How Does Circadian Rhythm Shape Host-Parasite Associations? A Comparative Study on Infection Patterns in Diurnal and Nocturnal Raptors
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Parasite Identification
2.3. Phylogenetic Analysis
2.4. Statistical Analysis
3. Results
3.1. Differences in Haemosporidian Prevalence between Diurnal and Nocturnal Raptors
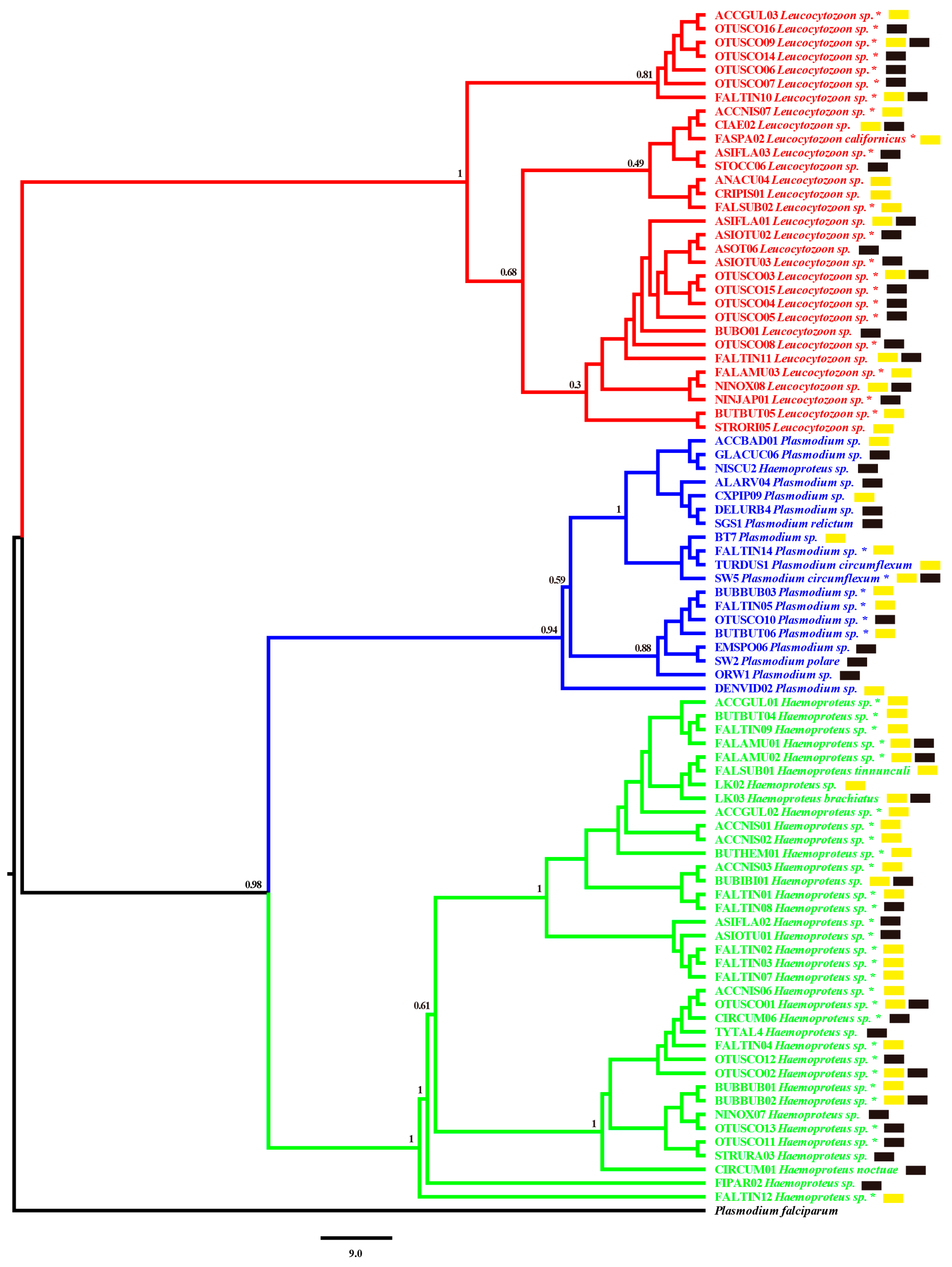
3.2. Phylogenetic Relationship of Haemosporidian Parasites in Diurnal and Nocturnal Raptors
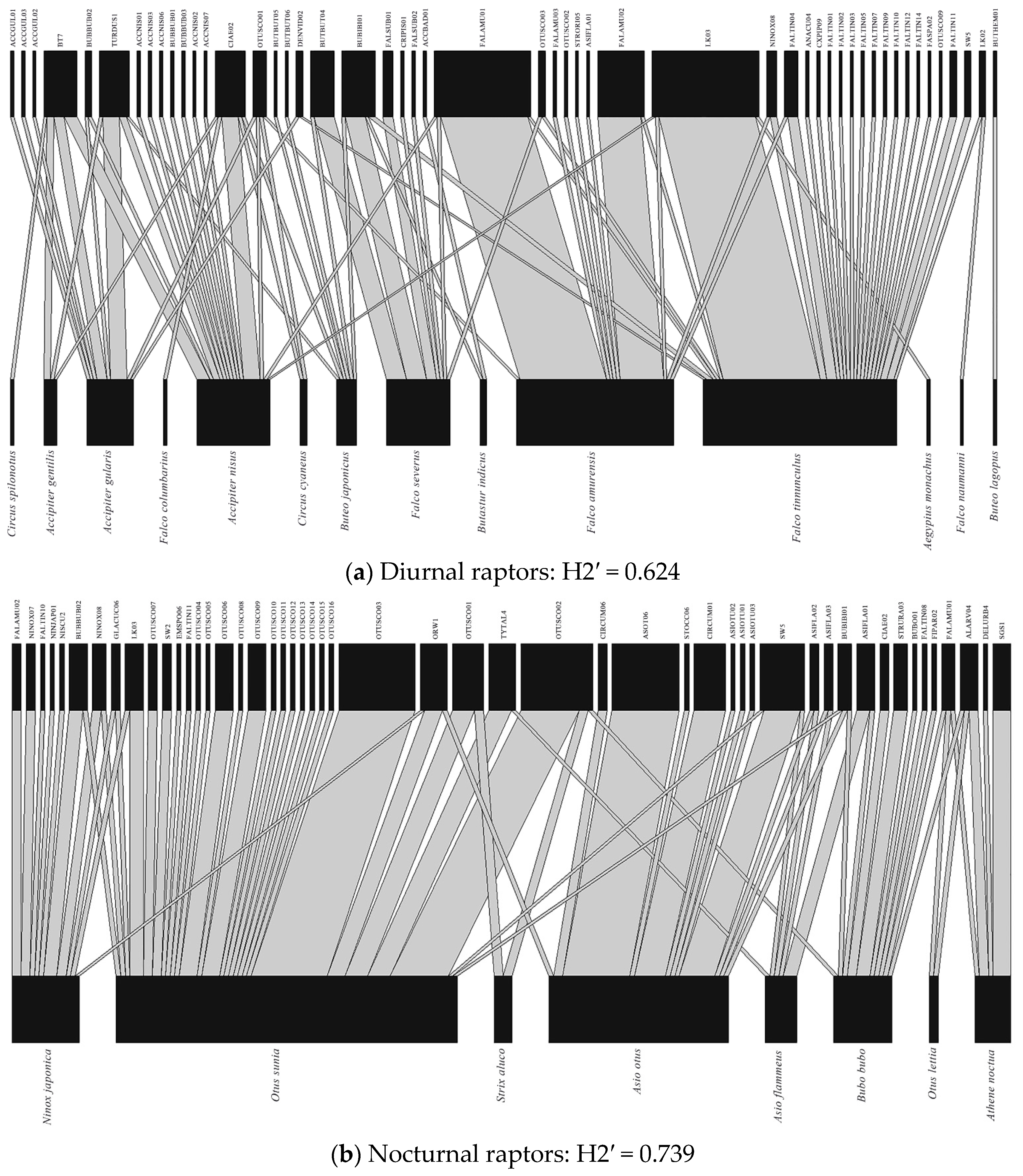
3.3. Avian Haemosporidian Network Structures in Diurnal and Nocturnal Raptors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Merino, S.; Moreno, J.; Sanz, J.J.; Arriero, E. Are avian blood parasites pathogenic in the wild? A medication experiment in blue tits (Parus caeruleus). Proc. R. Soc. B Biol. Sci. 2000, 267, 2507–2510. [Google Scholar] [CrossRef]
- Marzal, A.; de Lope, F.; Navarro, C.; Møller, A.P. Malarial parasites decrease reproductive success: An experimental study in a passerine bird. Oecologia 2005, 142, 541–545. [Google Scholar] [CrossRef]
- Knowles, S.C.L.; Wood, M.J.; Alves, R.; Wilkin, T.A.; Bensch, S.; Sheldon, B.C. Molecular epidemiology of malaria prevalence and parasitaemia in a wild bird population. Mol. Ecol. 2010, 20, 1062–1067. [Google Scholar] [CrossRef]
- Asghar, M.; Hasselquist, D.; Bensch, S. Are chronic avian haemosporidian infections costly in wild birds? J. Avian Biol. 2011, 42, 530–537. [Google Scholar] [CrossRef]
- Poulin, R. Evolutionary Ecology of Parasites; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Gardner, S.L.; Galbreath, K.E.; Herczeg, D.; Mejía-Madrid, H.H.; Rácz, S.E.; Dursahinhan, A.T. Finding them before they find us: Informatics, parasites, and environments in accelerating climate change. Comp. Parasitol. 2014, 81, 155–164. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; de Roode, J.C.; Fenton, A. Why infectious disease research needs community ecology. Science 2015, 349, 1259504. