Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genomic Analysis and Statistaical Analysis of Molecular Data
2.3. Morpho-Physiological Evaluation
2.4. Statistical Analysis of Morpho-Physiological Data
3. Results
3.1. Phylogenetical Allocation of Uncertain Accessions by Molecular Markers
3.2. Morpho-Physiological Differences between TauL1 and TauL2
3.3. Morpho-Physiological Variation between ssp. tauschii Belonging to TauL1 and TauL2
3.4. Morpho-Physiological Variation between ssp. tauschii and ssp. strangulata
3.5. Morpho-Physiological Variation of Accessions in TauL3
4. Discussion
4.1. Geographical Clines of Morphological Variation in Subspecies and Lineage Classification
4.2. Potential for Adaptive Convergence in Ae. tauschii Evolution
4.3. Implications of Ae. tauschii Diversity in Wheat Breeding
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, X.G.; Wu, B.H.; Yan, Z.H.; Dai, S.F.; Zhang, L.Q.; Liu, D.C.; Zheng, Y.L. Characteristics and polymorphism of NAM gene from Aegilops section sitopsis species. Afr. J. Agric. Res. 2012, 7, 5252–5258. [Google Scholar] [CrossRef]
- Kimber, G.; Zhao, Y.H. The D genome of the Triticeae. Can. J. Genet. Cytol. 1983, 25, 581–589. [Google Scholar] [CrossRef]
- Kellogg, E.A.; Appels, R.; Mason-Gamer, R.J. When Genes Tell Different Stories: The Diploid Genera of Triticeae (Gramineae). Syst. Bot. 1996, 21, 321. [Google Scholar] [CrossRef]
- Petersen, G.; Seberg, O.; Yde, M.; Berthelsen, K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 2006, 39, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Alnaddaf, L.M.; Moualla, M.Y.; Haider, N. Resolving genetic relationships among Aegilops L. and Triticum L. species using analysis of chloroplast DNA by cleaved amplified polymorphic sequence (CAPS). Asian J. Agric. Sci. 2012, 4, 270–279. [Google Scholar]
- Hammer, K. Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze 1980, 28, 33–180. [Google Scholar] [CrossRef]
- Kilian, B.; Mammen, K.; Millet, E.; Sharma, R.; Graner, A.; Salamini, F.; Hammer, K.; Özkan, H. Aegilops. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–76. [Google Scholar]
- Van Slageren, M.W. Wild Wheats: A monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae); Agricultural University: Wageningen, The Netherlands, 1994. [Google Scholar]
