Complete Chloroplast Genome Sequence and Comparative and Phylogenetic Analyses of the Cultivated Cyperus esculentus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Genomic DNA Extraction
2.2. DNA Sequencing and Genome Assembly
2.3. Annotation and Analysis of C. esculentus Chloroplast DNA Sequence
2.4. Genome Comparison with Other Cyperus Species
2.5. Phylogenetic Analysis
3. Results and Discussion
3.1. Cyperus Esculentus Chloroplast Genome Assembly and Its Features
3.2. Long-Repeats and Simple Sequence Repeats (SSRs) in C. esculentus Chloroplast Genome
3.3. Comparative Analysis of Chloroplast Genomic Structure with Other Cyperus Species
3.4. Phylogenetic Analysis of C. esculentus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Castro, O.; Gargiulo, R.; Del Guacchio, E.; Caputo, P.; De Luca, P. A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, poales): Two sides of the same coin (weed vs. Crop). Ann. Bot. 2015, 115, 733–745. [Google Scholar] [CrossRef]
- Turesson, H.; Marttila, S.; Gustavsson, K.-E.; Hofvander, P.; Olsson, M.E.; Bülow, L.; Stymne, S.; Carlsson, A.S. Characterization of oil and starch accumulation in tubers of Cyperus esculentus var. Sativus (Cyperaceae): A novel model system to study oil reserves in nonseed tissues. Am. J. Bot. 2010, 97, 1884–1893. [Google Scholar] [CrossRef]
- Yang, Z.; Ji, H.; Liu, D. Oil biosynthesis in underground oil-rich storage vegetative tissue: Comparison of Cyperus esculentus tuber with oil seeds and fruits. Plant Cell Physiol. 2016, 57, 2519–2540. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.A. Effect of dietary supplementation with tigernut tubers on streptozotocin-induced diabetic rats. Egypt J. Hosp. Med. 2007, 29, 475–485. [Google Scholar] [CrossRef]
- Maritim, A.C.; Sanders, R.A.; Watkins, J.B., III. Diabetes, oxidative stress, and antioxidants: A review. J. Biochem. Mol. Toxicol. 2003, 17, 24–38. [Google Scholar] [CrossRef]
