Genetic Consequences of Forest Fragmentation in a Widespread Forest Bat (Natalus mexicanus, Chiroptera: Natalidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Mitochondrial DNA
2.3. Microsatellite Loci Amplification
2.4. MtDNA Data Analysis
2.4.1. Genealogical Analysis
2.4.2. Demographic Analysis
2.4.3. Historical Gene Flow
2.5. Microsatellite Data Analysis
2.5.1. Population Structure and Genetic Diversity
2.5.2. Contemporary Migration Rates
3. Results
3.1. Mitochondrial DNA Data Analysis
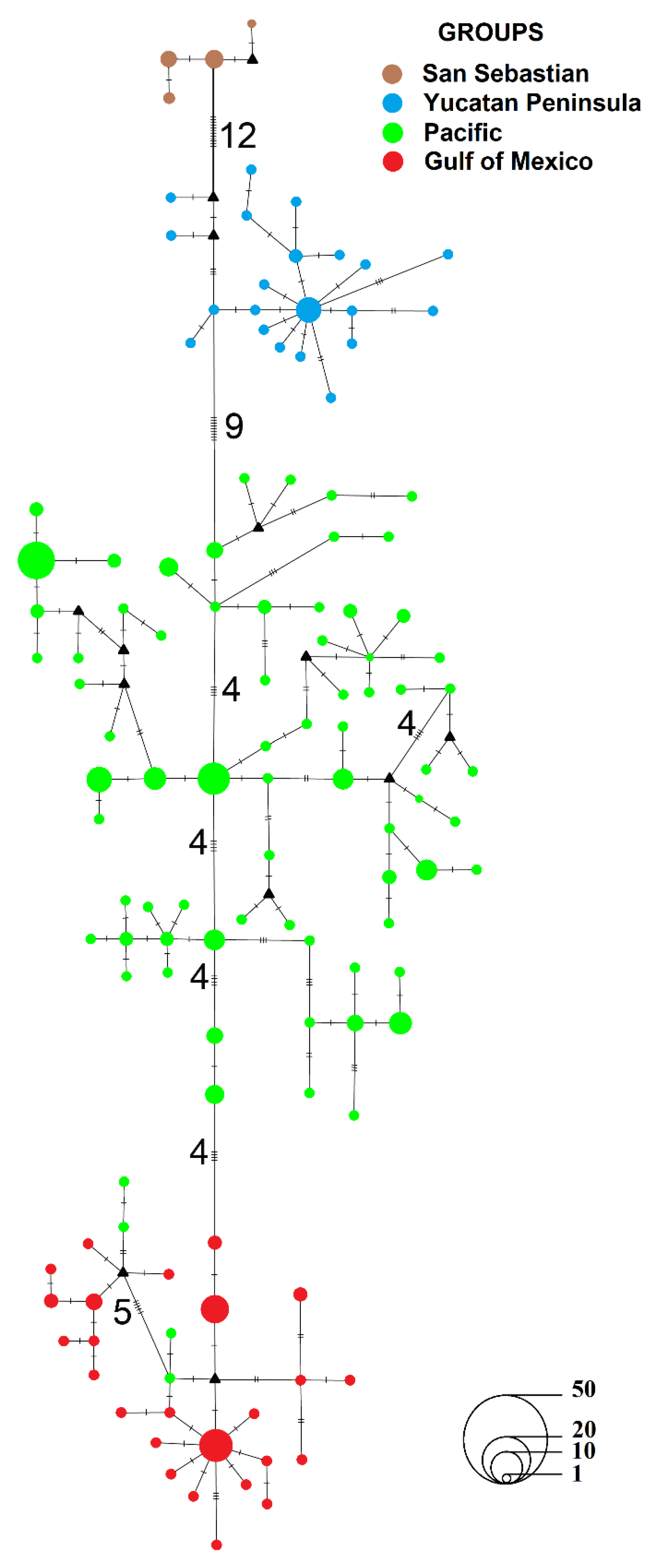
3.1.1. Genealogical Analysis
3.1.2. Population Structure and Genetic Diversity
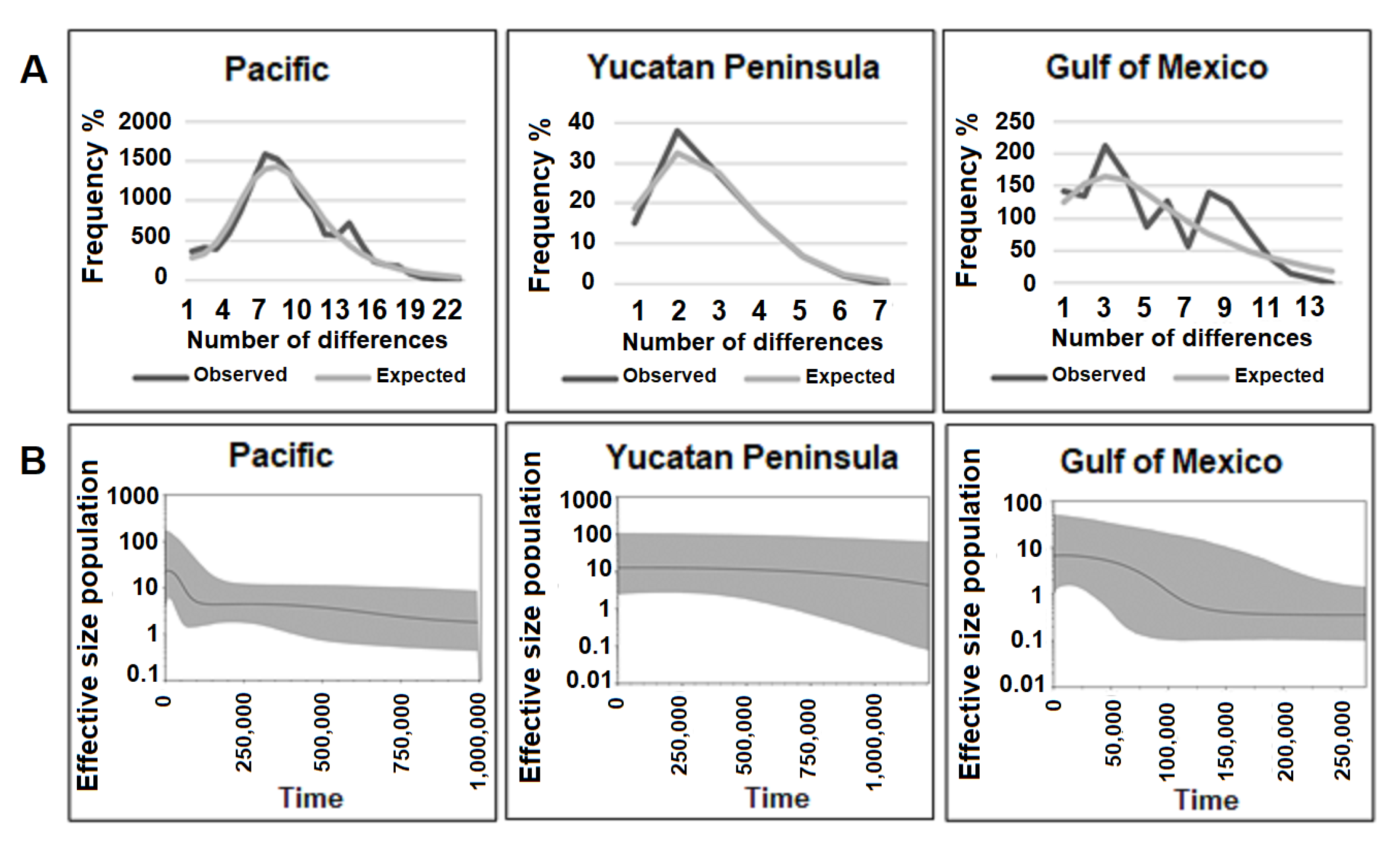
3.1.3. Demographic Analysis
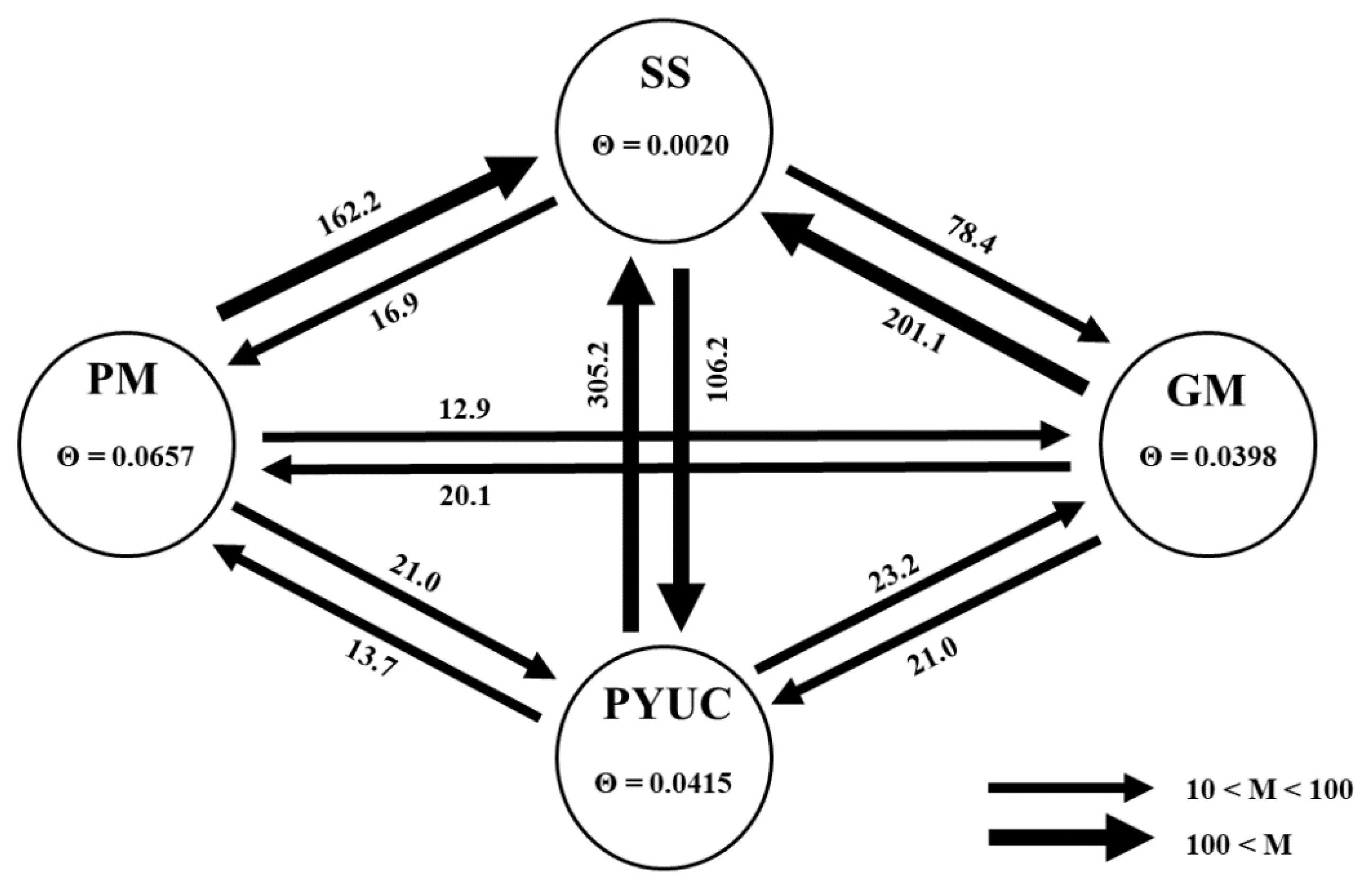
3.1.4. Historical Gene Flow
3.2. Microsatellite Data Analysis
3.2.1. Population Structure and Genetic Diversity
3.2.2. Contemporary Migration Rates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Gaines, M.S.; Diffendorfer, J.E.; Tamarin, R.H.; Whittam, T.S. The Effects of Habitat Fragmentation on the Genetic Structure of Small Mammal Populations. J. Hered. 1997, 88, 294–304. [Google Scholar] [CrossRef]
- Pereira, H.M.; Leadley, P.W.; Proença, V.; Alkemade, R.; Scharlemann, J.P.W.; Fernandez-Manjarrés, J.F.; Araújo, M.B.; Balvanera, P.; Biggs, R.; Cheung, W.W.L.; et al. Scenarios for Global Biodiversity in the 21st century. Science 2010, 330, 1496–1501. [Google Scholar] [CrossRef]
- Meyer, C.F.J.; Struebig, M.J.; Willig, M.R. Responses of Tropical Bats to Habitat Fragmentation, Logging, and Deforestation. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Metzler, J.B., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 63–103. [Google Scholar]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.-P.; Baur, B. Genetic Effects of Anthropogenic Habitat Fragmentation on Remnant Animal and Plant Populations: A Meta-analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of Habitat Fragmentation on Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Wilcove, D.S.; McLellan, C.H.; Dobson, A.P. Habitat Fragmentation in the Temperate Zone. In Conservation Biology—The Science of Scarcity and Diversity; Soulé, M.E., Ed.; Sinauer Associates: Sunderland, MA, USA, 1986; pp. 237–256. [Google Scholar]
- Galindo-González, J. Efectos de la Fragmentación del Paisaje Sobre Poblaciones de Mamíferos; el Caso de los Murciélagos de los Tuxtlas, Veracruz. In Tópicos en Sistemática, Biogeografía, Ecología y Conservación de Mamíferos; Sánchez-Rojas, G., Rojas-Martínez, A., Eds.; Universidad Autónoma del Estado de Hidalgo: Pachuca, México, 2007; pp. 97–114. [Google Scholar]
- Dibattista, J.D. Patterns of Genetic Variation in Anthropogenically Impacted Populations. Conserv. Genet. 2007, 9, 141–156. [Google Scholar] [CrossRef]
- Dixo, M.; Metzger, J.P.; Morgante, J.S.; Zamudio, K.R. Habitat Fragmentation Reduces Genetic Diversity and Connectivity Among Toad Populations in the Brazilian Atlantic Coastal Forest. Biol. Conserv. 2009, 142, 1560–1569. [Google Scholar] [CrossRef]