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Rivero, A.; Gandon, S. Evolutionary ecology of avian malaria: Past to present. Trends Parasitol. 2018, 34, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Bensch, S.; Åkesson, S. Temporal and spatial variation of Hematozoans in Scandinavian willow warblers. J. Parasitol. 2003, 89, 379–381. [Google Scholar] [CrossRef]
- Pérez-Tris, J.; Bensch, S. Dispersal increases local transmission of avian malarial parasites. Ecol. Lett. 2005, 8, 838–845. [Google Scholar] [CrossRef]
- Clark, N.J.; Clegg, S.M.; Lima, M.R. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int. J. Parasitol. 2014, 44, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Fecchio, A.; Wells, K.; Bell, J.A.; Tkach, V.V.; Lutz, H.L.; Weckstein, J.D.; Clegg, S.M.; Clark, N.J. Climate variation influences host specificity in avian malaria parasites. Ecol. Lett. 2019, 22, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Scheuerlein, A.; Ricklefs, R.E. Prevalence of blood parasites in European passeriform birds. Proc. R. Soc. B Biol. Sci. 2004, 271, 1363–1370. [Google Scholar] [CrossRef]
- Ellis, V.A.; Fecchio, A.; Ricklefs, R.E. Haemosporidian parasites of Neotropical birds: Causes and consequences of infection. Auk Ornithol. Adv. 2020, 137, 1–23. [Google Scholar] [CrossRef]
- Scordato, S.C.; Kardish, M.R. Prevalence and beta diversity in avian malaria communities: Host species is better predictor than geography. J. Anim. Ecol. 2014, 6, 1387–1397. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.S.; Piersma, T.; Thieltges, D.W. Micro- and macroparasite species richness in birds: The role of host life history and ecology. J. Anim. Ecol. 2019, 88, 1226–1239. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Alarcon, D.; Palinauskas, V.; Schaefer, H.M. Diptera vectors of avian Haemosporidian parasites: Untangling parasite life cycles and their taxonomy. Biol. Rev. Camb. Philos. Soc. 2012, 87, 928–964. [Google Scholar] [CrossRef]