- Olabiyi, A.A.; Oboh, G.; Adefegha, S.A. Effect of dietary supplementation of tiger nut (Cyperus esculentus L.) and walnut (tetracarpidium conophorum müll. Arg.) on sexual behavior, hormonal level, and antioxidant status in male rats. J. Food Biochem. 2017, 41, e12351. [Google Scholar] [CrossRef]
- Sabiu, S.; Oladipo Ajani, E.; Sunmonu, T.O.; Tom Ashafa, A.O. Kinetics of modulatory role of Cyperus esculentus L. On the specific activity of key carbohydrate metabolizing enzymes. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 46–53. [Google Scholar] [CrossRef][Green Version]
- Sánchez-Zapata, E.; Fernández-López, J.; Angel Pérez-Alvarez, J. Tiger nut (Cyperus esculentus) commercialization: Health aspects, composition, properties, and food applications. Compr. Rev. Food Sci. Food Saf. 2012, 11, 366–377. [Google Scholar] [CrossRef]
- Elshebini, S.M.; Moaty, M.; Tapozada, S.T.; Hanna, L.M.; Mohamed, H.I.; Raslan, H.M. Effect of regular consumption of tiger nut (Cyperus esculentus) on insulin resistance and tumor necrosis factor-alpha in obese type 2 diabetic egyptian women. Med. J. Cairo Univ. 2010, 78, 607–614. [Google Scholar]
- Gray, M.W. The evolutionary origins of organelles. Trends Genet. 1989, 5, 294–299. [Google Scholar] [CrossRef]
- Howe, C.J.; Barbrook, A.C.; Koumandou, V.L.; Nisbet, R.; Symington, H.A.; Wightman, T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. Lond. B Biol. 2003, 358, 99–107. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Sugiura, M. History of chloroplast genomics. Photosynth. Res. 2003, 76, 371–377. [Google Scholar] [CrossRef]
- Shetty, S.M.; Md Shah, M.U.; Makale, K.; Mohd-Yusuf, Y.; Khalid, N.; Othman, R.Y. Complete chloroplast genome sequence of musa balbisiana corroborates structural heterogeneity of inverted repeats in wild progenitors of cultivated bananas and plantains. Plant Genom. 2016, 9. [Google Scholar] [CrossRef]
