Genetic Diversity and Population Structure of Wild Beets (Beta spp.) from the Western Iberian Peninsula and the Azores and Madeira Islands
Abstract
:1. Introduction
2. Material and Methods
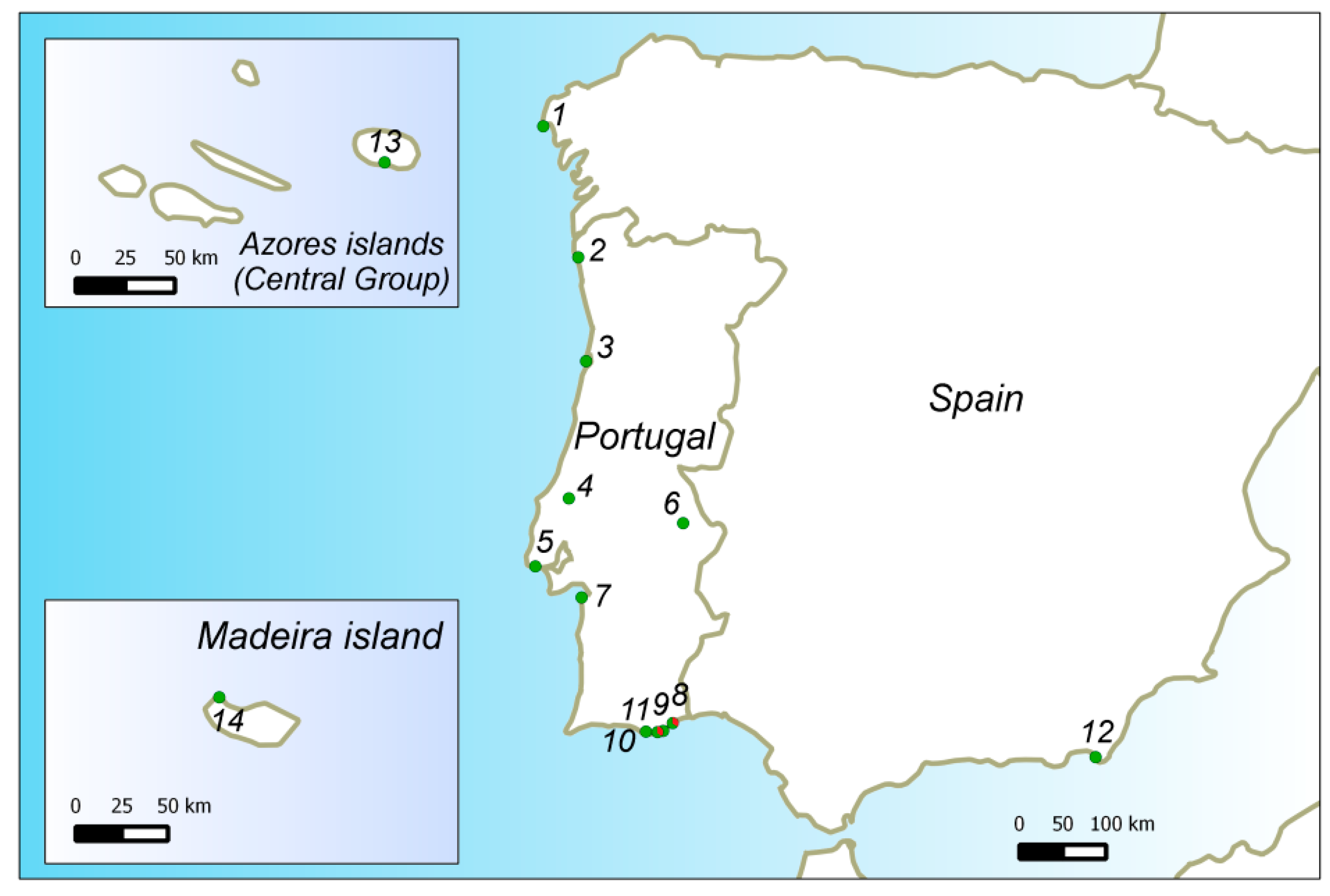
2.1. Plant Material and Sampling
2.2. DNA Extraction, PCR Amplification, and Fragments Sizing
2.3. Data Analysis
3. Results
3.1. Overall Genetic Diversity
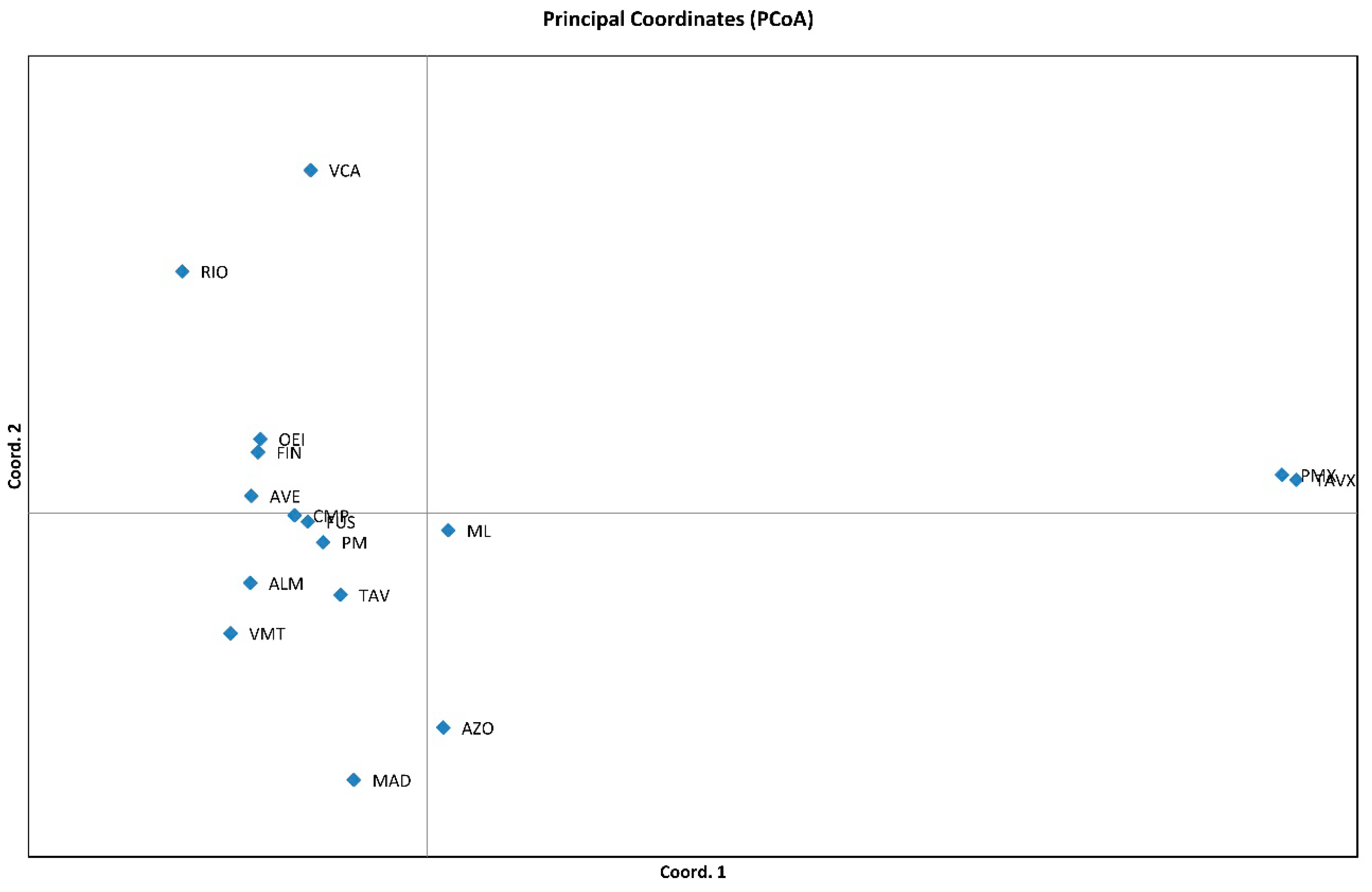
3.2. Genetic Relationships among Genotypes
3.3. Differentiation of the Populations
3.4. Genetic Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leys, M.; Petit, E.J.; El-Bahloul, Y.; Liso, C.; Fournet, S.; Arnaud, J. Spatial genetic structure in Beta vulgaris subsp. maritima and Beta macrocarpa reveals the effect of contrasting mating system, influence of marine currents, and footprints of postglacial recolonization routes. Ecol. Evol. 2014, 4, 1828–1852. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G. The Genetic Legacy of the Quaternary Ice Ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Frese, L.; Desprez, B.; Ziegler, D. Potential