- Fallon, S.M.; Bermingham, E.; Ricklefs, R.E. Island and taxon effects in parasitism revisited: Avian malaria in the Lesser Antilles. Evolution 2003, 57, 606–615. [Google Scholar] [CrossRef]
- Fallon, S.M.; Bermingham, E.; Ricklefs, R.E. Host specialization and geographic localization of avian malarial parasites: A regional analysis in the Lesser Antilles. Am. Naturalist. 2005, 165, 466–480. [Google Scholar] [CrossRef]
- Njabo, K.Y.; Cornel, A.J.; Bonneaud, C.; Toffelmier, E.; Sehgal, R.N.M.; Valkiūnas, G.; Russell, A.F.; Smith, T.B. Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol. Ecol. 2011, 20, 1049–1061. [Google Scholar] [CrossRef]
- Gupta, P.; Vishnudas, C.K.; Robin, V.V.; Dharmarajan, G. Host phylogeny matters: Examining sources of variation in infection risk by blood parasites across a tropical montane bird community in India. Parasit Vectors 2020, 13, 1–13. [Google Scholar] [CrossRef]
- Streicker, D.G.; Turmelle, A.S.; Vonhof, M.J.; Kuzmin, I.V.; McCracken, G.F.; Rupprecht, C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 2010, 329, 676–679. [Google Scholar] [CrossRef]
- Eggert, L.S.; Terwilliger, L.A.; Woodworth, B.L.; Hart, P.J.; Palmer, D.; Fleischer, R.C. Genetic structure along an elevational gradient in Hawaiian honeycreepers reveals contrasting evolutionary responses to avian malaria. BMC Evol. Biol. 2008, 8, 315. [Google Scholar] [CrossRef]
- Marzal, A.; Ricklefs, R.E.; Valkiūnas, G.; Albayrak, T.; Arriero, E.; Bonneaud, C.; Bensch, S. Diversity, loss, and gain of malaria parasites in a globally invasive bird. PLoS ONE 2011, 6, e21905. [Google Scholar] [CrossRef] [PubMed]
- Ewen, J.G.; Bensch, S.; Blackburn, T.M.; Bonneaud, C.; Brown, R.; Cassey, P.; Clarke, R.H.; Pérez-Tris, J. Establishment of exotic parasites: The origins and characteristics of an avian malaria community in an isolated island avifauna. Ecol. Lett. 2012, 15, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Bodawatta, K.H.; Synek, P.; Bos, N.; Garcia-del-Rey, E.; Koane, B.; Marki, P.Z.; Albrecht, T.; Lifjeld, J.; Poulsen, M.; Munclinger, P.; et al. Spatiotemporal patterns of avian host-parasite interactions in the face of biogeographical range expansions. Mol. Ecol. 2020, 29, 2431–2448. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G. Avian Malaria Parasites and Other Haemosporidia; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Fritz, M.L.; Walker, E.D.; Yunker, A.J.; Dworkin, I. Daily blood feeding rhythms of laboratory-reared North American Culex pipiens. J. Circadian Rhythm. 2014, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Grillet, M.-E.; Villamizar, N.J.; Cortez, J.; Frontado, H.L.; Escalona, M.; Vivas-Martínez, S.; Basáñez, M.-G. Diurnal biting periodicity of parous Simulium (Diptera: Simuliidae) vectors in the onchocerciasis Amazonian focus. Acta Trop. 2005, 94, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Lehane, M. The Biology of Blood-Sucking in Insects, 2nd ed.; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Bennett, G.F. On some ornithophilic blood-sucking diptera in Algonquin Park, Ontario, Canada. Can. J. Zool. 1960, 38, 377–389. [Google Scholar] [CrossRef]
- Marquardt, W.C.; Demaree, R.S.; Grieve, R.B. Parasitology and Vector Biology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569–573. [Google Scholar] [CrossRef]
- Xie, S.L.; Lu, F.; Cao, L.; Zhou, W.Q.; Ouyang, Z.Y. Multi-scale factors influencing the characteristics of avian communities in urban parks across Beijing during the breeding season. Sci. Rep. 2016, 6, 29350. [Google Scholar] [CrossRef]
- Healy, K.; Guillerme, T.; Finlay, S.; Kane, A.; Kelly, S.B.; McClean, D.; Kelly, D.J.; Donohue, L.; Jackson, A.L.; Cooper, N. Ecology and mode-of-life explain lifespan variation in birds and mammals. Proc. R. Soc. B Biol. Sci. 2014, 281, 1–7. [Google Scholar] [CrossRef]
- Hellgren, O.; Waldenström, J.; Bensch, S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J. Parasitol. 2004, 90, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Bensch, S.; Hellgren, O.; Pérez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v1.4.3. 2016. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 17 March 2021).