- Jackson, N.D.; Fahrig, L. Habitat Amount, Not Habitat Configuration, Best Predicts Population Genetic Structure in Fragmented Landscapes. Landsc. Ecol. 2016, 31, 951–968. [Google Scholar] [CrossRef]
- Wan, H.Y.; Cushman, S.A.; Ganey, J.L. Habitat Fragmentation Reduces Genetic Diversity and Connectivity of the Mexican Spotted Owl: A Simulation Study Using Empirical Resistance Models. Genes 2018, 9, 403. [Google Scholar] [CrossRef]
- Frankham, R. Relationship of Genetic Variation to Population Size in Wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar] [CrossRef]
- Kunz, T.H.; Fenton, M.B. (Eds.) Bat Ecology; University of Chicago Press: Chicago, IL, USA, 2005; p. 779. [Google Scholar]
- Kunz, T.H.; de Torrez, E.B.; Bauer, D.M.; Lobova, T.; Fleming, T.H. Ecosystem Services Provided by Bats. Ann. N. Y. Acad. Sci. 2011, 1223, 1–38. [Google Scholar] [CrossRef]
- Meyer, C.F.J.; Kalko, E.K.V.; Kerth, G. Small-Scale Fragmentation Effects on Local Genetic Diversity in Two Phyllostomid Bats with Different Dispersal Abilities in Panama. Biotropica 2009, 41, 95–102. [Google Scholar] [CrossRef]
- Struebig, M.J.; Kingston, T.; Petit, E.J.; le Comber, S.C.; Zubaid, A.; Mohd-Adnan, A.; Rossiter, S.J. Parallel Declines in Species and Genetic Diversity in Tropical Forest Fragments. Ecol. Lett. 2011, 14, 582–590. [Google Scholar] [CrossRef]
- Silva, S.M.; Ferreira, G.; Pamplona, H.; Carvalho, T.L.; Cordeiro, J.; Trevelin, L.C. Effects of Landscape Heterogeneity on Population Genetic Structure and Demography of Amazonian Phyllostomid Bats. Mammal. Res. 2021, 66, 217–225. [Google Scholar] [CrossRef]
- Galindo-Gonzalez, J.; Sosa, V.J. Frugivorous Bats in Isolated Trees and Riparian Vegetation Associated with Human-Made Pastures in a Fragmented Tropical Landscape. Southwest. Nat. 2003, 48, 579–589. [Google Scholar] [CrossRef]
- Montiel, S.; Estrada, A.; León, P. Bat Assemblages in a Naturally Fragmented Ecosystem in the Yucatan Peninsula, Mexico: Species Richness, Diversity and Spatio-Temporal Dynamics. J. Trop. Ecol. 2006, 22, 267–276. [Google Scholar] [CrossRef]
- Arroyo-Rodríguez, V.; Rojas, C.; Saldaña-Vázquez, R.A.; Stoner, K.E. Landscape Composition is More Important Than Landscape Configuration for Phyllostomid Bat Assemblages in a Fragmented Biodiversity Hotspot. Biol. Conserv. 2016, 198, 84–92. [Google Scholar] [CrossRef]
- García-García, J.L.; Santos-Moreno, A.; Kraker-Castañeda, C. Ecological Traits of Phyllostomid Bats Associated with Sensitivity to Tropical Forest Fragmentation in Los Chimalapas, Mexico. Trop. Conserv. Sci. 2014, 7, 457–474. [Google Scholar] [CrossRef]
- Lucho, I.R.; Coates, R.; González-Christen, A. The Understory Bat Community in a Fragmented Landscape in the Lowlands of the Los Tuxtlas, Veracruz, Mexico. Therya 2017, 8, 99–107. [Google Scholar] [CrossRef]
- Estrada-Villegas, S.; Meyer, C.F.; Kalko, E.K. Effects of Tropical Forest Fragmentation on Aerial Insectivorous Bats in a Land-bridge Island System. Biol. Conserv. 2010, 143, 597–608. [Google Scholar] [CrossRef]
- Treitler, J.T.; Heim, O.; Tschapka, M.; Jung, K. The Effect of Local Land Use and Loss of Forests on Bats and Nocturnal Insects. Ecol. Evol. 2016, 6, 4289–4297. [Google Scholar] [CrossRef]