- Čerepanov, S.K. Vascular Plants of Russia and Adjacent Countries; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Feldman, M. Origin of cultivated wheat. In The World Wheat Book: A history of Wheat Breeding; Bonjean, A.P., Angus, W.J., Eds.; Lavoisier Publishing: Paris, France, 2001; pp. 3–56. [Google Scholar]
- McFadden, E.S.; Sears, E.R. The artificial synthesis of Triticum spelta. Proc. Records Genet. Soc. Am. 1944, 13, 26–27. [Google Scholar]
- Kihara, H. Discovery of the DD-analyzer, one of the ancestors of Triticum vulgare. Agric. Hortic. 1944, 19, 13–14. [Google Scholar]
- Eig, A. Monographisch-kritische Übersicht der Gattung Aegilops. Rep. Spec. Nov. Reg. Veget. Berh. 1929, 55, 1–228. [Google Scholar]
- Matsuoka, Y.; Nishioka, E.; Kawahara, T.; Takumi, S. Genealogical analysis of subspecies divergence and spikelet-shape diversification in central Eurasian wild wheat Aegilops tauschii Coss. Plant Syst. Evol. 2009, 279, 233–244. [Google Scholar] [CrossRef]
- Dvorak, J.; Luo, M.C.; Yang, Z.L.; Zhang, H.B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar] [CrossRef]
- Gill, K.S.; Lubbers, E.L.; Gill, B.S.; Raupp, W.J.; Cox, T.S. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D genome of bread wheat (AABBDD). Genome 1991, 34, 362–374. [Google Scholar] [CrossRef]
- Pestsova, E.; Korzun, V.; Goncharov, N.P.; Hammer, K.; Ganal, M.W.; Röder, M.S. Microsatellite analysis of Aegilops tauschii germplasm. Theor. Appl. Genet. 2000, 101, 100–106. [Google Scholar] [CrossRef]
- Lelley, T.; Stachel, M.; Grausgruber, H.; Vollmann, J. Analysis of relationships between Aegilops tauschii and the D genome of wheat utilizing microsatellites. Genome 2000, 43, 661–668. [Google Scholar] [CrossRef]
- Saeidi, H.; Rahiminejad, M.R.; Vallian, S.; Heslop-Harrison, J.S. Biodiversity of diploid D-genome Aegilops tauschii Coss. in Iran measured using microsatellites. Genet. Resour. Crop Evol. 2006, 53, 1477–1484. [Google Scholar] [CrossRef]
- Dudnikov, A.J.; Kawahara, T. Aegilops tauschii: Genetic variation in Iran. Genet. Resour. Crop Evol. 2006, 53, 579–586. [Google Scholar] [CrossRef]