- Wu, F.-H.; Chan, M.-T.; Liao, D.-C.; Hsu, C.-T.; Lee, Y.-W.; Daniell, H.; Duvall, M.R.; Lin, C.-S. Complete chloroplast genome of oncidium gower ramsey and evaluation of molecular markers for identification and breeding in oncidiinae. BMC Plant Biol. 2010, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lu, R.-S.; Xu, W.-Q.; Ohi-Toma, T.; Cai, M.-Q.; Qiu, Y.-X.; Cameron, K.M.; Fu, C.-X. Comparative genomics and phylogenomics of east asian tulips (amana, liliaceae). Front. Plant Sci. 2017, 8, 451. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genom. Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Zhang, M.-F.; Xue, J.; Dong, R.; Du, Y.-P.; Zhang, X.-H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef]
- Eguiluz, M.; Rodrigues, N.F.; Guzman, F.; Yuyama, P.; Margis, R. The chloroplast genome sequence from eugenia uniflora, a myrtaceae from neotropics. Plant Syst. Evol. 2017, 303, 1199–1212. [Google Scholar] [CrossRef]
- Du, Y.-P.; Bi, Y.; Yang, F.-P.; Zhang, M.-F.; Chen, X.-Q.; Xue, J.; Zhang, X.-H. Complete chloroplast genome sequences of lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 5751. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, T.; Bai, G.; Zhao, Y. Complete chloroplast genome sequence of fagopyrum dibotrys: Genome features, comparative analysis and phylogenetic relationships. Sci. Rep. 2018, 8, 12379. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Patel, R.K.; Jain, M. Ngs qc toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. Spades: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Qu, X.-J.; Moore, M.J.; Li, D.-Z.; Yi, T.-S. Pga: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellargenomedraw (ogdraw): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. Vista: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. Dnasp 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting est databases for the development