- Keenan, R.J.; Reams, G.A.; Achard, F.; de Freitas, J.V.; Grainger, A.; Lindquist, E. Dynamics of Global Forest Area: Results from the FAO Global Forest Resources Assessment 2015. For. Ecol. Manag. 2015, 352, 9–20. [Google Scholar] [CrossRef]
- Kolb, M.; Galicia, L. Challenging the Linear Forestation Narrative in the Neotropic: Regional Patterns and Processes of Deforestation and Regeneration in Southern Mexico. Geogr. J. 2012, 178, 147–161. [Google Scholar] [CrossRef]
- Watson, D.M. Long-term Consequences of Habitat Fragmentation—Highland Birds in Oaxaca, Mexico. Biol. Conserv. 2003, 111, 283–303. [Google Scholar] [CrossRef]
- Arroyo-Rodríguez, V.; Aguirre, A.; Benítez-Malvido, J.; Mandujano, S. Impact of Rain Forest Fragmentation on the Population Size of a Structurally Important Palm Species: Astrocaryum mexicanum at Los Tuxtlas, Mexico. Biol. Conserv. 2007, 138, 198–206. [Google Scholar] [CrossRef]
- Cristóbal-Azkarate, J.; Arroyo-Rodríguez, V. Diet and Activity Pattern of Howler Monkeys (Alouatta palliata) in Los Tuxtlas, Mexico: Effects of Habitat Fragmentation and Implications for Conservation. Am. J Primatol. Off. J. Am. Soc. Primatol. 2007, 69, 1013–1029. [Google Scholar] [CrossRef] [PubMed]
- Mas, J.-F.; Velázquez, A.; Díaz-Gallegos, J.R.; Mayorga-Saucedo, R.; Alcántara, C.; Bocco, G.; Castro, R.; Fernández, T.; Pérez-Vega, A. Assessing Land Use/Cover Changes: A Nationwide Multidate Spatial Database for Mexico. Int. J. Appl. Earth Obs. Geoinformation 2004, 5, 249–261. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). China—Global Forest Resources Assessment 2015—Country Report. In Global Forest Resources Assessment 2015; UN Food and Agriculture Organization: Rome, Italy, 2015. [Google Scholar]
- Trejo, I.; Dirzo, R. Deforestation of Seasonally Dry Tropical Forest: A National and Local Analysis in Mexico. Biol. Conserv. 2000, 94, 133–142. [Google Scholar] [CrossRef]
- Maass, J.M. Conversion of Tropical Dry Forest to Pasture and Agriculture. In Seasonally Dry Forest; Bullock, S.H., Mooney, H.A., Medina, E., Eds.; Cambridge University Press: Cambridge, UK, 1995; pp. 9–32. [Google Scholar]
- Miranda, R.P.; Romero-Sánchez, M.; González-Hernández, A.; Moreno-Sánchez, F.; Acosta-Mireles, M.; Carrillo-Anzures, F. Temporary Analysis of Land Use Changes in Pine and Mixed Forests in Mexico. AGRO Product. 2020, 13, 91–98. [Google Scholar] [CrossRef]
- Arriaga, L. Implicaciones del Cambio de Uso de Suelo en la Biodiversidad de los Matorrales Xerófilos: Un Enfoque Multiescalar. Investig. Ambient. 2009, 1, 6–16. [Google Scholar]
- Webala, P.W.; Mwaura, J.; Mware, J.M.; Ndiritu, G.G.; Patterson, B.D. Effects of Habitat Fragmentation on the Bats of Kakamega Forest, Western Kenya. J. Trop. Ecol. 2019, 35, 260–269. [Google Scholar] [CrossRef]
- Gamboa Alurralde, S.; Díaz, M.M. Assemblage-Level Responses of Neotropical Bats to Forest Loss and Fragmentation. Basic Appl. Ecol. 2021, 50, 57–66. [Google Scholar] [CrossRef]