- Okuno, A.; Tamemoto, H.; Tobe, K.; Ueki, K.; Mori, Y.; Iwamoto, K.; Umesono, K.; Akanuma, Y.; Fujiwara, T.; Horikoshi, H.; et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Invest. 1998, 101, 1354–1361. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Mori, N.; Kawahara, T. Genealogical use of chloroplast DNA variation for intraspecific studies of Aegilops tauschii Coss. Theor. Appl. Genet. 2005, 111, 265–271. [Google Scholar] [CrossRef]
- Mizuno, N.; Yamasaki, M.; Matsuoka, Y.; Kawahara, T.; Takumi, S. Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: Implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 2010, 19, 999–1013. [Google Scholar] [CrossRef]
- Naghavi, M.R.; Mardi, M. Characterization of genetic variation among accessions of Aegilops tauschii. Asia Pac. J. Mol. Biol. Biotechnol. 2010, 18, 91–94. [Google Scholar]
- Sohail, Q.; Shehzad, T.; Kilian, A.; Eltayeb, A.E.; Tanaka, H.; Tsujimoto, H. Development of diversity array technology (DArT) markers for assessment of population structure and diversity in Aegilops tauschii. Breed. Sci. 2012, 62, 38–45. [Google Scholar] [CrossRef]
- Arora, S.; Singh, N.; Kaur, S.; Bains, N.S.; Uauy, C.; Poland, J.; Chhuneja, P. Genome-wide association study of grain architecture in wild wheat Aegilops tauschii. Front. Plant Sci. 2017, 8, 886. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Nishijima, R.; Okamoto, Y.; Hatano, H.; Takumi, S. Quantitative trait locus analysis for spikelet shape-related traits in wild wheat progenitor Aegilops tauschii: Implications for intraspecific diversification and subspecies differentiation. PLoS ONE 2017, 12, e0173210. [Google Scholar] [CrossRef]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, M.C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Wang, N.; Sakuma, S.; Pourkheirandish, M.; Koba, T.; Komatsuda, T. Variation in the wheat AP2 homoeologs, the genes underlying lodicule development. Breed. Sci. 2013, 63, 255–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naghavi, M.R.; Aghaei, M.J.; Taleei, A.R.; Omidi, M.; Mozafari, J.; Hassani, M.E. Genetic diversity of the D-genome in T. aestivum and Aegilops species using SSR markers. Genet. Resour. Crop Evol. 2009, 56, 499–506. [Google Scholar] [CrossRef]
- Jaaska, V. NADP-dependent aromatic alcohol dehydrogenase in polyploid wheats and their diploid relatives. On the origin and phylogeny of polyploid wheats. Theor. Appl. Genet. 1978, 53, 209–217. [Google Scholar] [CrossRef]