- Rambaut, A.; Drummond, A. Tracer v1.4. 2007. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 17 March 2021).
- Blüthgen, N.; Menzel, F.; Bluthgen, N. Measuring specialization in species interaction networks. BMC Ecol. 2006, 6, 9. [Google Scholar] [CrossRef]
- Dormann, C.F.; Gruber, B.; Fründ, J. Introducing the bipartite package: Analysing ecological networks. R News 2008, 8, 8–11. [Google Scholar]
- Svensson-Coelho, M.; Ellis, V.A.; Loiselle, B.A.; Blake, J.G.; Ricklefs, R.E. Reciprocal specialization in multihost malaria parasite communities of Birds: A Temperate-Tropical comparison. Am. Nat. 2014, 184, 624–635. [Google Scholar] [CrossRef]
- Wood, M.J.; Cosgrove, C.L.; Wilkin, T.A.; Knowles, S.C.L.; Day, K.P.; Sheldon, B.C. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol. Ecol. 2007, 16, 3263–3273. [Google Scholar] [CrossRef] [PubMed]
- Chasar, A.; Loiseau, C.; Valkiūnas, G.; Iezhova, T.; Smith, T.B.; Sehgal, R.N. Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Mol. Ecol. 2009, 18, 4121–4133. [Google Scholar] [CrossRef] [PubMed]
- Hager, S.B. Human-related threats to urban raptors. J. Raptor Res. 2009, 43, 210–226. [Google Scholar] [CrossRef]
- Gaynor, K.M.; Hojnowski, C.E.; Carter, N.H.; Brashares, J.S. The influence of human disturbance on wildlife nocturnality. Science 2018, 360, 1232–1235. [Google Scholar] [CrossRef]
- Berenger, J.-M.; Parola, P. Arthropod Vectors of Medical Importance. In Infect Dis (Auckl), 4th ed.; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Illera, J.C.; Padilla, L.G.; Lopez, G.; Angel, M. Factors governing the prevalence and richness of avian haemosporidian communities within and between temperate mountains. PLoS ONE 2017, 12, e0184587. [Google Scholar] [CrossRef]
- Harrigan, R.J.; Sedano, R.; Chasar, A.C.; Chaves, J.A.; Nguyen, J.T.; Whitaker, A.; Smith, T.B. New host and lineage diversity of avian haemosporidia in the northern Andes. Evol. Appl. 2014, 7, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Chakarov, N.; Kampen, H.; Wiegmann, A.; Werner, D.; Bensch, S. Blood parasites in vectors reveal a united blackfly community in the upper canopy. Parasites Vectors 2020, 13, 309. [Google Scholar] [CrossRef]
- Ishak, H.D.; Dumbacher, J.P.; Anderson, N.L.; Keane, J.J.; Valkiūnas, G.; Haig, S.M.; Tell, L.A.; Sehgal, R.N.M. Blood parasites in owls with conservation implications for the spotted owl (Strix occidentalis). PLoS ONE 2008, 3, e2304. [Google Scholar] [CrossRef]
- Ortego, J.; Cordero, P.J. PCR-based detection and genotyping of haematozoa (Protozoa) parasitizing eagle owls, Bubo bubo. Parasitol. Res. 2009, 104, 467–470. [Google Scholar] [CrossRef]