- Sánchez-Hernández, C.; Romero-Almaraz, M.L.; Gurrola-Hidalgo, M.A. Natalus stramineus saturatus (Dalquest and Hall, 1949). In Historia Natural de Chamela; Noguera, F.A., Vega, J.H., Rivera-García-Aldrete, A.N., Quesada-Avendaño, M., Eds.; Instituto de Biología; Universidad Nacional Autónoma de México: México City, México, 2002; pp. 403–405. [Google Scholar]
- Torres-Flores, J.W. Dinámica Poblacional, Patrón Reproductivo, Dieta, Selección de Condiciones Microclimáticas y Hábitos de Percha de Natalus mexicanus (Chiroptera: Natalidae) en la Parte Central de Colima, México. Ph.D. Thesis, Universidad Autónoma Metropolitana, Ciudad de México, México, 2013. [Google Scholar]
- López-Wilchis, R.; Torres-Flores, J.W.; Arroyo-Cabrales, J. Natalus mexicanus (Chiroptera: Natalidae). Mamm. Species 2020, 52, 27–39. [Google Scholar] [CrossRef]
- Iucn Natalus mexicanus. IUCN Red List of Threatened Species; Solari, S., Ed.; IUCN: Gland, Switzerlnad, 2019. [Google Scholar]
- Reid, F.A. A Field Guide to the Mammals of Central America and Southeast Mexico; Oxford University Press: New York, NY, USA, 2009; p. 346. [Google Scholar]
- Castro-Luna, A.A.; Sosa, V.J.; Castillo-Campos, G. Bat Diversity and Abundance Associated with the Degree of Secondary Succession in a Tropical Forest Mosaic in South-Eastern Mexico. Anim. Conserv. 2007, 10, 219–228. [Google Scholar] [CrossRef]
- Galindo-González, J. Clasificación de los Murciélagos de la Región de Los Tuxtlas, Veracruz, Respecto a su Respuesta a la Fragmentación del Hábitat. Acta Zool. Mex. 2004, 20, 239–243. [Google Scholar]
- Medellín, R.; Arita, H.; Sánchez, O. Identificación de los Murciélagos de México, Clave de Campo; Asociación Mexicana de Mastozoología, A.C.: Distrito Federal, México, 1997. [Google Scholar]
- Hall, E.R. The Mammals of North America; John Wiley & Sons: New York, NY, USA, 1981. [Google Scholar]
- Secretaría de Medio Ambiente y Recursos Naturales (SEMARNAT). Norma Oficial Mexicana NOM-059, Protección Ambiental-Especies Nativas de México de Flora y Fauna Silvestres. Categorías de Riesgo y Especificaciones Para su Inclusión, Exclusión o Cambio, Lista de Especies en Riesgo; Diario oficial de la Federación 2008: Ciudad de México, México, 2010. [Google Scholar]
- Arroyo-Cabrales, J.; van den Bussche, R.A.; Sigler, K.H.; Chesser, R.K.; Baker, R.J. Genic Variation of Mainland and Island Populations of Natalus stramineus (Chiroptera: Natalidae). Occas. Pap. Mus. Texas Tech. Univ. 1997, 171, 1–9. [Google Scholar]
- López-Wilchis, R.; Guevara-Chumacero, L.M.; Ángeles Pérez, N.; Juste, J.; Ibáñez, C.; Barriga-Sosa, I.D.L.A. Taxonomic Status Assessment of the Mexican Populations of Funnel-Eared Bats, Genus Natalus (Chiroptera: Natalidae). Acta Chiropterologica 2012, 14, 305. [Google Scholar] [CrossRef]
- Guevara-Chumacero, L.M.; López-Wilchis, R.; Pedroche, F.F.; Juste, J.; Ibáñez, C.; Barriga-Sosa, I.D.L.A. Molecular Phylogeography of Pteronotus davyi (Chiroptera: Mormoopidae) in Mexico. J. Mammal. 2010, 91, 220–232. [Google Scholar] [CrossRef]