- Jaaska, V. Aspartate aminotransferase and alcohol dehydrogenase isoenzymes: Intraspecific differentiation in Aegilops tauschii and the origin of the D genome polyploids in the wheat group. Plant Syst. Evol. 1981, 137, 259–273. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef]
- Londo, J.P.; Chiang, Y.C.; Hung, K.H.; Chiang, T.Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar] [CrossRef]
- Morrell, P.L.; Clegg, M.T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 2007, 104, 3289–3294. [Google Scholar] [CrossRef]
- Saisho, D.; Purugganan, M.D. Molecular phylogeography of domesticated barley traces expansion of agriculture in the old world. Genetics 2007, 177, 1765–1776. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, J.G.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Williams, B.L.; Selegue, J.E.; Carroll, S.B. Drosophila pigmentation evolution: Divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. USA 2003, 100, 1808–1813. [Google Scholar] [CrossRef]
- Pascoal, S.; Cezard, T.; Eik-Nes, A.; Gharbi, K.; Majewska, J.; Payne, E.; Ritchie, M.G.; Zuk, M.; Bailey, N.W. Rapid convergent evolution in wild crickets. Curr. Biol. 2014, 24, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Aardema, M.L.; Medina, E.M.; Schumer, M.; Andolfatto, P. Parallel molecular evolution in an herbivore community. Science 2012, 337, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Orgogozo, V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 2013, 67, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Ralph, P.L.; Coop, G. Convergent evolution during local adaptation to patchy landscapes. PLoS Genet. 2015, 11, 1–31. [Google Scholar] [CrossRef]
- Kishii, M. An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef]
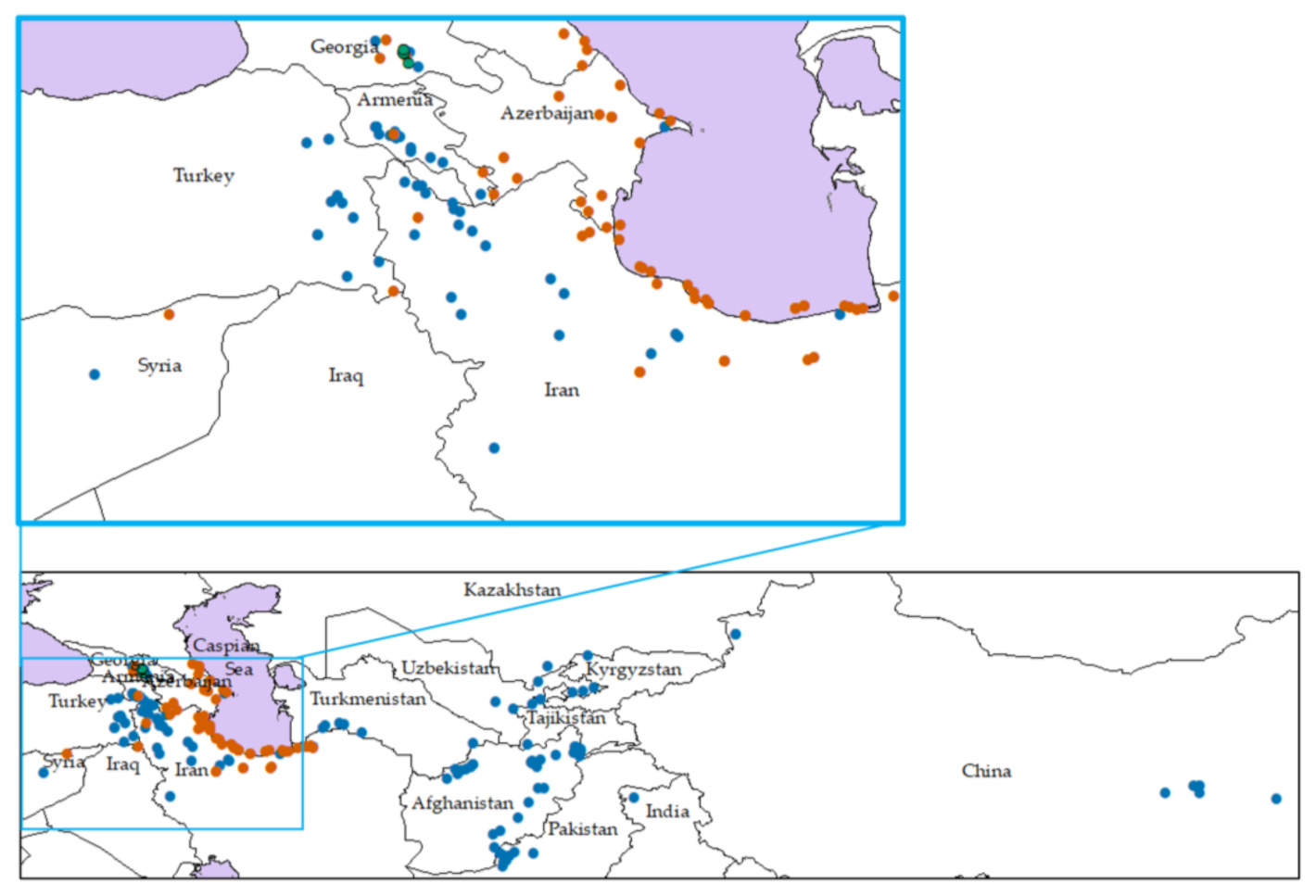
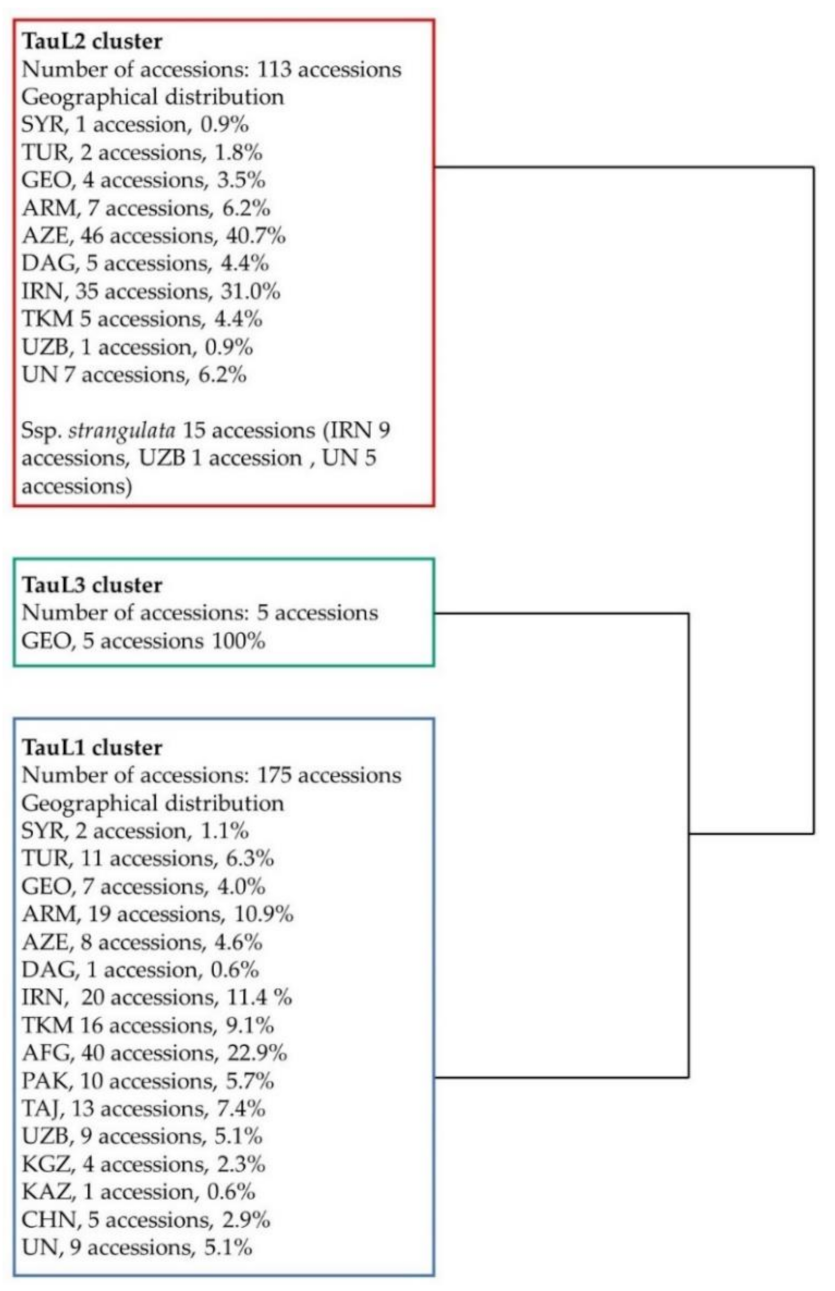
| Origin | TauL1 | TauL2 | TauL3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Syria | AE 1069 | IG 47259 | IG 46623 | |||||||||
| Turkey | KU-2131 | KU-2132 | KU-2133 | KU-2136 | KU-2137 | PI 486267 | PI 486274 | |||||
| KU-2138 | KU-2140 | KU-2141 | PI 486270 | PI 486277 | ||||||||
| PI 554319 | ||||||||||||
