Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers
Abstract
1. Introduction
2. Materials and Methods
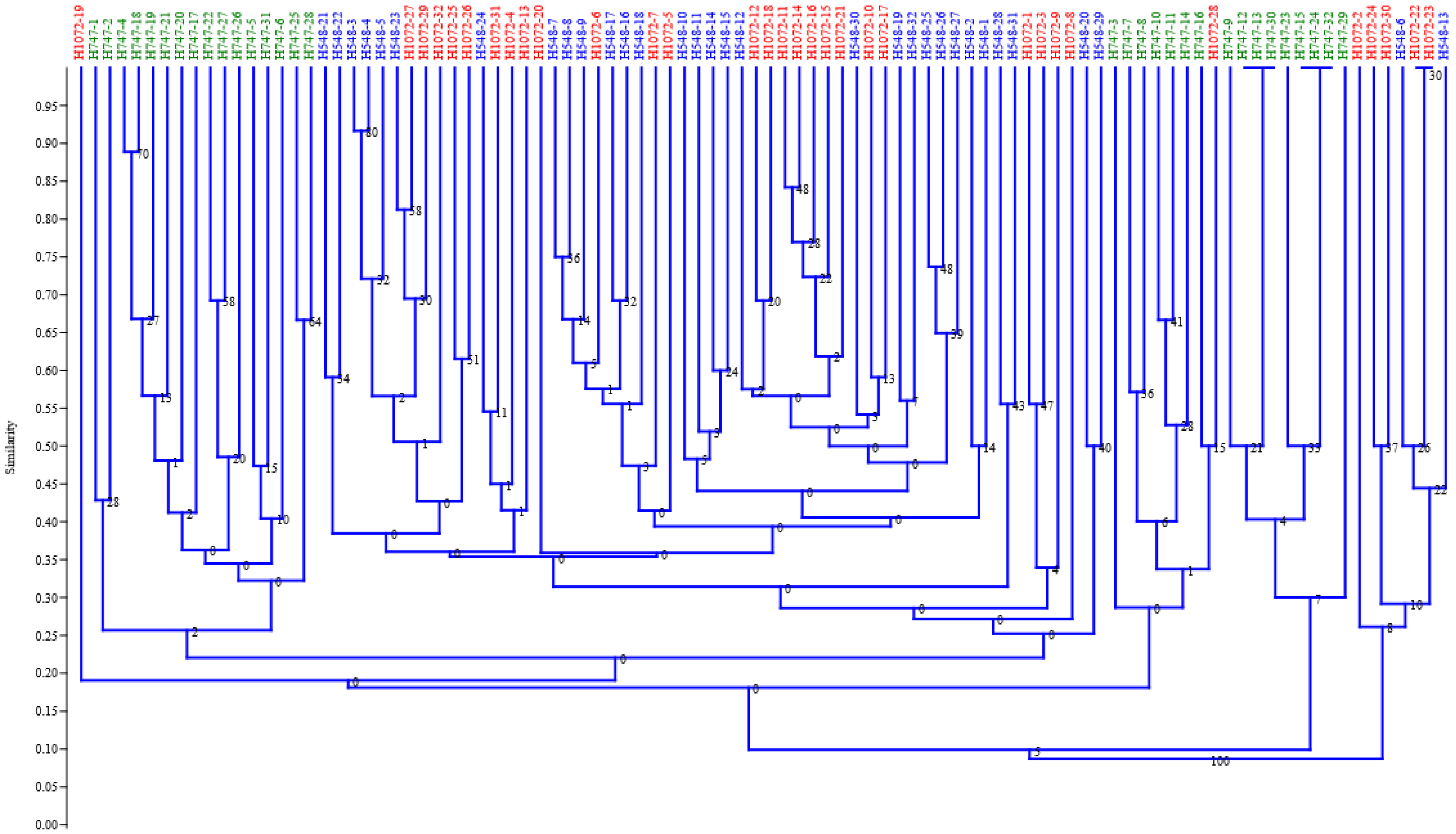
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rick, C.; Simmonds, N. Tomato Lycopersicon escultentum (Solanaceae). Evo. Crop Plants 1976, 268–273. [Google Scholar]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Conti, V.; Mareri, L.; Faleri, C.; Nepi, M.; Romi, M.; Cai, G.; Cantini, C. Drought Stress Affects the Response of Italian Local Tomato (Solanum lycopersicum L.) Varieties in a Genotype-Dependent Manner. Plants 2019, 8, 336. [Google Scholar] [CrossRef] [PubMed]
- Conesa, M.À.; Fullana-Pericàs, M.; Granell, A.; Galmés, J. Mediterranean Long Shelf-Life Landraces: An Untapped Genetic Resource for Tomato Improvement. Front. Plant Sci. 2020, 10, 1651. [Google Scholar] [CrossRef] [PubMed]
- Scarano, A.; Olivieri, F.; Gerardi, C.; Liso, M.; Chiesa, M.; Chieppa, M.; Frusciante, L.; Barone, A.; Santino, A.; Rigano, M.M. Selection of tomato landraces with high fruit yield and nutritional quality under elevated temperatures. J. Sci. Food Agric. 2020, 100, 2791–2799. [Google Scholar] [CrossRef] [PubMed]
- Mazzucato, A.; Papa, R.; Bitocchi, E.; Mosconi, P.; Nanni, L.; Negri, V.; Picarella, M.E.; Siligato, F.; Soressi, G.P.; Tiranti, B.; et al. Genetic diversity.; structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L.) landraces. Theor. Appl. Genet. 2008, 116, 657–669. [Google Scholar] [CrossRef]
- Bota, J.; Conesa, M.; Ochogavia, J.; Medrano, H.; Francis, D.M.; Cifre, J. Characterization of a landrace collection for Tomàtiga de Ramellet (Solanum lycopersicum L.). from the Balearic Islands. Genet. Res. Crop Evo. 2014, 61, 1131–1146. [Google Scholar] [CrossRef]
- Cooke, R.; Bredemeijer, G.; Ganal, M.; Peeters, R.; Isaac, P.; Rendell, S.; Jackson, J.; Röder, M.; Korzun, V.; Wendehake, K.; et al. Assessment of the uniformity of wheat and tomato varieties at DNA microsatellite loci. Euphytica 2003, 132, 331–341. [Google Scholar] [CrossRef]
- Ruiz, J.J.; García-Martínez, S.; Picó, B.; Gao, M.; Quiros, C.F. Genetic variability and relationship of closely related Spanish traditional cultivars of tomato as detected by SRAP and SSR markers. J. Am. Soc. Hortic. Sci. 2005, 130, 88–94. [Google Scholar] [CrossRef]
- Comlekcioglu, N.; Simsek, O.; Boncuk, M.; Aka-Kacar, Y. Genetic characterization of heat tolerant tomato (Solanum lycopersicon). genotypes by SRAP and RAPD markers. Genet. Mol. Res. 2010, 9, 2263–2274. [Google Scholar] [CrossRef]
- Mane, R.; Sridevi, O.; Nishani, S.; Salimath, P. Evaluation of genetic diversity and relationships among tomato genotypes using morphological parameters and SRAP markers. Indian J. Hortic. 2013, 70, 357–363. [Google Scholar]