- Synek, P.; Popelková, A.; Koubínová, D.; Šťastný, K.; Langrová, I.; Votýpka, J.; Munclinger, P. Haemosporidian infections in the Tengmalm’s owl (Aegolius funereus) and potential insect vectors of their transmission. Parasitol. Res. 2016, 115, 291–298. [Google Scholar] [CrossRef]
- Inumaru, M.; Murata, K.; Sato, Y. Prevalence of avian haemosporidia among injured wild birds in Tokyo and environs, Japan. Int. J. Parasitol. Parasites Wildl. 2017, 6, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Vishnudas, C.K.; Ramakrishnan, U.; Robin, V.V.; Dharmarajan, G. Geographical and host species barriers differentially affect generalist and specialist parasite community structure in a tropical sky-island archipelago. Proc. R. Soc. B Biol. Sci. 2019, 286, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nieberding, C.; Morand, S.; Libois, R.; Michaux, J.R. Parasites and the island syndrome: The colonization of the western Mediterranean islands by Heligmosomoides polygyrus (Dujardin, 1845). J. Biogeogr. 2006, 33, 1212–1222. [Google Scholar] [CrossRef]
- Garamszegi, L.Z. Climate change increases the risk of malaria in birds. Glob. Chang. Biol. 2011, 17, 1751–1759. [Google Scholar] [CrossRef]
- Piersma, T.; van der Velde, M. Dutch House Martins Delichon urbicum gain blood parasite infections over their lifetime, but do not seem to suffer. J. Ornithol. 2012, 153, 907–912. [Google Scholar] [CrossRef]
- Zuk, M.; McKean, K.A. Sex differences in parasite infections: Patterns and processes. Int. J. Parasitol. 1996, 26, 1009–1024. [Google Scholar] [CrossRef]
- Jeffries, M.I.; Miller, R.; Laskowski, M.; Carlisle, J. High prevalence of Leucocytozoon parasites in nestling northern goshawks (Accipiter gentilis) in the northern great basin, USA. J. Raptor Res. 2015, 3, 294–302. [Google Scholar] [CrossRef]
- Dubiec, A.; Podmokła, E.; Gustafsson, L. Intra-individual changes in haemosporidian infections over the nesting period in great tit females. Parasitol. Res. 2017, 116, 2385–2392. [Google Scholar] [CrossRef]
- Shurulinkov, P.; Chakarov, N.; Daskalova, G. Blood parasites, body condition, and wing length in two subspecies of yellow wagtail (Motacilla flava) during migration. Parasitol. Res. 2012, 110, 2043–2051. [Google Scholar] [CrossRef]
| Prevalence | Host Biotic Traits | β | SE | Standardized β | t | p | 95% CI for β | |
|---|---|---|---|---|---|---|---|---|
| Lower | Upper | |||||||
| Total | (Constant) | 173.717 | 70.669 | 2.458 | 0.028 | 22.146 | 325.287 | |
| Activity pattern (ref = nocturnal) | −42.444 | 16.544 | −0.873 | −2.566 | 0.022 | −77.928 | −6.960 | |
| Phylogenetic signal | −256.190 | 172.178 | −0.580 | −1.488 | 0.159 | −625.475 | 113.095 | |
| Weight | −0.003 | 0.003 | −0.268 | −1.149 | 0.270 | −0.009 | 0.003 | |
| Longevity | 0.066 | 0.272 | 0.064 | 0.243 | 0.811 | −0.517 | 0.649 | |