| Georgia | AE 254 | AE 461 | GE12-28-O-2 | KU-20-2 | KU-2826 | AE 1037 | GE12-14-O-1 | KU-2827 | KU-2835B | AE 929 | AE 454 | |
| KU-2828 | KU-2834 | KU-2829A | KU-2832 | |||||||||
| AE 929a | ||||||||||||
| Armenia | AE 245 | AE 253 | AE 476 | AE 721 | CGN 10734 | AE 229 | AE 231 | AE 940 | AE 941 | IG 126991 | ||
| IG 126273 | IG 126280 | IG 126293 | IG 126353 | IG 48748 | IG 127015 | KU-2811 | ||||||
| IG 48758 | KU-2809 | KU-2810 | KU-2814 | KU-2816 | ||||||||
| KU-2821 | KU-2822A | KU-2823 | KU-2824 | |||||||||
| Azerbaijan | AE 143 | AE 220 | AE 251 | AE 723 | AE 724 | AE 144 | AE 191 | AE 194 | AE 195 | AE 197 | ||
| AE 725 | AE 1055 | IG 47196 | AE 198 | AE 199 | AE 200 | AE 202 | AE 203 | |||||
| AE 204 | AE 205 | AE 206 | AE 207 | AE 210 | ||||||||
| AE 211 | AE 216 | AE 217 | AE 218 | AE 219 | ||||||||
| AE 221 | AE 222 | AE 223 | AE 224 | AE 226 | ||||||||
| AE 230 | AE 255 | AE 260 | AE 261 | AE 262 | ||||||||
| AE 263 | AE 264 | AE 267 | AE 270 | AE 272 | ||||||||
| AE 273 | AK 228 | IG 47182 | IG 47186 | IG 47188 | ||||||||
| IG 47193 | IG 47199 | IG 47202 | IG 47203 | KU-2801 | ||||||||
| KU-2806 | ||||||||||||
| Dagestan | AE 234 | AE 498 | IG 120863 | IG 120866 | IG 48274 | KU-20-1 | ||||||
| Iran | AE 183 | AE 184 | AE 541 | IG 49095 | KU-2082 | AE 525 * | AE 526 | KU-20-8 | KU-20-9 * | KU-20-10 | ||
| KU-2109 | KU-2113 | KU-2115 | KU-2116 | KU-2120 | KU-2069 | KU-2075 * | KU-2079 * | KU-2080 * | KU-2083 | |||
| KU-2121 | KU-2142 | KU-2143 | KU-2144 | KU-2148 | KU-2086 | KU-2088 * | KU-2090 * | KU-2092 * | KU-2093 * | |||
| KU-2152 | KU-2153 | KU-2154 | KU-2157 | KU-2158 | KU-2096 | KU-2097 | KU-2098 | KU-2100 | KU-2101 | |||
| KU-2102 | KU-2103 | KU-2104 | KU-2105 | KU-2106 | ||||||||
| KU-2110 | KU-2111 | KU-2112 | KU-2118 | KU-2124 | ||||||||
| KU-2126 | KU-2155 | KU-2156 | KU-2159 | KU-2160 | ||||||||
| Turkmenistan | AE 141 | AE 146 | AE 242 | AE 248 | AE 249 | AE 192 | AE 213 | AE 250 | CGN 10733 | IG 120735 | ||
| AE 291 | AE 398 | AE 472 | AE 473 | AE 499 | ||||||||
| AE 637 | AE 964 | IG 126387 | IG 126489 | IG 48508 | ||||||||
| IG 48518 | ||||||||||||
| Afghanistan | AE 193 | AE 275 | AE 276 | AE 277 | AE 279 | |||||||
| AE 280 | AE 281 | AE 1087 | KU-2010 | KU-2012 | ||||||||