- Vargas, J.E.; Aguirre, N.C.; Coronado, Y.M. Study of the genetic diversity of tomato (Solanum spp.) with ISSR markers. Rev. Ceres 2020, 67, 199–206. [Google Scholar] [CrossRef]
- Brake, M.; Al-Gharaibeh, M.; Hamasha, H.; Al Sakarneh, N.; Alshomali, I.; Migdadi, H.; Qaryouti, M.; Haddad, N. Assessment of genetic variability among Jordanian tomato landrace using inter-simple sequence repeats markers. JJBS 2021, 14, 91–95. [Google Scholar]
- Sharifova, S.S.; Mehdiyeva, S.P.; Abbasov, M.A. Analysis of genetic diversity among different tomato genotypes using ISSR DNA marker. Genetika 2017, 49, 31–42. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence-related amplified polymorphism (SRAP): A new marker system based on a simple PCR reaction, its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- AlShaye, N.; Migdadi, H.; Charbaji, A.; Alsayegh, S.; Daoud, S.; Wala, A.A.; Alghamdi, S. Genetic variation among Saudi tomato (Solanum lycopersicum L.) landraces studied using SDS-page and SRAP markers. Saudi J. Biol. Sci. 2018, 25, 1007–1015. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST, Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Anderson, J.A.; Churchill, G.; Autrique, J.; Tanksley, S.; Sorrells, M. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Khierallah, H.; Bader, S.; Baum, M.; Hamwieh, A. Assessment of genetic diversity for some Iraqi date palms (Phoenix dactylifera L.) using amplified fragment length polymorphisms (AFLP) markers. Afr. J. Biotechnol. 2011, 10, 9570–9576. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GenAlEx 6.503: Genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Da Costa, J.P.; Rodríguez, G.; Pratta, G.; Picardi, L.; Zorzoli, R. Pericarp polypeptides and SRAP markers associated with fruit quality traits in an interspecific tomato backcross. Genet. Mol. Res. 2014, 13, 2539–2547. [Google Scholar] [CrossRef]
- Park, Y.H.; West, M.A.; St. Clair, D.A. Evaluation of AFLPs for germplasm fingerprinting and assessment of genetic diversity in cultivars of tomato (Lycopersicon esculentum L.). Genome 2004, 47, 510–518. [Google Scholar] [CrossRef]
- Henareh, M.; Dursun, A.; Abdollahi-Mandoulakani, B.; Haliloğlu, K. Assessment of genetic diversity in tomato landraces using ISSR markers. Genetika 2016, 48, 25–35. [Google Scholar] [CrossRef]
- Sharifova, S.; Mehdiyeva, S.; Theodorikas, K.; Roubos, K. Assessment of genetic diversity in cultivated tomato (Solanum lycopersicum L.) genotypes using RAPD primers. J. Hortic. Res. 2013, 21, 83–89. [Google Scholar] [CrossRef][Green Version]
- Thamir, A.; Al-Saadi, A.; Abbass, M. Genetic diversity of some tomato Lycopersicon esculentum Mill varieties in Iraq using random amplified polymorphism DNA (RAPD) markers. J. Baby. Univ. Pure Appl. Sci. 2014, 9, 2342–2351. [Google Scholar]