- Zárate-Martínez, D.G.; López-Wilchis, R.; Ruiz-Ortíz, J.D.; Barriga-Sosa, I.D.L.A.; Serrato-Díaz, A.; Ibáñez, C.; Juste, J.; Guevara-Chumacero, L.M. Intraspecific Evolutionary Relationships and Diversification Patterns of the Wagner’s Mustached Bat, Pteronotus personatus (Chiroptera: Mormoopidae). Acta Chiropterologica 2018, 20, 51–58. [Google Scholar] [CrossRef]
- Sikes, R.S. The Animal Care and Use Committee of the American Society of Mammalogists 2016 Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Anonymous. Lineamientos para la Conducción ética de la Investigación, la Docencia y la Difusión de la División de Ciencias Biológicas y de la Salud; Universidad Autónoma Metropolitana, Iztapalapa: Ciudad de México, México, 2010; p. 39. [Google Scholar]
- Fumagalli, L.; Taberlet, P.; Favre, L.; Hausser, J. Origin and Evolution of Homologous Repeated Sequences in the Mitochondrial DNA Control Region of Shrews. Mol. Biol. Evol. 1996, 13, 31–46. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Rodríguez, A.; López-Wilchis, R.; Serrato-Díaz, A.; del Río-Portilla, M.A.; Guevara-Chumacero, L.M. Isolation and Characterization of Microsatellite Markers for Funnel-Eared Bats Natalus mexicanus (Chiroptera: Natalidae) and Cross-Amplification Using Next-Generation Sequencing. Biochem. Syst. Ecol. 2015, 62, 69–72. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Pfenninger, M.; Posada, D. Phylogeographic History of the Land Snail Candidula unifasciata (Helicellinae, Stylommatophora): Fragmentation, Corridor Migration, and Secondary Contact. Evolution 2002, 56, 1776–1788. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Manni, F.; Guérard, E.; Heyer, E. Geographic Patterns of (Genetic, Morphologic, Linguistic) Variation: How Barriers can be Detected by “Monmonier´s Algorithm”. Hum. Biol. 2004, 76, 173–190. [Google Scholar] [CrossRef]
- Slatkin, M.; Hudson, R.R. Pairwise Comparisons of Mitochondrial DNA Sequences in Stable and Exponentially Growing Populations. Genetics 1991, 129, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Harpending, R.C. Signature of Ancient Population Growth in a Low-Resolution Mitochondrial DNA Mismatch Distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar]
- Tajima, F. The Effect of Change in Population Size on DNA Polymorphism. Genetics 1989, 123, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-X. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef]
- Clare, E.L.; Adams, A.M.; Maya-Simões, A.Z.; Eger, J.L.; Hebert, P.D.; Fenton, M.B. Diversification and Reproductive Isolation: Cryptic Species in the Only New World High-Duty Cycle Bat, Pteronotus parnellii. BMC Evol. Biol. 2013, 13, 26. [Google Scholar] [CrossRef]
- Beerli, P.; Palczewski, M. Unified Framework to Evaluate Panmixia and Migration Direction Among Multiple Sampling Locations. Genetics 2010, 185, 313–326. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICROCHECKER: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Chapuis, M.-P.; Estoup, A. Microsatellite Null Alleles and Estimation of Population Differentiation. Mol. Biol. Evol. 2006, 24, 621–631. [Google Scholar] [CrossRef]
- Hintze, J. PASS 11; NCSS, LLC.: Kaysville, UT, USA, 2011; Available online: www.ncss.com.