| KU-2016 | KU-2018 | KU-2022 | KU-2025 | KU-2027 | ||||||||
| KU-2035 | KU-2039 | KU-2042 | KU-2043 | KU-2044 | ||||||||
| KU-2050 | KU-2051 | KU-2056 | KU-2059 | KU-2061 | ||||||||
| KU-2063 | KU-2066 | KU-2616 | KU-2617 | KU-2619 | ||||||||
| KU-2621 | KU-2624 | KU-2630 | KU-2632 | KU-2633 | ||||||||
| KU-2635 | KU-2636 | KU-2638 | KU-2639 | PI 476874 | ||||||||
| Pakistan | CGN 10767 | CGN 10768 | CGN 10769 | CGN 10771 | IG 108561 | |||||||
| IG 46663 | IG 46666 | KU-2003 | KU-2006 | KU-2008 | ||||||||
| Tajikistan | AE 189 | AE 233 | AE 647 | AE 817 | AE 858 | |||||||
| AE 955 | AE 956 | AE 1038 | AE 1039 | AE 1040 | ||||||||
| IG 48554 | IG 48559 | IG 48564 | ||||||||||
| Uzbekistan | AE 3 | AE 239 | AE 469 | AE 560 | IG 120736 | AE 692 * | ||||||
| IG 123910 | IG 48539 | IG 48565 | IG 48567 | |||||||||
| Kyrgyzstan | AE 256 | AE 257 | AE 1180 | IG 131606 | ||||||||
| Kazakhstan | AE 1090 | |||||||||||
| China | AT 55 | AT 60 | AT 76 | PI 499262 | PI 508262 | |||||||
| Unknown location | AE 32 | AE 67 | AE 147 | AE 150 | AE 422 | AE 426 * | AE 428 * | AE 429 * | AE 430 * | AE 431 | ||
| AE 427 | AE 433 | AE 432 | AE 434 * | |||||||||
| Trait | Abbreviation (Unit) | Measurement/Definition |
|---|---|---|
| Flag leaf length | FLL (cm) | Measured from three tillers per accession. |
| Flag leaf width | FLW (mm) | Measured from three tillers per accession. |
| Spike length | SPL (cm) | Measured from the middle five spikes after maturity stage. |
| Spike width | SPW (cm) | Measured from the middle of five spikes after maturity stage. |
| Seed number/spike | SN/SP | Counted from five spikes at harvesting. |
| Spike weight | SPWg (g) | Weighed from five spikes (one per tiller) using a sensitive scale. |
| Days to heading | DH | Recorded when the whole spike above the flag leaf fully emerged on the earliest tiller in each plant of each accession. |
| Biomass weight | Bio (g) | Weighed after harvesting and drying of five plants in a glasshouse. |
| Normalized Difference Vegetation Index | NDVI | A vegetative index that compares reflectance in the red and near-infrared regions. Measured during flowering using a handheld optical sensor unit (Green Seeker), NTech Industries, Inc., Ukiah, CA, USA. |
| Canopy temperature | CT (°C) | Measured during flowering using an infrared thermometer AD-5611A. |