and characterization of gene-derived ssr-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. Reputer: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. Iq-tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Zhu, Z. The complete chloroplast genome of pioneering plant Cyperus difformis(Cyperaceae) in ecological restoration. Mitochondrial DNA B Resour. 2019, 4, 1988–1989. [Google Scholar] [CrossRef]
- Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete chloroplast genome sequence and phylogenetic analysis of paeonia ostii. Molecules 2018, 23, 246. [Google Scholar] [CrossRef]
- Biju, V.C.; P.R., S.; Vijayan, S.; Rajan, V.S.; Sasi, A.; Janardhanan, A.; Nair, A.S. The complete chloroplast genome of Trichopus zeylanicus, and phylogenetic analysis with dioscoreales. Plant Genom. 2019, 12, 190032. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zang, M.; Li, M.; Fang, Y. Complete chloroplast genome sequence and phylogenetic analysis of quercus acutissima. Int. J. Mol. Sci. 2018, 19, 2443. [Google Scholar] [CrossRef]
- Weng, M.-L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the ancestral plastid genome in geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol. Biol. Evol. 2013, 31, 645–659. [Google Scholar] [CrossRef]
- Park, I.; Yang, S.; Choi, G.; Kim, W.J.; Moon, B.C. The complete chloroplast genome sequences of aconitum pseudolaeve and aconitum longecassidatum, and development of molecular markers for distinguishing species in the aconitum subgenus lycoctonum. Molecules 2017, 22, 2012. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative analysis of the complete chloroplast genomes of five quercus species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef]
- Kim, K.-J.; Lee, H.-L. Complete chloroplast genome sequences from korean ginseng ( panax schinseng nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.V.; Miller, J.T.; Small, I.; Nevill, P.G.; Boykin, L.M. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in acacia. Mol. Biol. Evol. 2016, 96, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef]
