||||
| Sitta europaea | SD | 5 (2/2/1) | 4 | 1 | 20 (1/5) | - | 0.32 | - | P: GRW11 (1) |
| Certhiidae | |||||||||
| Certhia familiaris | SD | 9 (2/3/3) | 2 | 7 | 22 (2/9) | - | 0.07 | - | P: SGS1 (1) |
| Troglodytidae | |||||||||
| T. troglodytes | SD | 29 (12/4/13) | 10 | 19 | 59 (16/27) | 0.00 | 0.20 | 0.54 | H: SYAT03 (1), P: BT7 (2), TURDUS1 (8), SGS1 (1), L: PARUS20 (1) |
| Turdidae | |||||||||
| Turdus iliacus | SD | 1 (0/0/1) | 1 | 0 | 100 (1/1) | 0.05 | - | - | H: ROBIN1 (1) |
| Turdus merula | SD | 96 (26/33/37) | 45 | 51 | 67 (64/95) | 0.94 | 0.50 | 1.00 | TURDUS2 (17), TURMER08 (1), P: LINN1 (9), SYAT05 (16), TURDUS1 (4), TURMER09 (1), L: NEVE01 (2), TUMER03 (2), TUMER10 (1), TURMER10 (1), TURMER11 (1), TURMER12 (1), TURMER13 (1), TURMER14 (1) |
| Turdus philomelos | SD | 40 (8/10/22) | 22 | 18 | 78 (31/40) | 0.74 | 0.30 | 1.00 | H: TUPHI01 (6), TURDUS2 (1), P: LINN1 (8), SYAT05 (3), TURDUS1 (3), L: EUSE2 (1), SANG02 (1), STUR1 (2), TUPHI06 (7), TUPHI13 (4), TUPHI14 (1) |
| Muscicapidae | |||||||||
| Muscicapa striata | LD | 1 (0/0/1) | 1 | 0 | - (0/0) | - | - | - | |
| Ficedula albicollis | LD | 29 (6/7/16) | 15 | 13 | 14 (4/29) | 0.74 | - | - | H: COLL2 (2), COLL3 (1), SYAT03 (1) |
| Ficedula hypoleuca | LD | 3 (1/1/1) | 1 | 1 | 33 (1/3) | - | - | ||
| Luscinia megarhynchos | LD | 17 (6/6/5) | 8 | 11 | 24 (4/17) | 0.00 | 0.53 | - | H: SYAT02 (1), P: AFTRU4 (1), LINN1 (2) |
| Erithacus rubecula | SD | 354 (78/171/135) | 225 | 125 | 25 (85/336) | 0.65 | 0.13 | 0.68 | H: ERIRUB02 (1), ERIRUB03 (1), PARUS1 (2), PHSIB1 (2), ROBIN1 (22), SYAT03 (4), P: BT7 (1), LINN1 (10), SGS1 (5), SYAT05 (2), SYBOR2 (1), TURDUS1 (7), L: BT2 (4), PARUS25 (1), SFC8 (2) |
| Phoenicurus phoenicurus | LD | 6 (1/3/2) | 3 | 3 | 17 (1/6) | - | - | - | |
| Phoenicurus ochruros | SD | 5 (1/1/3) | 2 | 3 | 20 (1/5) | - | 0.07 | - | P: SGS1 (1) |
| Prunellidae | |||||||||
| Prunella modularis | SD | 75 (33/21/21) | 54 | 21 | 38 (26/69) | 0.60 | 0.18 | - | H: DUNNO01 (1), PRUMOD04 (1), ROBIN1 (4), P: BT7 (4), COLL1 (1), SGS1 (3), SYAT05 (1), SYBOR2 (1), TURDUS1 (8) |
| Passeridae | |||||||||
| Passer montanus | SD | 5 (1/4/0) | 2 | 3 | 60 (3/5) | - | 0.07 | - | P: SGS1 (1) |
| Motacillidae | |||||||||
| Anthus trivialis | LD | 8 (0/7/1) | 5 | 3 | 25 (2/8) | 1.00 | 0.16 | 1.00 | H: YWT2 (1), P: BT7 (1), TURDUS1 (1), L: TRPIP1 (1) |
| Fringillidae | |||||||||
| Fringilla coelebs | SD | 31 (8/7/16) | 5 | 26 | 61 (19/31) | 0.97 | 0.08 | 1.00 | H: CCF2 (1), CCF3 (1), CCF6 (3), CCF23 (1), PHSIB2 (1), P: SGS1 (2), TURDUS1 (3), L: BRAM3 (2) |
| F. montifringilla | SD | 2 (0/0/2) | 0 | 2 | 0 (0/2) | - | - | - | |