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Wilson, G.A.; Rannala, B. Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [PubMed]
- Collevatti, R.G.; Vitorino, L.C.; Vieira, T.B.; Oprea, M.; Telles, M.P. Landscape Changes Decrease Genetic Diversity in the Pallas’ Long-Tongued Bat. Perspect. Ecol. Conserv. 2020, 18, 169–177. [Google Scholar] [CrossRef]
- Petit, E.; Balloux, F.; Goudet, J. Sex-Biased Dispersal in a Migratory Bat: A Characterization Using Sex-Specific Demographic Parameters. Evolution 2001, 53, 635–640. [Google Scholar] [CrossRef]
- Moussy, C.; Hosken, D.; Mathews, F.; Smith, G.; Aegerter, J.; Bearhop, S. Migration and Dispersal Patterns of Bats and Their Influence on Genetic Structure. Mammal. Rev. 2012, 43, 183–195. [Google Scholar] [CrossRef]
- Flores, V.; Carter, G.G.; Halczok, T.K.; Kerth, G.; Page, R.A. Social Structure and Relatedness in the Fringe-Lipped Bat (Trachops cirrhosus). R. Soc. Open Sci. 2020, 7, 192256. [Google Scholar] [CrossRef]
- Torres-Flores, J.W.; López-Wilchis, R.; Soto-Castruita, A. Dinámica Poblacional, Selección de Sitios de Percha y Patrones Reproductivos de Algunos Murciélagos Cavernícolas en el Oeste de México. Rev. Biol. Trop. 2012, 60, 1369–1389. [Google Scholar] [CrossRef]
- Mitchell, G.C. Population Study of the Funnel-Eared Bat (Natalus stramineus) in Sonora. Southwest. Nat. 1967, 12, 172. [Google Scholar] [CrossRef]
- Tejedor, A. A New Species of Funnel-Eared Bat (Natalidae: Natalus) from Mexico. J. Mammal. 2005, 86, 1109–1120. [Google Scholar] [CrossRef]
- Tejedor, A. Systematics of Funnel-Eared Bats (Chiroptera: Natalidae). Bull. Am. Mus. Nat. Hist. 2011, 353, 1–140. [Google Scholar] [CrossRef]
- Arstens, B.C.C.; Sullivan, J.; Avalos, L.M.D.; Arsen, P.A.L.; Pedersen, S.C. Exploring Population Genetic Structure in Three Species of Lesser Antillean Bats. Mol. Ecol. 2004, 13, 2557–2566. [Google Scholar] [CrossRef]
- Russell, A.L.; Medellín, R.A.; McCracken, G.F. Genetic Variation and Migration in the Mexican Free-Tailed Bat (Tadarida brasiliensis mexicana). Mol. Ecol. 2005, 14, 2207–2222. [Google Scholar] [CrossRef]
- Dong, J.; Mao, X.; Sun, H.; Irwin, D.M.; Zhang, S.; Hua, P. Introgression of Mitochondrial DNA Promoted by Natural Selection in the Japanese Pipistrelle Bat (Pipistrellus abramus). Genetica 2014, 142, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, M.C.; Piñero, D.; Eguiarte, L.E.; Gasca, J.; Medellín, R.A. Genetic Structure and Diversity of the Nine-Banded Armadillo in Mexico. J. Mammal. 2012, 93, 547–559. [Google Scholar] [CrossRef]
- Ruiz, E.A.; Vargas-Miranda, B.; Zúñiga, G. Late-Pleistocene Phylogeography and Demographic History of Two Evolutionary Lineages of Artibeus jamaicensis (Chiroptera: Phyllostomidae) in Mexico. Acta Chiropterologica 2013, 15, 19–33. [Google Scholar] [CrossRef]
- Metcalfe, S.E.; O’Hara, S.L.; Caballero, M.; Davies, S.J. Records of Late Pleistocene–Holocene Climatic Change in Mexico—A Review. Quat. Sci. Rev. 2000, 19, 699–721. [Google Scholar] [CrossRef]
- Hofreiter, M.; Stewart, J. Ecological Change, Range Fluctuations and Population Dynamics During the Pleistocene. Curr. Biol. 2009, 19, R584–R594. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Méndez, A.; Sánchez-González, L.A.; González, C.; Navarro-Sigüenza, A.G. The Geography of Evolutionary Divergence in the Highly Endemic Avifauna from the Sierra Madre del Sur, Mexico. BMC Evol. Biol. 2019, 19, 237. [Google Scholar] [CrossRef] [PubMed]
- Barber, B.R.; Klicka, J. Two Pulses of Diversification Across the Isthmus of Tehuantepec in a Montane Mexican Bird Fauna. Proc. Royal Soc. B Biol. Sci. 2010, 277, 2675–2681. [Google Scholar] [CrossRef]
- Mason, N.A.; Olvera-Vital, A.; Lovette, I.J.; Navarro-Sigüenza, A.G. Hidden Endemism, Deep Polyphyly, and Repeated Dispersal Across the Isthmus of Tehuantepec: Diversification of the White-Collared Seedeater Complex (Thraupidae: Sporophila torqueola). Ecol. Evol. 2018, 8, 1867–1881. [Google Scholar] [CrossRef]
- Guevara-Chumacero, L.M.; López-Wilchis, R.; Juste, J.; Ibáñez, C.; Martínez-Méndez, L.A.; Barriga-Sosa, I.D.L.A. Conservation Units of Pteronotus davyi (Chiroptera: Mormoopidae) in Mexico Based on Phylogeographical Analysis. Acta Chiropterologica 2013, 15, 353–363. [Google Scholar] [CrossRef]
- Hernández-Soto, M.; Licona-Vera, Y.; Lara, C.; Ornelas, J.F. Molecular and Climate Data Reveal Expansion and Genetic Differentiation of Mexican Violet-Ear Colibri thalassinus thalassinus (Aves: Trochilidae) Populations Separated by the Isthmus of Tehuantepec. J. Ornithol. 2018, 159, 687–702. [Google Scholar] [CrossRef]
- Zamudio-Beltrán, L.E.; Licona-Vera, Y.; E Hernández-Baños, B.; Klicka, J.; Ornelas, J.F. Phylogeography of the Widespread White-Eared Hummingbird (Hylocharis leucotis): Pre-glacial Expansion and Genetic Differentiation of Populations Separated by the Isthmus of Tehuantepec. Biol. J. Linn. Soc. 2020, 130, 247–267. [Google Scholar] [CrossRef]