- Kaushal, A.; Singh, A.; Singh, A.J. Genetic diversity in tomato (Solanum lycopersicum L.) genotypes revealed by simple sequence repeats (SSR) markers. J. Appl. Nat. Sci. 2017, 9, 966–973. [Google Scholar] [CrossRef]
- Marin-Montes, I.M.; Lobato-Ortiz, R.; Carrillo-Castañeda, G.; Rodríguez-Pérez, J.E.; García-Zavala, J.J.; Velasco-García, A.M. Riqueza alélica de poblaciones nativas de jitomate (Solanum lycopersicum L.) para el mejoramiento genético. Agrociencia 2019, 53, 355–370. [Google Scholar]
- DeWoody, J.; Honeycutt, R.; Skow, L. Microsatellite markers in white-tailed deer. J. Hered. 1995, 86, 317–319. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Williams, C.E.; Clair, D.A. Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random amplified polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome 1993, 36, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Van de Wiel, C.; Smulders, M.; Vosman, B. Use of microsatellites to evaluate genetic diversity and species relationships in the genus Lycopersicon. Theor. App. Genet. 2001, 103, 1283–1292. [Google Scholar] [CrossRef]
- Corrado, G.; Caramante, M.; Piffanelli, P.; Rao, R. Genetic diversity in Italian tomato landraces, Implications for the development of a core collection. Sci. Hortic. 2014, 168, 138–144. [Google Scholar] [CrossRef]
- Gonias, E.D.; Ganopoulos, I.; Mellidou, I.; Bibi, A.; Kalivas, A.; Mylona, P.V.; Osanthanunkul, M.; Tsaftaris, A.; Madesis, P.; Doulis, A.G. Exploring genetic diversity of tomato (Solanum lycopersicum L.). germplasm of genebank collection employing SSR and SCAR markers. Genet. Res. Crop Evo. 2019, 66, 1295–1309. [Google Scholar] [CrossRef]
- Carelli, V.; Achilli, A.; Valentino, M.; Rengo, C.; Semino, O.; Pala, M.; Olivieri, A.; Mattiazzi, M.; Pallotti, F.; Carrara, F.; et al. Haplogroup effects and recombination of mitochondrial DNA, novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am. J. Hum. Genet. 2006, 78, 564–574. [Google Scholar] [CrossRef]
- Sacco, A.; Ruggieri, V.; Parisi, M.; Festa, G.; Rigano, M.; Picarella, M.E.; Mazzucato, A.; Barone, A. Exploring a tomato landraces collection for fruit-related traits by the aid of a high-throughput genomic platform. PLoS ONE 2015, 10, e0137139. [Google Scholar]
- Castellana, S.; Ranzino, L.; Beritognolo, I.; Cherubini, M.; Luneia, R.; Villani, F.; Mattioni, C. Genetic characterization and molecular fingerprint of traditional Umbrian tomato (Solanum lycopersicum L.) landraces through SSR markers and application for varietal identification. Genet. Res. Crop Evo. 2020, 67, 1807–1820. [Google Scholar] [CrossRef]
- Cattáneo, R.A.; McCarthy, A.N.; Feingold, S.E. Evidence of genetic diversity within Solanum lycopersicum L.’Platense’ landrace and identification of various subpopulations. Genet. Res. Crop Evo. 2020, 67, 2057–2069. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Z.; Cao, X.; Jiang, F. Genetic diversity of cultivated and wild tomatoes revealed by morphological traits and SSR markers. Genet. Mol. Res. 2015, 14, 13868–13879. [Google Scholar] [CrossRef]