| Coccothraustes coccothraustes | SD | 44 (16/12/16) | 19 | 25 | 60 (25/42) | 1.00 | 0.07 | 1.00 | H: HAWF1 (2), HAWF2 (6), HAWF6 (10), P: SGS1 (1), L: HAWF7 (8) |
| Linaria cannabina | SD | 3 (2/1/0) | 2 | 1 | 33 (1/3) | - | - | - | |
| Carduelis carduelis | SD | 47 (27/20/0) | 29 | 18 | 13 (6/47) | - | 0.13 | - | P: GRW11 (1), LINN1 (1), SGS1 (2) |
| Chloris chloris | SD | 12 (10/2/0) | 7 | 5 | 0 (0/11) | - | - | - | |
| Serinus serinus | SD | 1 (1/0/0) | 1 | 0 | - (0/0) | - | - | - | |
| Pyrrhula pyrrhula | SD | 39 (3/11/25) | 20 | 8 | 42 (16/38) | 1.00 | 0.09 | 0.91 | H: SISKIN1 (2), P: GRW11 (1), LINN1 (1), TURDUS1 (1), L: PARUS25 (5), PICVIR01 (1), PRUMOD01 (2), PYRPYR01 (1) |
| Emberizidae | |||||||||
| Emberiza citrinella | SD | 17 (4/9/4) | 3 | 14 | 53 (8/15) | 1.00 | 0.13 | - | H: EMCIR01 (1), P: GRW11 (2), SGS1 (4), SYAT05 (1), TURDUS1 (1) |
| Emberiza cia | SD | 1 (1/0/0) | 0 | 1 | 0 (0/1) | - | - | - | |
| Emberiza schoeniclus | SD | 3 (0/1/2) | 1 | 2 | 0 (0/2) | - | - | - |
| Time of Year | S | Exp (H′) | |
|---|---|---|---|
| Avian hosts (n = 61) | spring | 38 | 12.6 |
| summer | 30 | 10.9 | |
| autumn | 50 | 17 | |
| Haemosporidian lineages | spring | 60 | 38.0 |
| summer | 44 | 28.4 | |
| autumn | 65 | 24.4 | |
| Haemoproteus (n = 53) | spring | 27 | 17.2 |
| summer | 30 | 18.4 | |
| autumn | 26 | 13 | |
| Plasmodium (n = 14) | spring | 8 | 6 |
| summer | 5 | 4.2 | |
| autumn | 13 | 5.8 | |
| Leucocytozoon (n = 44) | spring | 25 | 22.3 |
| summer | 9 | 7.6 | |
| autumn | 26 | 15.2 |
| Infection Type | ||||
|---|---|---|---|---|
| Time of Year | Single | Double | >Double | Total |
| Haemoproteus | ||||
| spring | 85 | 12 | 5 | 102 |
| summer | 70 | 19 | 8 | 97 |
| autumn | 97 | 10 | 4 | 111 |
| Plasmodium | ||||
| spring | 29 | 6 | 0 | 35 |
| summer | 16 | 6 | 0 | 22 |
| autumn | 180 | 17 | 1 | 198 |
| Leucocytozoon | ||||
| spring | 28 | 6 | 4 | 38 |
| summer | 11 | 2 | 7 | 20 |
| autumn | 56 | 10 | 7 | 73 |
| Infection Type | ||
|---|---|---|
| Time of Year | Intra-Generic | with Leucocytozoon |
| Haemoproteus | ||
| spring | 90 | 12 |
| summer | 87 | 10 |
| autumn | 100 | 11 |
| Plasmodium | ||
| spring | 30 | 5 |
| summer | 17 | 5 |
| autumn | 155 | 43 |
| Intra-generic | with Haemoproteus | |
| Leucocytozoon | ||
| spring | 26 | 12 |
| summer | 10 | 10 |
| autumn | 62 | 11 |
| Intra-generic | with Plasmodium | |
| Leucocytozoon | ||
| spring | 33 | 5 |
| summer | 15 | 5 |
| autumn | 30 | 43 |
| Parameter | Estimate | Est. Error | L-95% CI | U-95% CI | Rhat | Bulk ESS | Tail ESS | pMCMC |