| Plasmodium | (Constant) | −4.632 | 24.272 | −0.191 | 0.851 | −56.689 | 47.426 | |
| Activity pattern (ref = nocturnal) | −0.969 | 5.682 | −0.066 | −0.171 | 0.867 | −13.156 | 11.218 | |
| Phylogenetic signal | 48.4 | 59.135 | 0.361 | 0.818 | 0.427 | −78.433 | 175.232 | |
| Weight | −0.001 | 0.001 | −0.191 | −0.723 | 0.481 | −0.003 | 0.001 | |
| Longevity | −0.037 | 0.093 | −0.120 | −0.400 | 0.695 | −0.238 | 0.163 | |
| Haemoproteus | (Constant) | 131.521 | 45.35 | 2.9 | 0.012 | 34.254 | 228.788 | |
| Activity pattern (ref = nocturnal) | −24.340 | 10.617 | −0.808 | −2.293 | 0.038 | −47.111 | −1.569 | |
| Phylogenetic signal | −271.565 | 110.491 | −0.993 | −2.458 | 0.028 | −508.545 | −34.585 | |
| Weight | −0.002 | 0.002 | −0.332 | −1.374 | 0.191 | −0.006 | 0.001 | |
| Longevity | 0.191 | 0.175 | 0.301 | 1.097 | 0.291 | −0.183 | 0.566 | |
| Leucocytozoon | (Constant) | 66.516 | 37.685 | 1.765 | 0.099 | −14.311 | 147.343 | |
| Activity pattern (ref = nocturnal) | −22.620 | 8.822 | −0.843 | −2.564 | 0.023 | −41.542 | −3.698 | |
| Phylogenetic signal | −59.497 | 91.816 | −0.244 | −0.648 | 0.527 | −256.423 | 137.429 | |
| Weight | 0.001 | 0.001 | −0.017 | −0.077 | 0.939 | −0.003 | 0.003 | |
| Longevity | −0.125 | 0.145 | −0.22 | −0.859 | 0.405 | −0.436 | 0.187 | |
| H2′ | d′ | |||||||
|---|---|---|---|---|---|---|---|---|
| Observed | Null Mean ± SD | p | SES (H2′) | Observed Mean ± SD | Null Mean ± SD | p | SES (d’) | |
| Diurnal raptors | 0.624 | 0.171 ± 0.026 | <0.001 | 17.3 | 0.5295 ± 0.313 | 0.202 ± 0.231 | 0.202 | 1.419 |
| Nocturnal raptors | 0.739 | 0.260 ± 0.323 | <0.001 | 14.9 | 0.306 ± 0.257 | 0.170 ± 0.176 | 0.385 | 0.774 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, K.; Zhou, B.; Yang, L.-X.; Dong, L.; Huang, X.; Deng, W.-H. How Does Circadian Rhythm Shape Host-Parasite Associations? A Comparative Study on Infection Patterns in Diurnal and Nocturnal Raptors. Diversity 2021, 13, 338. https://doi.org/10.3390/d13080338
Gao K, Zhou B, Yang L-X, Dong L, Huang X, Deng W-H. How Does Circadian Rhythm Shape Host-Parasite Associations? A Comparative Study on Infection Patterns in Diurnal and Nocturnal Raptors. Diversity. 2021; 13(8):338. https://doi.org/10.3390/d13080338
Chicago/Turabian StyleGao, Kai, Bing Zhou, Li-Xing Yang, Lu Dong, Xi Huang, and Wen-Hong Deng. 2021. "How Does Circadian Rhythm Shape Host-Parasite Associations? A Comparative Study on Infection Patterns in Diurnal and Nocturnal Raptors" Diversity 13, no. 8: 338. https://doi.org/10.3390/d13080338
APA StyleGao, K., Zhou, B., Yang, L.-X., Dong, L., Huang, X., & Deng, W.-H. (2021). How Does Circadian Rhythm Shape Host-Parasite Associations? A Comparative Study on Infection Patterns in Diurnal and Nocturnal Raptors. Diversity, 13(8), 338. https://doi.org/10.3390/d13080338