- Arroyo-Cabrales, J.; Álvarez, T. A Preliminary Report of the Late Quaternary Mammal Fauna from Loltún Cave, Yucatán, México. In Ice Age Cave Faunas of North America; Shubert, B.W., Mead, J.I., Graham., R.W., Eds.; Indiana University Press y Denver Museum of Nature and Science: Bloomington, IN, USA, 2003; pp. 262–272. [Google Scholar]
- León-Tapia, M.A. DNA Barcoding and Demographic History of Peromyscus yucatanicus (Rodentia: Cricetidae) Endemic to the Yucatan Peninsula, Mexico. J. Mamm. Evol. 2020, 1–15. [Google Scholar] [CrossRef]
- Castella, V.; Ruedi, M.; Excoffier, L. Contrasted Patterns of Mitochondrial and Nuclear Structure Among Nursery Colonies of the Bat Myotis myotis. J. Evol. Biol. 2008, 14, 708–720. [Google Scholar] [CrossRef]
- Flanders, J.; Jones, G.; Benda, P.; Dietz, C.; Zhang, S.; Li, G.; Sharifi, M.; Rossiter, S.J. Phylogeography of the Greater Horseshoe Bat, Rhinolophus ferrumequinum: Contrasting Results from Mitochondrial and Microsatellite data. Mol. Ecol. 2009, 18, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, T.; Schoeman, M.C.; Goodman, S.M.; Taylor, P.J.; Lamb, J.M. Discordance Between Mitochondrial and Nuclear Genetic Structure in the Bat Chaerephon pumilus (Chiroptera: Molossidae) from Southern Africa. Mamm. Biol. 2016, 81, 115–122. [Google Scholar] [CrossRef]
- Brito, P.H. Contrasting Patterns of Mitochondrial and Microsatellite Genetic Structure Among Western European Populations of Tawny Owls (Strix aluco). Mol. Ecol. 2007, 16, 3423–3437. [Google Scholar] [CrossRef]
- Zink, R.M. Microsatellite and Mitochondrial DNA Differentiation in the Fox Sparrow. Condor 2008, 110, 482–492. [Google Scholar] [CrossRef]
- Zarza, E.; Reynoso, V.H.; Emerson, B.C. Discordant Patterns of Geographic Variation Between Mitochondrial and Microsatellite Markers in the Mexican Black Iguana (Ctenosaura pectinata) in a Contact Zone. J. Biogeogr. 2011, 38, 1394–1405. [Google Scholar] [CrossRef]
- Monsen, K.J.; Blouin, M.S. Genetic Structure in a Montane Ranid Frog: Restricted Gene Flow and Nuclear–Mitochondrial Discordance. Mol. Ecol. 2003, 12, 3275–3286. [Google Scholar] [CrossRef]
- Chen, Z.; Li, H.; Zhai, X.; Zhu, Y.; He, Y.; Wang, Q.; Li, Z.; Jiang, J.; Xiong, R.; Chen, X. Phylogeography, Speciation and Demographic History: Contrasting Evidence from Mitochondrial and Nuclear Markers of the Odorrana graminea sensu lato (Anura, Ranidae) in China. Mol. Phylogenetics Evol. 2020, 144, 106701. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.W.; Murphy, R.W. Paleobiogeography and Genetic Differentiation of the Baja California Herpetofauna. Occas. Papers Calif. Acad. Sci. 1983, 137, 1–48. [Google Scholar]
- Riddle, B.R.; Hafner, D.J.; Alexander, L.F.; Jaeger, J.R. Cryptic Vicariance in the Historical Assembly of a Baja California Peninsular Desert Biota. Proc. Natl. Acad. Sci. USA 2000, 97, 14438–14443. [Google Scholar] [CrossRef]
- López-Wilchis, R.; Flores-Romero, M.; Guevara-Chumacero, L.M.; Serrato-Díaz, A.; Díaz-Larrea, J.; Salgado-Mejia, F.; Ibáñez, C.; Salles, L.O.; Juste, J. Evolutionary Scenarios Associated with the Pteronotus parnellii Cryptic Species-Complex (Chiroptera: Mormoopidae). Acta Chiropterologica 2016, 18, 91–116. [Google Scholar] [CrossRef]
- Hernández-Canchola, G.; León-Paniagua, L. Genetic and Ecological Processes Promoting Early Diversification in the Lowland Mesoamerican Bat Sturnira parvidens (Chiroptera: Phyllostomidae). Mol. Phylogenetics Evol. 2017, 114, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Zárate-Martínez, D.G. Relaciones Filogenéticas y Filogeográficas en el Murciélago Pteronotus personatus (Chiroptera: Mormoopidae). Ph.D. Thesis, Universidad Autónoma Metropolitana, Ciudad de México, México, 2019. [Google Scholar]
- Sullivan, J.; Markert, J.A.; Kilpatrick, C.W. Phylogeography and Molecular Systematics of the Peromyscus aztecus Species Group (Rodentia: Muridae) Inferred Using Parsimony and Likelihood. Syst. Biol. 1997, 46, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Hafner, M.S.; Spradling, T.A.; Light, J.E.; Hafner, D.J.; Demboski, J.R. Systematic Revision of Pocket Gophers of the Cratogeomys gymnurus Species Group. J. Mammal. 2004, 85, 1170–1183. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística y Geografía (INEGI). Carta de Uso del Suelo y Vegetación, Escala 1:250,000, 2016, serie VI (Continuo Nacional); Instituto Nacional de Estadística y Geografía: Aguascalientes, México, 2016. [Google Scholar]
- Burg, T.M.; Gaston, A.J.; Winker, K.; Friesen, V.L. Rapid Divergence and Postglacial Colonization in Western North American Steller’s Jays (Cyanocitta stelleri). Mol. Ecol. 2005, 14, 3745–3755. [Google Scholar] [CrossRef]