- Larridon, I.; Bauters, K.; Reynders, M.; Huygh, W.; Muasya, A.M.; Simpson, D.A.; Goetghebeur, P. Towards a new classification of the giant paraphyletic genus Cyperus (Cyperaceae): Phylogenetic relationships and generic delimitation in c4 Cyperus. Bot. J. Linn. Soc. 2013, 172, 106–126. [Google Scholar] [CrossRef]
- Whitney, K.D.; Baack, E.J.; Hamrick, J.L.; Godt, M.J.W.; Barringer, B.C.; Bennett, M.D.; Eckert, C.G.; Goodwillie, C.; Kalisz, S.; Leitch, I.J.; et al. A role for nonadaptive processes in plant genome size evolution? Evolution 2010, 64, 2097–2109. [Google Scholar] [CrossRef]

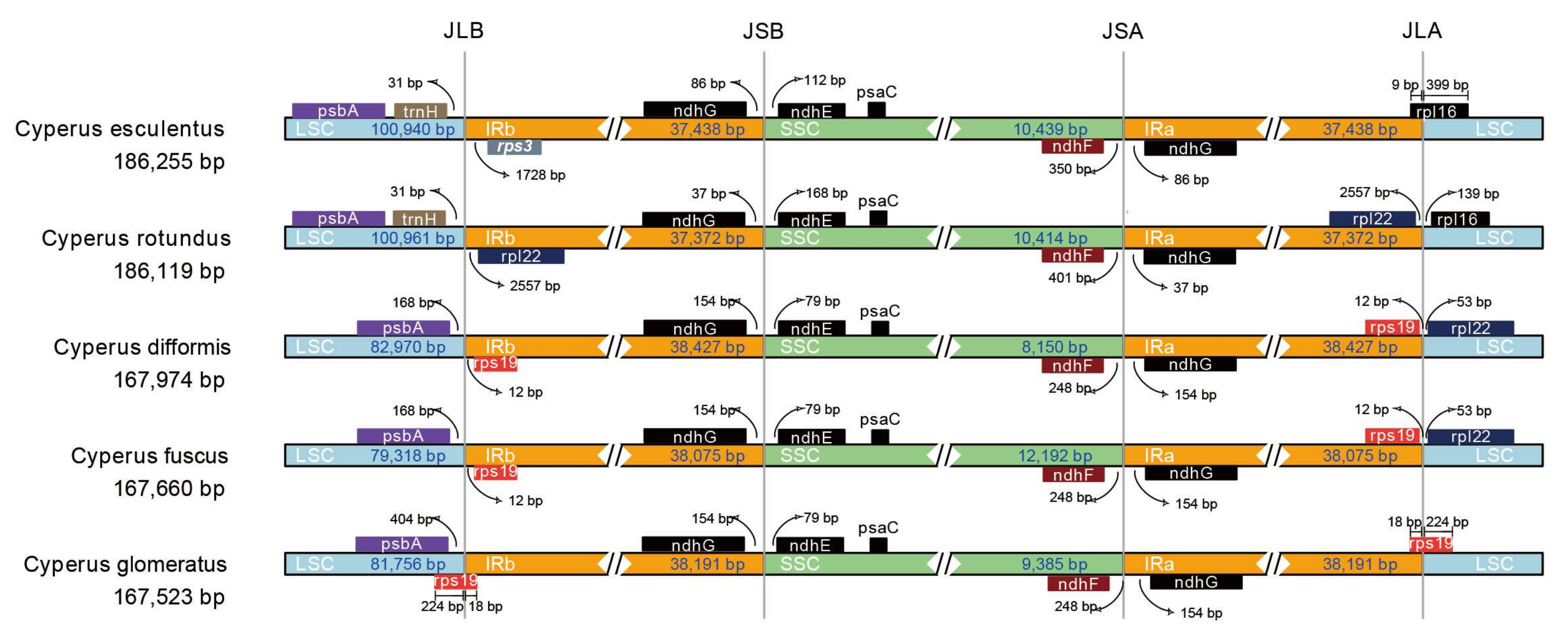
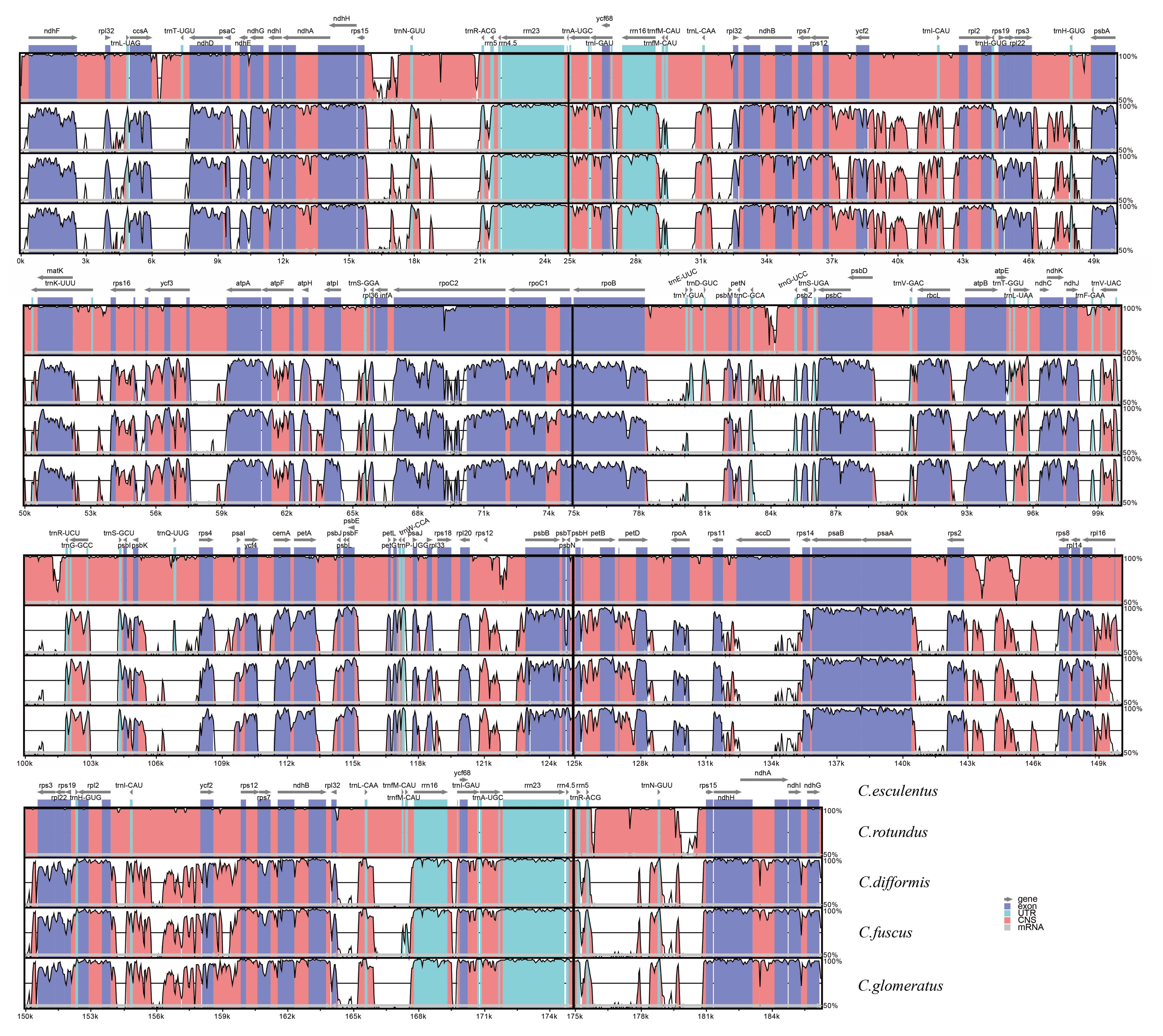

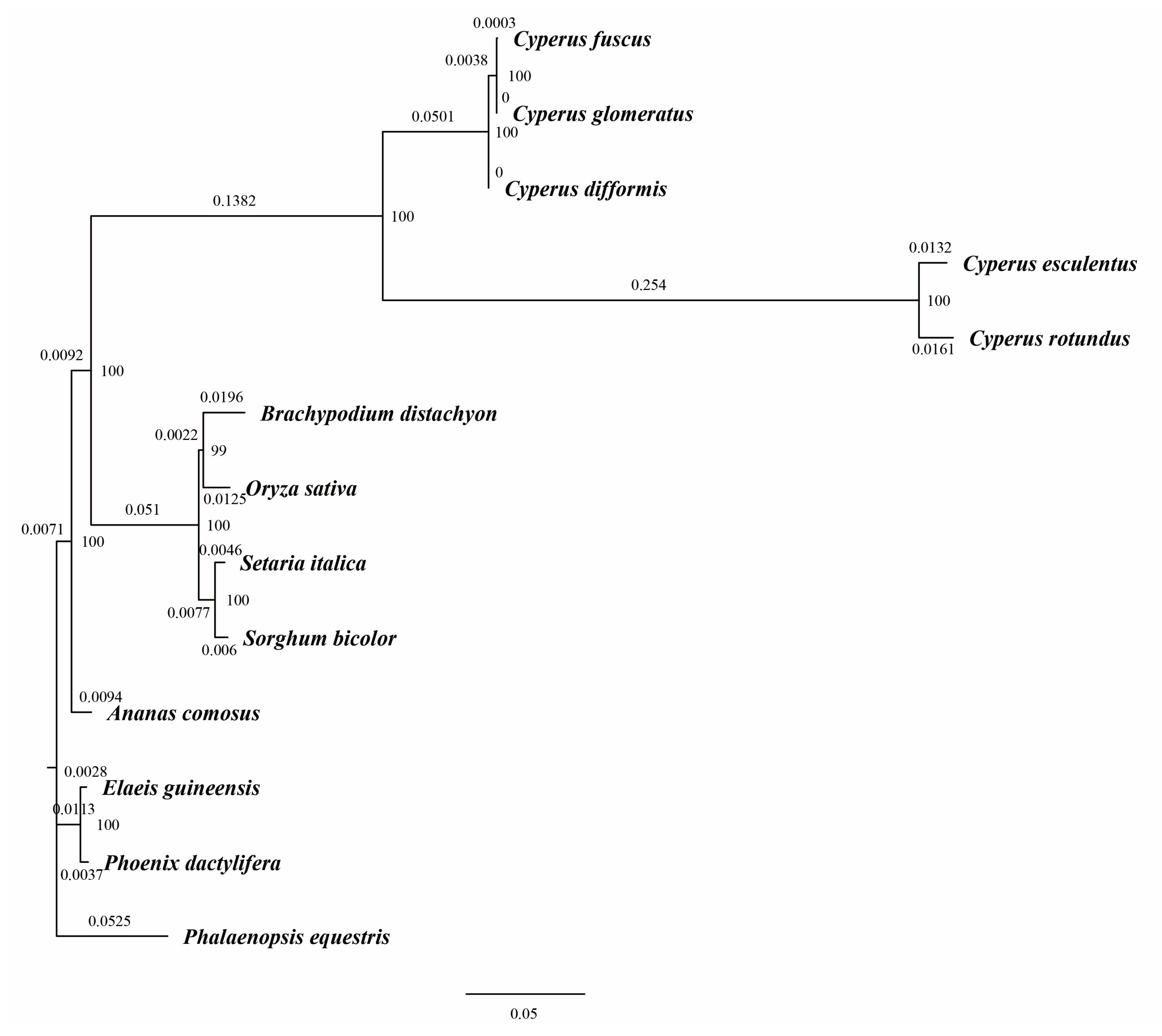
| Genome Features | C. esculentus | C. fuscus | C. glomeratus | C. difformis | C. rotundus |
|---|---|---|---|---|---|
| Genome size (bp) | 186,255 | 167,660 | 167,523 | 167,974 | 186,119 |
| LSC length (bp) | 100,940 | 79,318 | 81,756 | 82,970 | 100,961 |
| SSC length (bp) | 10,439 | 12,192 | 9385 | 8150 | 10,414 |
| IR length (bp) | 37,438 | 38,075 | 38,191 | 38,427 | 37,372 |
| Overall GC content, % | 33.19 | 36.13 | 36.34 | 36.06 | 33.19 |
| GC content in LSC, % | 30.96 | 35.11 | 35.19 | 34.61 | 30.91 |
| GC content in SSC, % | 25.11 | 28.32 | 28.73 | 28.15 | 25.64 |
| GC content in IR, % | 37.32 | 38.45 | 38.5 | 38.48 | 37.37 |
| Number of genes | 141 | 137 | 135 | 137 | 132 |
| Number of PCGs | 93 | 94 | 93 | 91 | 84 |
| Number of tRNA genes | 40 | 35 | 34 | 38 | 40 |
| Number of rRNA genes | 8 | 8 | 8 | 8 | 8 |
| Category | Function | Genes |
|---|---|---|
| RNA genes | Transfer RNA | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnfM-CAU, trnG-GCC *, trnG-UCC, trnH-GUG, trnI-CAU, trnI-GAU *, trnK-UUU *, trnL-CAA, trnL-UAA *, trnL-UAG, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC *, trnW-CCA, trnY-GUA |