- Meng, F.J.; Xu, X.Y.; Huang, F.l.; Li, J.F. Analysis of genetic diversity in cultivated and wild tomato varieties in Chinese market by RAPD and SSR. Agric. Sci. China 2010, 9, 1430–1437. [Google Scholar] [CrossRef]
- Parmar, P.; Oza, V.P.; Chauhan, V.; Patel, A.; Kathiria, K.; Subramanian, R. Genetic diversity and DNA fingerprint study of tomato discerned by SSR markers. Inter. J. Biotech. Biochem. 2010, 6, 657–667. [Google Scholar]
- Hassan, N.; Mostafa, S.; Twfik, A. Assessment of genetic diversity of tomato (Lycopersicon esculentum L.). germplasm using molecular markers (RAPD and ISSR). Egypt. J. Genet. Cytol. 2013, 42, 163–182. [Google Scholar] [CrossRef]
- Kohpayegani, J.A.; Behbahani, M. Genetic diversity of some populations of Iranian melon using SSR markers. Biotechnology 2008, 7, 19–26. [Google Scholar] [CrossRef][Green Version]
- Schltterer, C. Evolutionary dynamics of microsatellite DNA. Chromosoma 2000, 109, 365–371. [Google Scholar] [CrossRef]
- Igwe, D.O.; Afiukwa, C.A.; Acquaah, G.; Ude, G. Genetic diversity and structure of Capsicum annuum as revealed by start codon targeted and directed amplified minisatellite DNA markers. Heredity 2019, 156, 32. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.; Teixeira da Silva, J.; Edris, S.; Younis, R.A. Comparative assessment of genetic diversity in tomato cultivars using IRAP, ISSR and RAPD molecular markers Focus on Tree Genetics and Genomics. Genes Genomes Genom. 2010, 4, 41–47. [Google Scholar]
| Primers Name | Primer Sequence 5′-3′ | # Alleles | # Bands | Polymorphism% | PIC | DP |
|---|---|---|---|---|---|---|
| SRAP1 | F-TGAGTCCAAACCGGAGA R-GACTGCGTACGAATTGAA | 9 | 121 | 100 | 0.73 | 16.36 |
| SRAP2 | F-TGAGTCCAAACCGGAGA R-GACTGCGTACGAATTTGC | 8 | 187 | 100 | 0.82 | 14.55 |
| SRAP3 | F-TGAGTCCAAACCGGACC R-GACTGCGTACGAATTATG | 3 | 75 | 100 | 0.17 | 5.45 |
| SRAP4 | F-TGAGTCCAAACCGGACT R-GACTGCGTACGAATTGAC | 8 | 153 | 100 | 0.76 | 14.55 |
| SRAP5 | F-TGAGTCCAAACCGGTAA R-GACTGCGTACGAATTACG | 10 | 122 | 100 | 0.77 | 18.18 |
| SRAP6 | F-TGAGTCCAAACCGGTAA R-GACTGCGTACGAATTCAA | 9 | 147 | 100 | 0.79 | 16.36 |
| SRAP7 | F-TGAGTCCAAACCGGAGA R-GACTGCGTACGAATTTGA | 8 | 222 | 100 | 0.72 | 14.55 |
| Total | 55 | 1027 | ----- | ----- | ---- | |
| Min | 3 | 75 | 100 | 0.17 | 5.45 | |
| Max | 10 | 222 | 100 | 0.82 | 18.18 | |
| Mean | 7.86 | 146.71 | 100 | 0.68 | 14.29 | |
| Population | N | Na | Ne | I | He | Polymorphism% | Number of Private Bands | |
|---|---|---|---|---|---|---|---|---|
| Hail 548 | Mean | 32 | 1.791 | 1.485 | 0.442 | 0.291 | 88.37% | 1.0 |
| SE | 0.091 | 0.051 | 0.034 | 0.025 | ||||