|---|---|---|---|---|---|---|---|---|
| Random effects (SD) | ||||||||
| Phylogeny (35 levels) | 0.05 | 0.03 | 0.01 | 0.11 | 1 | 1126 | 1665 | |
| Year (3 levels) | 0.55 | 0.62 | 0.03 | 2.2 | 1 | 784 | 633 | |
| Year:Species (67 levels) | 0.24 | 0.15 | 0.01 | 0.55 | 1 | 812 | 3163 | |
| Fixed effects | ||||||||
| Intercept | −0.7 | 0.68 | −2.05 | 0.65 | 1 | 2553 | 3620 | 0.134 |
| Date | 0.02 | 0.01 | 0 | 0.03 | 1 | 7369 | 21,192 | 0.013 |
| Date^2 | ~0 | ~0 | ~0 | ~0 | 1 | 6810 | 20,008 | 0.006 |
| Date/species | ~0 | ~0 | ~0 | ~0 | 1 | 72,435 | 103,455 | 0.466 |
| Age_HY | −0.25 | 0.16 | −0.56 | 0.06 | 1 | 19,968 | 51,277 | 0.057 |
| Residual effect (SD) | ||||||||
| Sigma | 1.18 | 0.05 | 1.09 | 1.28 | 1 | 15,783 | 48,731 |
| Parameter | Estimate | Est. Error | L-95% CI | U-95% CI | Rhat | Bulk ESS | Tail ESS | pMCMC |
|---|---|---|---|---|---|---|---|---|
| Random effects (SD) | ||||||||
| Phylogeny (58 levels) | 0.12 | 0.05 | 0.04 | 0.25 | 1 | 38,921 | 35,894 | |
| Year (3 levels) | 0.72 | 0.72 | 0.03 | 2.69 | 1 | 27,292 | 18,348 | |
| Year:Species (130 levels) | 0.23 | 0.19 | 0.01 | 0.71 | 1 | 48,274 | 57,250 | |
| Fixed effects | ||||||||
| Intercept | −0.06 | 0.95 | −1.99 | 1.9 | 1 | 34,464 | 26,486 | 0.469 |
| Season_Spring | −0.29 | 0.41 | −1.11 | 0.51 | 1 | 80,281 | 68,270 | 0.242 |
| Season_Summer | −0.83 | 0.38 | −1.59 | −0.09 | 1 | 103,349 | 80,439 | 0.014 |
| d′ | SESMPD | H2 | d′ | SESMPD | H2 | d′ | SESMPD | H2 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haemoproteus | 0.70 | Plasmodium | 0.31 | Leucocytozoon | 0.69 | ||||||
| ARW1 | 0.57 | −1.67 | AFTRU4 | 0.72 | - | BRAM3 | 0.48 | - | |||
| CIRCUM05 | 0.59 | - | BT7 | 0.19 | 0.51 | BT1 | 0.25 | −0.25 | |||
| CCF23 | 0.42 | - | COLL1 | 0.27 | 0.54 | BT2 | 0.16 | −0.15 | |||
| CCF2 | 0.42 | GRW06 | 0.18 | - | COCOR02 | 0.59 | - | ||||
| CCF3 | 0.42 | - | GRW11 | 0.20 | −2.64 * | EUSE2 | 0.38 | - | |||
| CCF6 | 0.49 | - | LINN1 | 0.18 | −1.87 * | GAGLA06 | 0.64 | - | |||
| COLL2 | 0.49 | - | PADOM02 | 0.26 | - | HAWF7 | 0.46 | - | |||
| COLL3 | 0.43 | - | SGS1 | 0.12 | −1.00 | NEVE01 | 0.31 | - | |||
| CWT2 | 0.58 | - | SW5 | 1.00 | - | PARUS14 | 0.19 | - | |||
| CWT4 | 0.50 | - | SYAT05 | 0.26 | −1.00 | PARUS15 | 0.19 | - | |||
| DUNNO01 | 0.27 | - | SYBOR2 | 0.16 | 0.40 | PARUS16 | 0.12 | −1.06 | |||
| EMCIR01 | 0.52 | - | TUPHI08 | 0.26 | - | PARUS18 | 0.19 | - | |||
| GAGLA02 | 0.59 | - | TURDUS1 | 0.13 | 0.26 | PARUS19 | 0.13 | - | |||
| HAWF1 | 0.43 | - | TURMER09 | 0.31 | - | PARUS20 | 0.19 | −1.69 * | |||
| HAWF2 | 0.45 | - | PARUS22 | 0.28 | −2.99 * | ||||||