| Ribosomal RNA | rrn23, rrn16, rrn5, rrn4.5 | |
| Transcription and translation related genes | Transcription and splicing | rpoC1 *, rpoC2, rpoA, rpoB |
| Translation, ribosomal proteins | rps2, rps3, rps4, rps7, rps8, rps11, rps12 **, rps14, rps15, rps16 *, rps18, rps19, rpl2 *, rpl14, rpl16 *, rpl20, rpl22, rpl32, rpl33, rpl36 | |
| Photosynthesis | ATP synthase | atpE, atpB, atpA, atpF *, atpH, atpI |
| Photosystem I | psaI, psaB, psaA, psaC, psaJ, ycf3 **, ycf4 | |
| Photosystem II | psbD, psbC, psbZ, psbT, psbH, psbK, psbI, psbJ, psbF, psbE, psbM, psbN, psbL, psbA, psbB | |
| Calvin cycle | rbcL | |
| Cytochrome complex | petN, petA, petL, petG, petB *, petD * | |
| NADH dehydrogenase | ndhB*, ndhI, ndhK, ndhC, ndhF, ndhD, ndhG, ndhE, ndhA *, ndhH, ndhJ | |
| Other genes | Conserved reading frames | infA, ycf68 *, ycf2, accD, cemA, ccsA, matK |
| ID | Repeat Start I | Type | Size (bp) | Repeat Start 2 | Mismatch (bp) | E-Value | Gene | Region |
|---|---|---|---|---|---|---|---|---|
| 1 | 2942 | F | 1175 | 31,558 | 0 | 0 | IGS | SSC; IRB |
| 2 | 2942 | P | 1175 | 163,961 | 0 | 0 | IGS | SSC; IRA |
| 3 | 120,442 | F | 697 | 143,897 | 0 | 0 | IGS | LSC |
| 4 | 18,319 | P | 385 | 76,067 | 0 | 1.57 × 10−222 | IGS; rpoB | IRB; LSC |
| 5 | 76,067 | F | 385 | 177,990 | 0 | 1.57 × 10−222 | rpoB; IGS | LSC; IRA |
| 6 | 46,192 | P | 235 | 147,126 | 0 | 3.2 × 10−132 | IGS | IRB; LSC |
| 7 | 147,126 | F | 235 | 150,267 | 0 | 3.2 × 10−132 | IGS | LSC; IRA |
| 8 | 69,193 | F | 216 | 69,337 | 0 | 8.8 × 10−121 | rpoC2 | LSC |
| 9 | 65,231 | F | 209 | 122,316 | 0 | 1.44 × 10−116 | IGS | LSC |
| 10 | 15,879 | P | 206 | 20,605 | 0 | 9.22 × 10−115 | IGS | IRB |
| 11 | 15,879 | F | 206 | 175,883 | 0 | 9.22 × 10−115 | IGS | IRB; IRA |
| 12 | 20,605 | F | 206 | 180,609 | 0 | 9.22 × 10−115 | IGS | IRB; IRA |