| Chlorophyll content | SPAD | Measured at the flowering stage from the middle of the flag leaf of three tillers using a Minolta brand chlorophyll meter (Model SPAD-502; Spectrum Technologies Inc., Plainfield, IL, USA). |
| Trait | TauL1 | TauL2 | p-Value (TauL1 versus TauL2) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | STD | Min | Max | Mean | STD | ||
| FLL | 5.35 | 20.65 | 13.74 | 2.48 | 5.77 | 20.32 | 12.96 | 2.84 | 0.052 |
| FLW | 4.80 | 11.00 | 8.10 | 1.20 | 4.20 | 10.90 | 7.80 | 1.10 | 0.145 |
| SPL | 9.08 | 17.55 | 12.61 | 1.50 | 8.80 | 17.27 | 12.03 | 1.55 | 0.325 |
| SPW | 0.40 | 0.71 | 0.53 | 0.06 | 0.40 | 0.75 | 0.58 | 0.07 | 0.011 |
| SN/SP | 15.82 | 29.67 | 22.00 | 2.32 | 15.42 | 29.93 | 19.51 | 2.05 | 0.081 |
| SPWg | 0.35 | 0.67 | 0.50 | 0.06 | 0.34 | 0.71 | 0.54 | 0.07 | 0.005 |
| DH | 150.78 | 184.03 | 169.19 | 5.78 | 159.77 | 191.45 | 174.39 | 4.04 | 0.000 |
| Bio | 60.53 | 189.78 | 99.24 | 23.61 | 73.90 | 227.09 | 134.50 | 37.11 | 0.000 |
| NDVI | 0.60 | 0.63 | 0.62 | 0.01 | 0.60 | 0.64 | 0.62 | 0.01 | 0.389 |
| CT | 15.11 | 25.14 | 18.34 | 1.91 | 14.49 | 24.50 | 17.91 | 1.84 | 0.303 |
| SPAD | 40.92 | 45.37 | 43.50 | 0.73 | 42.06 | 45.46 | 43.69 | 0.71 | 0.413 |
| Trait | TauL1T | TauL2T | p-Value (TauL1T versus TauL2T) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | STD | Min | Max | Mean | STD | ||
| FLL | 5.35 | 20.65 | 13.74 | 2.48 | 5.77 | 20.32 | 12.78 | 2.89 | 0.040 |
| FLW | 4.80 | 11.00 | 8.10 | 1.20 | 4.20 | 1.90 | 7.80 | 1.20 | 0.239 |
| SPL | 9.08 | 17.55 | 12.61 | 1.50 | 8.80 | 16.75 | 12.19 | 1.41 | 0.271 |
| SPW | 0.40 | 0.71 | 0.53 | 0.06 | 0.40 | 0.72 | 0.57 | 0.07 | 0.145 |
| SN/SP | 15.82 | 29.67 | 22.00 | 2.32 | 16.13 | 29.93 | 19.72 | 2.07 | 0.106 |
| SPWg | 0.35 | 0.67 | 0.50 | 0.06 | 0.37 | 0.67 | 0.53 | 0.06 | 0.091 |
| DH | 150.78 | 184.03 | 169.19 | 5.78 | 159.77 | 191.45 | 174.46 | 4.28 | 0.001 |
| Bio | 60.53 | 189.78 | 99.24 | 23.61 | 73.90 | 227.09 | 135.39 | 37.28 | 0.000 |
| NDVI | 0.60 | 0.63 | 0.62 | 0.01 | 0.60 | 0.64 | 0.62 | 0.01 | 0.327 |
| CT | 15.11 | 25.14 | 18.34 | 1.91 | 14.49 | 24.50 | 17.84 | 1.87 | 0.377 |
| SPAD | 40.92 | 45.37 | 43.50 | 0.73 | 42.31 | 45.46 | 43.67 | 0.69 | 0.278 |