of genetic resources and breeding strategies for base-broadening in Beta. In Broadening the Genetic Base of Crop Production; Cooper, H.D., Spillane, C., Hodgkin, T., Eds.; CABI: Wallingford, UK, 2001; pp. 295–309. ISBN 978-0-85199-411-6. [Google Scholar]
- Biancardi, E.; Panella, L.W.; Lewellen, R.T. Beta maritima; Springer: New York, NY, USA, 2012; ISBN 978-1-4614-0841-3. [Google Scholar]
- Mörchen, M.; Cuguen, J.; Michaelis, G.; Hänni, C.; Saumitou-Laprade, P. Abundance and Length Polymorphism of Microsatellite Repeats in Beta vulgaris L. Theor. Appl. Genet. 1996, 92, 326–333. [Google Scholar] [CrossRef]
- Desplanque, B.; Boudry, P.; Broomberg, K.; Saumitou-Laprade, P.; Cuguen, J.; Van Dijk, H. Genetic Diversity and Gene Flow between Wild, Cultivated and Weedy Forms of Beta vulgaris L. (Chenopodiaceae), Assessed by RFLP and Microsatellite Markers. Theor. Appl. Genet. 1999, 98, 1194–1201. [Google Scholar] [CrossRef]
- Arnaud, J.-F.; Fénart, S.; Godé, C.; Deledicque, S.; Touzet, P.; Cuguen, J. Fine-Scale Geographical Structure of Genetic Diversity in Inland Wild Beet Populations. Mol. Ecol. 2009, 18, 3201–3215. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, I.C.; Pinheiro, C.; Ribeiro, C.M.; Veloso, M.M.; Simoes-Costa, M.C.; Evaristo, I.; Paulo, O.S.; Ricardo, C.P. Genetic Diversity and Physiological Performance of Portuguese Wild Beet (Beta vulgaris spp. maritima) from Three Contrasting Habitats. Front. Plant Sci. 2016, 7, 1293. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, F.; Romeiras, M.M.; Batista, D.; Duarte, M.C. Biodiversity Assessment of Sugar Beet Species and Its Wild Relatives: Linking Ecological Data with New Genetic Approaches. Am. J. Plant Sci. 2013, 04, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Carapeto, A.; Francisco, A.; Pereira, P.; Porto, M. (Eds.) Lista Vermelha Da Flora Vascular de Portugal Continental. Sociedade Portuguesa de Botânica, Associação Portuguesa de Ciência Da Vegetação PHYTOS e Instituto Da Conservação Da Natureza e Das Florestas (Coord.). Coleção «Botânica Em Português»; Imprensa Nacional: Lisboa, Portugal, 2000; Volume 7, ISBN 978-972-27-2876-8.