| 13 | 175,883 | P | 206 | 180,609 | 0 | 9.22 × 10−115 | IGS | IRB |
| 14 | 6115 | F | 135 | 6246 | 0 | 5.14 × 10−72 | IGS | SSC |
| 15 | 143,213 | F | 117 | 144,757 | 0 | 3.53 × 10−61 | IGS | LSC |
| 16 | 66,316 | F | 116 | 146,776 | 0 | 1.41 × 10−60 | infA; IGS | LSC |
| 17 | 69,266 | F | 106 | 69,746 | 0 | 1.48 × 10−54 | rpoC2 | LSC |
| 18 | 69,410 | F | 106 | 69,746 | 0 | 1.48 × 10−54 | rpoC2 | LSC |
| 19 | 41,590 | F | 95 | 122,865 | 0 | 6.22 × 10−48 | IGS | IRB; LSC |
| 20 | 122,865 | P | 95 | 155,009 | 0 | 6.22 × 10−48 | IGS | LSC; IRA |
| 21 | 64,509 | F | 92 | 142,999 | 0 | 3.98 × 10−46 | IGS | LSC |
| 22 | 44,316 | F | 89 | 47,901 | 0 | 2.55 × 10−44 | IGS | IRB; LSC |
| 23 | 47,901 | P | 89 | 152,289 | 0 | 2.55 × 10−44 | IGS | LSC; IRA |
| 24 | 69,673 | F | 89 | 69,721 | 0 | 2.55 × 10−44 | rpoC2 | LSC |
| 25 | 143,776 | F | 89 | 145,353 | 0 | 2.55 × 10−44 | IGS | LSC |
| 26 | 69,529 | F | 88 | 69,577 | 0 | 1.02 × 10−43 | rpoC2 | LSC |
| 27 | 20,178 | P | 81 | 58,390 | 0 | 1.67 × 10−39 | IGS | IRB; LSC |
| 28 | 29,232 | F | 81 | 29,406 | 0 | 1.67 × 10−39 | IGS | IRB |
| 29 | 29,232 | P | 81 | 167,207 | 0 | 1.67 × 10−39 | IGS | IRB; IRA |
| 30 | 29,406 | P | 81 | 167,381 | 0 | 1.67 × 10−39 | IGS | IRB; IRA |
| 31 | 58,390 | F | 81 | 176,435 | 0 | 1.67 × 10−39 | IGS | LSC; IRA |
| 32 | 167,207 | F | 81 | 167,381 | 0 | 1.67 × 10−39 | IGS | IRA |
| 33 | 143,058 | F | 79 | 145,560 | 0 | 2.67 × 10−38 | IGS | LSC |
| 34 | 69,193 | F | 72 | 69,481 | 0 | 4.38 × 10−34 | rpoC2 | LSC |
| 35 | 66,236 | F | 69 | 146,696 | 0 | 2.8 × 10−32 | infA; IGS | LSC |
| 36 | 16,930 | P | 68 | 118,533 | 0 | 1.12 × 10−31 | IGS; rpl33 | IRB; LSC |
| 37 | 118,533 | F | 68 | 179,696 | 0 | 1.12 × 10−31 | rpl33; IGS | LSC; IRA |
| 38 | 145,380 | F | 68 | 146,273 | 0 | 1.12 × 10−31 | IGS | LSC |
| 39 | 41,417 | F | 67 | 122,682 | 0 | 4.48 × 10−31 | IGS | IRB; LSC |