| HAWF6 | 0.46 | - | PARUS25 | 0.29 | −0.09 | ||||||
| LWT1 | 0.40 | - | PARUS4 | 0.17 | −1.06 | ||||||
| PARUS1 | 0.13 | −3.00 * | PARUS7 | 0.13 | - | ||||||
| PHSIB1 | 0.11 | - | PARUS81 | 0.13 | - | ||||||
| PHSIB2 | 0.53 | 0.34 | PERATE06 | 0.61 | - | ||||||
| RBS2 | 0.76 | - | PYRPYR01 | 0.38 | - | ||||||
| ROBIN1 | 0.11 | −0.18 | PRUMOD01 | 0.44 | - | ||||||
| SISKIN1 | 0.44 | - | PICVIR01 | 0.38 | - | ||||||
| SYAT01 | 0.18 | −3.35 * | REB11 | 0.40 | - | ||||||
| SYAT02 | 0.18 | 0.37 | RECOB3 | 0.40 | - | ||||||
| SYAT03 | 0.07 | −1.09 | SANG02 | 0.38 | - | ||||||
| SYAT07 | 0.16 | - | SFC8 | 0.14 | 1.77 | ||||||
| SYAT10 | 0.00 | - | STUR1 | 0.44 | - | ||||||
| SYAT11 | 0.10 | - | SYAT22 | 0.10 | - | ||||||
| SYAT12 | 0.10 | - | SYBOR23 | 0.46 | - | ||||||
| SYAT13 | 0.16 | - | SYCON05 | 0.00 | - | ||||||
| SYAT14 | 0.00 | - | TRPIP1 | 0.65 | |||||||
| SYAT16 | 0.17 | - | TUMER03 | 0.31 | |||||||
| SYAT17 | 0.00 | - | TUPHI06 | 0.47 | |||||||
| SYAT28 | 0.12 | - | TUPHI13 | 0.46 | |||||||
| SYAT33 | 0.00 | - | TUMER10 | 0.23 | |||||||
| SYAT44 | 0.13 | - | TURMER10 | 0.23 | |||||||
| SYBOR35 | 0.00 | - | TURMER11 | 0.23 | |||||||
| TUPHI01 | 0.47 | - | TUPHI14 | 0.38 | |||||||
| TURDUS2 | 0.27 | −0.73 | PARUS93 | 0.13 | |||||||
| WW1 | 0.50 | - | TURMER12 | 0.23 | |||||||
| WW2 | 0.20 | −3.35 * | TURMER13 | 0.23 | |||||||
| YWT2 | 0.65 | - | TURMER14 | 0.23 | |||||||
| ERIRUB02 | 0.01 | PARUS94 | 0.13 | ||||||||
| PRUMOD04 | 0.27 | ||||||||||
| TURMER08 | 0.23 | ||||||||||
| ERIRUB03 | 0.01 | ||||||||||
| CYACAE09 | 0.19 | ||||||||||
| CYACAE08 | 0.19 | ||||||||||
| SYAT50 | 0.00 | ||||||||||
| SYAT51 | 0.00 | ||||||||||
| SYAT52 | 0.00 | ||||||||||
| SYCUR03 | 0.40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šujanová, A.; Špitalská, E.; Václav, R. Seasonal Dynamics and Diversity of Haemosporidians in a Natural Woodland Bird Community in Slovakia. Diversity 2021, 13, 439. https://doi.org/10.3390/d13090439
Šujanová A, Špitalská E, Václav R. Seasonal Dynamics and Diversity of Haemosporidians in a Natural Woodland Bird Community in Slovakia. Diversity. 2021; 13(9):439. https://doi.org/10.3390/d13090439
Chicago/Turabian StyleŠujanová, Alžbeta, Eva Špitalská, and Radovan Václav. 2021. "Seasonal Dynamics and Diversity of Haemosporidians in a Natural Woodland Bird Community in Slovakia" Diversity 13, no. 9: 439. https://doi.org/10.3390/d13090439
APA StyleŠujanová, A., Špitalská, E., & Václav, R. (2021). Seasonal Dynamics and Diversity of Haemosporidians in a Natural Woodland Bird Community in Slovakia. Diversity, 13(9), 439. https://doi.org/10.3390/d13090439