| Trait | Ssp. tauschii | Ssp. strangulata | p-Value (tauschii versus strangulata) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | STD | Min | Max | Mean | STD | ||
| FLL | 5.35 | 20.65 | 13.40 | 2.68 | 11.04 | 17.97 | 14.15 | 2.17 | 0.228 |
| FLW | 4.20 | 11.00 | 8.00 | 1.20 | 6.40 | 9.50 | 7.90 | 0.90 | 0.123 |
| SPL | 8.80 | 17.55 | 12.46 | 1.48 | 8.82 | 17.27 | 11.00 | 1.97 | 0.027 |
| SPW | 0.40 | 0.72 | 0.54 | 0.07 | 0.58 | 0.75 | 0.66 | 0.06 | 0.432 |
| SN/SP | 15.82 | 29.93 | 21.18 | 2.48 | 15.42 | 20.42 | 18.13 | 1.30 | 0.006 |
| SPWg | 0.35 | 0.67 | 0.51 | 0.06 | 0.34 | 0.71 | 0.58 | 0.09 | 0.004 |
| DH | 150.78 | 191.45 | 171.08 | 5.86 | 170.37 | 178.51 | 173.94 | 1.72 | 0.000 |
| Bio | 60.53 | 227.09 | 112.21 | 34.01 | 89.70 | 223.05 | 128.67 | 35.40 | 0.294 |
| NDVI | 0.60 | 0.64 | 0.62 | 0.01 | 0.60 | 0.63 | 0.62 | 0.01 | 0.088 |
| CT | 14.49 | 25.14 | 18.16 | 1.91 | 16.11 | 21.55 | 18.37 | 1.55 | 0.280 |
| SPAD | 40.92 | 45.46 | 43.56 | 0.72 | 42.06 | 44.83 | 43.82 | 0.84 | 0.151 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahjoob, M.M.M.; Chen, T.-S.; Gorafi, Y.S.A.; Yamasaki, Y.; Kamal, N.M.; Abdelrahman, M.; Iwata, H.; Matsuoka, Y.; Tahir, I.S.A.; Tsujimoto, H. Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat. Diversity 2021, 13, 217. https://doi.org/10.3390/d13050217
Mahjoob MMM, Chen T-S, Gorafi YSA, Yamasaki Y, Kamal NM, Abdelrahman M, Iwata H, Matsuoka Y, Tahir ISA, Tsujimoto H. Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat. Diversity. 2021; 13(5):217. https://doi.org/10.3390/d13050217
Chicago/Turabian StyleMahjoob, Mazin Mahjoob Mohamed, Tai-Shen Chen, Yasir Serag Alnor Gorafi, Yuji Yamasaki, Nasrein Mohamed Kamal, Mostafa Abdelrahman, Hiroyoshi Iwata, Yoshihiro Matsuoka, Izzat Sidahmed Ali Tahir, and Hisashi Tsujimoto. 2021. "Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat" Diversity 13, no. 5: 217. https://doi.org/10.3390/d13050217
APA StyleMahjoob, M. M. M., Chen, T.-S., Gorafi, Y. S. A., Yamasaki, Y., Kamal, N. M., Abdelrahman, M., Iwata, H., Matsuoka, Y., Tahir, I. S. A., & Tsujimoto, H. (2021). Traits to Differentiate Lineages and Subspecies of Aegilops tauschii, the D Genome Progenitor Species of Bread Wheat. Diversity, 13(5), 217. https://doi.org/10.3390/d13050217