| Hail 747 | Mean | 32 | 1.698 | 1.344 | 0.346 | 0.217 | 83.72% | 0.0 |
| SE | 0.108 | 0.050 | 0.034 | 0.025 | ||||
| Hail 1072 | Mean | 32 | 1.558 | 1.414 | 0.367 | 0.242 | 76.74% | 2.0 |
| SE | 0.126 | 0.059 | 0.040 | 0.029 | ||||
| Mean | 1.682 | 1.287 | 0.385 | 0.250 | 82.95% | |||
| SE | 0.063 | 0.025 | 0.021 | 0.015 |
| (A) Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among landraces | 2 | 65.354 | 32.677 | 0.848 | 13% |
| Within accessions | 93 | 515.813 | 5.546 | 5.546 | 87% |
| Total | 95 | 581.167 | 6.394 | 100% | |
| (B) Pair-wise Population PhiPT Values | |||||
| Hail 548 | Hail 747 | ||||
| Hail 747 | 0.114 ** | 0.000 | |||
| Hail1072 | 0.179 ** | 0.094 ** | |||
| Mean PhiPT | 0.133 | p < 0.001 | |||
| Hail 548 | Hail 747 | Hail 1072 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primer | #Alleles | #Bands | PIC | DP | #Alleles | #Bands | PIC | DP | #Alleles | #Bands | PIC | DP |
| SRAP1 | 8 | 66 | 0.79 | 18.600 | 4 | 21 | 0.46 | 10.26 | 5 | 34 | 0.72 | 11.63 |
| SRAP2 | 8 | 79 | 0.84 | 18.60 | 7 | 46 | 0.79 | 17.95 | 7 | 62 | 0.78 | 16.28 |
| SRAP3 | 3 | 34 | 0.47 | 6.98 | 3 | 21 | 0.54 | 7.69 | 3 | 20 | 0.27 | 6.98 |
| SRAP4 | 7 | 63 | 0.78 | 16.28 | 7 | 34 | 0.71 | 17.95 | 7 | 56 | 0.74 | 16.28 |
| SRAP5 | 4 | 44 | 0.53 | 9.30 | 6 | 25 | 0.71 | 15.38 | 10 | 53 | 0.79 | 23.26 |
| SRAP6 | 9 | 65 | 0.84 | 20.93 | 7 | 40 | 0.78 | 17.95 | 4 | 42 | 0.56 | 9.30 |
| SRAP7 | 4 | 82 | 0.68 | 9.30 | 5 | 58 | 0.62 | 12.82 | 7 | 82 | 0.74 | 16.28 |
| Total | 43 | 433 | --- | --- | 39 | 245 | --- | --- | 43 | 349 | --- | --- |
| Min | 3 | 34 | 0.47 | 6.98 | 3 | 21 | 0.46 | 7.69 | 3 | 20 | 0.27 | 6.98 |
| Max | 9 | 82 | 0.84 | 20.93 | 7 | 58 | 0.79 | 17.95 | 10 | 82 | 0.79 | 23.26 |
| Mean | 6.144 | 61.86 | 0.70 | 14.29 | 5.57 | 35.00 | 0.66 | 14.28 | 6.14 | 49.86 | 0.66 | 14.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alzahib, R.H.; Migdadi, H.M.; Ghamdi, A.A.A.; Alwahibi, M.S.; Afzal, M.; Elharty, E.H.; Alghamdi, S.S. Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers. Diversity 2021, 13, 135. https://doi.org/10.3390/d13030135
Alzahib RH, Migdadi HM, Ghamdi AAA, Alwahibi MS, Afzal M, Elharty EH, Alghamdi SS. Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers. Diversity. 2021; 13(3):135. https://doi.org/10.3390/d13030135
Chicago/Turabian StyleAlzahib, Reem H., Hussein M. Migdadi, Abdullah A. Al Ghamdi, Mona S. Alwahibi, Muhammad Afzal, Ehab H. Elharty, and Salem S. Alghamdi. 2021. "Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers" Diversity 13, no. 3: 135. https://doi.org/10.3390/d13030135
APA StyleAlzahib, R. H., Migdadi, H. M., Ghamdi, A. A. A., Alwahibi, M. S., Afzal, M., Elharty, E. H., & Alghamdi, S. S. (2021). Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers. Diversity, 13(3), 135. https://doi.org/10.3390/d13030135