- Adams, R.V.; Burg, T.M. Influence of Ecological and Geological Features on Rangewide Patterns of Genetic Structure in a WideSpread Passerine. Heredity 2015, 114, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.A.; Räikkönen, J.; Vucetich, L.M.; Vucetich, J.A.; Peterson, R.O.; Lohmueller, K.E.; Wayne, R.K. Genomic Signatures of Extensive Inbreeding in Isle Royale Wolves, a Population on the Threshold of Extinction. Sci. Adv. 2019, 5, eaau0757. [Google Scholar] [CrossRef] [PubMed]
- Santymire, R.M.; Lonsdorf, E.V.; Lynch, C.M.; Wildt, D.E.; Marinari, P.E.; Kreeger, J.S.; Howard, J.G. Inbreeding Causes Decreased Seminal Quality Affecting Pregnancy and Litter Size in the Endangered Black-Footed Ferret. Anim. Conserv. 2019, 22, 331–340. [Google Scholar] [CrossRef]
- Hinkson, K.M.; Poo, S. Inbreeding Depression in Sperm Quality in a Critically Endangered Amphibian. Zoo Biol. 2020, 39, 197–204. [Google Scholar] [CrossRef]
- Lawson, L.P.; Fessl, B.; Vargas, F.H.; Farrington, H.L.; Cunninghame, H.F.; Mueller, J.C.; Nemeth, E.; Sevilla, P.C.; Petren, K. Slow Motion Extinction: Inbreeding, Introgression, and Loss in the Critically Endangered Mangrove Finch (Camarhynchus heliobates). Conserv. Genet. 2016, 18, 159–170. [Google Scholar] [CrossRef]
- Gómez-Sánchez, D.; Olalde, I.; Sastre, N.; Enseñat, C.; Carrasco, R.; Marques-Bonet, T.; Lalueza-Fox, C.; Leonard, J.A.; Vilà, C.; Ramírez, O. On the Path to Extinction: Inbreeding and Admixture in a Declining Grey Wolf Population. Mol. Ecol. 2018, 27, 3599–3612. [Google Scholar] [CrossRef]
- Torres-Flores, J.W.; López-Wilchis, R. Condiciones Microclimáticas, Hábitos de Percha y Especies Asociadas a los Refugios de Natalus stramineus en México. Acta Zoológica Mex. 2010, 26, 191–213. [Google Scholar] [CrossRef]


| Source of Variation | D. F | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices |
|---|---|---|---|---|---|
| No groups defined | AP | 855.293 | 3.565 | 71.07 | FST = 0.710 * |
| WP | 325.144 | 1.451 | 28.93 | ||
| PAC vs. GM vs. SS vs. PYUC | AG | 405.035 | 2.852 | 44.69 | FSC = 0.588 * |
| APWG | 450.257 | 2.0793 | 32.57 | FST = 0.772 | |
| WP | 325.144 | 1.451 | 22.74 | FCT = 0.446 |
| PAC | GM | PYUC | |
|---|---|---|---|
| Tajima’s D | −0.4897 | −0.8331 | −1.5258 * |
| Fu’s F | −24.599 * | −12.3845 * | −20.4019 * |
| Locality | N | Na | Np | HO | HE |
|---|---|---|---|---|---|
| Pe | 16 | Σ = 46 = 4.6 | 2 | 0.356 | 0.575 |
| Pa | 12 | Σ = 62 = 6.2 | 2 | 0.504 | 0.723 |
| Ca | 11 | Σ = 58 = 5.8 | 6 | 0.491 | 0.683 |
| VB | 16 | Σ = 67 = 6.7 | 10 | 0.535 | 0.649 |
| Co | 16 | Σ = 72 = 7.2 | 3 | 0.569 | 0.710 |
| SS | 16 | Σ = 59 = 5.9 | 3 | 0.404 | 0.585 |
| LV | 15 | Σ = 92 = 9.2 | 8 | 0.653 | 0.776 |
| TG | 23 | Σ = 97 = 9.7 | 3 | 0.617 | 0.759 |
| Bo | 16 | Σ = 87 = 8.7 | 4 | 0.681 | 0.750 |
| Cr | 14 | Σ = 78 = 7.8 | 5 | 0.532 | 0.787 |
| Cva | 16 | Σ = 74 = 7.4 | 2 | 0.563 | 0.696 |
| Source of Variation | D. F | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices |
|---|---|---|---|---|---|
| Among localities | 10 | 196.822 | 0.535 | 14.75 | FST = 0.147 |
| 10 | 21,396.3 | 66.817 | 49.02 | RST = 0.490 | |
| Within localities | 331 | 1024.137 | 3.094 | 85.25 | |
| 331 | 22,997.86 | 69.49 | 50.98 |
| Donor Group | ||||
|---|---|---|---|---|
| Recipient Group | Group 1 | Group 2 | Group 3 | Group 4 |
| Group 1 | 0.7 (0.682–0.717) | 0.002 (0.000–0.005) | 0.003 (0.001–0.008) | 0.001 (0.000–0.004) |
| Group 2 | 0.01 (0.001–0.025) | 0.99 (0.986–0.999) | 0.003 (0.001–0.008) | 0.001 (0.000–0.004) |
| Group 3 | 0.01 (0.001–0.025) | 0.002 (0.001–0.005) | 0.99 (0.981–0.998) | 0.003 (0.000–0.008) |
| Group 4 | 0.29 (0.264–0.315) | 0.002 (0.001–0.005) | 0.003 (0.000–0.007) | 0.99 (0.980–0.990) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Wilchis, R.; Méndez-Rodríguez, A.; Juste, J.; Serrato-Díaz, A.; Rodríguez-Gómez, F.; Guevara-Chumacero, L.M. Genetic Consequences of Forest Fragmentation in a Widespread Forest Bat (Natalus mexicanus, Chiroptera: Natalidae). Diversity 2021, 13, 140. https://doi.org/10.3390/d13040140
López-Wilchis R, Méndez-Rodríguez A, Juste J, Serrato-Díaz A, Rodríguez-Gómez F, Guevara-Chumacero LM. Genetic Consequences of Forest Fragmentation in a Widespread Forest Bat (Natalus mexicanus, Chiroptera: Natalidae). Diversity. 2021; 13(4):140. https://doi.org/10.3390/d13040140
Chicago/Turabian StyleLópez-Wilchis, Ricardo, Aline Méndez-Rodríguez, Javier Juste, Alejandra Serrato-Díaz, Flor Rodríguez-Gómez, and Luis Manuel Guevara-Chumacero. 2021. "Genetic Consequences of Forest Fragmentation in a Widespread Forest Bat (Natalus mexicanus, Chiroptera: Natalidae)" Diversity 13, no. 4: 140. https://doi.org/10.3390/d13040140
APA StyleLópez-Wilchis, R., Méndez-Rodríguez, A., Juste, J., Serrato-Díaz, A., Rodríguez-Gómez, F., & Guevara-Chumacero, L. M. (2021). Genetic Consequences of Forest Fragmentation in a Widespread Forest Bat (Natalus mexicanus, Chiroptera: Natalidae). Diversity, 13(4), 140. https://doi.org/10.3390/d13040140