| 40 | 65,220 | F | 67 | 142,805 | 0 | 4.48 × 10−31 | IGS | LSC |
| 41 | 122,682 | P | 67 | 155,210 | 0 | 4.48 × 10−31 | IGS | LSC; IRA |
| 42 | 19,762 | P | 66 | 29,760 | 0 | 1.79 × 10−30 | IGS | IRB |
| 43 | 19,762 | F | 66 | 166,868 | 0 | 1.79 × 10−30 | IGS | IRB; IRA |
| 44 | 29,760 | F | 66 | 176,866 | 0 | 1.79 × 10−30 | IGS | IRB; IRA |
| 45 | 53,694 | F | 66 | 143,045 | 0 | 1.79 × 10−30 | IGS | LSC |
| 46 | 166,868 | P | 66 | 176,866 | 0 | 1.79 × 10−30 | IGS | IRA |
| 47 | 64,721 | F | 64 | 143,253 | 0 | 2.87 × 10−29 | IGS | LSC |
| 48 | 64,721 | F | 64 | 144,797 | 0 | 2.87 × 10−29 | IGS | LSC |
| 49 | 69,266 | F | 64 | 69,698 | 0 | 2.87 × 10−29 | rpoC2 | LSC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, W.; Guo, D.; Xing, G.; Yang, C.; Zhang, Y.; Yang, J.; Niu, L.; Zhong, X.; Zhao, Q.; Cui, Y.; et al. Complete Chloroplast Genome Sequence and Comparative and Phylogenetic Analyses of the Cultivated Cyperus esculentus. Diversity 2021, 13, 405. https://doi.org/10.3390/d13090405
Ren W, Guo D, Xing G, Yang C, Zhang Y, Yang J, Niu L, Zhong X, Zhao Q, Cui Y, et al. Complete Chloroplast Genome Sequence and Comparative and Phylogenetic Analyses of the Cultivated Cyperus esculentus. Diversity. 2021; 13(9):405. https://doi.org/10.3390/d13090405
Chicago/Turabian StyleRen, Wei, Dongquan Guo, Guojie Xing, Chunming Yang, Yuanyu Zhang, Jing Yang, Lu Niu, Xiaofang Zhong, Qianqian Zhao, Yang Cui, and et al. 2021. "Complete Chloroplast Genome Sequence and Comparative and Phylogenetic Analyses of the Cultivated Cyperus esculentus" Diversity 13, no. 9: 405. https://doi.org/10.3390/d13090405
APA StyleRen, W., Guo, D., Xing, G., Yang, C., Zhang, Y., Yang, J., Niu, L., Zhong, X., Zhao, Q., Cui, Y., Zhao, Y., & Yang, X. (2021). Complete Chloroplast Genome Sequence and Comparative and Phylogenetic Analyses of the Cultivated Cyperus esculentus. Diversity, 13(9), 405. https://doi.org/10.3390/d13090405