- Frese, L. Rationale for in Situ Management of Wild Beta Species. Crop Wild Relat. 2004, 2, 4–7. [Google Scholar]
- Bilz, M.; Kell, S.P.; Maxeted, N.; Landsdown, R.V. European Red List of Vascular Plants; Publications Office of the European Union: Luxembourg, 2011; ISBN 978-92-79-20199-8. [Google Scholar] [CrossRef]
- Buttler, K.P. Variation in Wild Populations of Annual Beet (Beta, Chenopodiaceae). Plant Syst. Evol. 1977, 128, 123–136. [Google Scholar] [CrossRef]
- Villain, S.; Touzet, P.; Cuguen, J. Reconstructing the Evolutionary History of Beta Section Beta with Molecular Data: A Focus on the Canary Islands. In Report of a Working Group on Beta and the World Beta Network, Proceedings of the Third Joint Meeting, Puerto Cruz, Spain, 8–11 March 2006; Bioversity International: Rome, Italy, 2009; pp. 31–37. Available online: http://www.ecpgr.cgiar.org/fileadmin/bioversity/publications/pdfs/1353_Report_Working_Group_Beta_World_Beta_Network.pdf (accessed on 18 July 2020).
- Abe, J.; Tsuda, C. Distorted Segregation in the Backcrossed Progeny between Beta vulgaris L. and B. macrocarpa Guss. Ikushugaku Zasshi 1988, 38, 309–318. [Google Scholar] [CrossRef] [Green Version]
- McFarlane, J.S. Naturally Occurring Hybrids between Sugarbeet and Beta macrocarpa in the Imperial Valley of California. J. Am. Soc. Sugar Beet Technol. 1975, 18, 245–251. Available online: https://agris.fao.org/agris-search/search.do?recordID=US19760083471 (accessed on 9 December 2020). [CrossRef]
- Castro, S.; Romeiras, M.M.; Castro, M.; Duarte, M.C.; Loureiro, J. Hidden Diversity in Wild Beta Taxa from Portugal: Insights from Genome Size and Ploidy Level Estimations Using Flow Cytometry. Plant Sci. 2013, 207, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Warschefsky, E.; Penmetsa, R.V.; Cook, D.R.; Wettberg, E.J.B. von Back to the Wilds: Tapping Evolutionary Adaptations for Resilient Crops through Systematic Hybridization with Crop Wild Relatives. Am. J. Bot. 2014, 101, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Paesold, S.; Borchardt, D.; Schmidt, T.; Dechyeva, D. A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. Plant J. 2012, 72, 600–611. [Google Scholar] [CrossRef]
- Del Rio, A.R.; Minoche, A.E.; Zwickl, N.F.; Friedrich, A.; Liedtke, S.; Schmidt, T.; Himmelbauer, H.; Dohm, J.C. Genomes of the wild beets Beta patula and Beta vulgaris ssp. maritima. Plant J. 2019, 99, 1242–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohm, J.C.; Minoche, A.E.; Holtgrawe, D.; Capella-Gutierrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R.; et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505, 546–549. [Google Scholar] [CrossRef] [Green Version]
- Funk, A.; Galewski, P.; McGrath, J.M. Nucleotide-binding resistance gene signatures in sugar beet, insights from a new reference genome. Plant J. 2018, 95, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Galewski, P.; McGrath, J.M. Genetic Diversity among Cultivated Beets (Beta vulgaris) Assessed via Population-Based Whole Genome Sequences. BMC Genom. 2020, 21, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrello, M.; Henry, K.; Devaux, P.; Verdelet, D.; Desprez, B.; Manel, S. Insights into the Genetic Relationships among Plants of Beta Section Beta Using SNP Markers. Theor. Appl. Genet. 2017, 130, 1857–1866. [Google Scholar] [CrossRef]
- Abou-Elwafa, S.F.; Amin, A.E.A.; Eujayl, I. Genetic diversity of sugar beet under heat stress and deficit irrigation. Agron. J. 2020, 112, 3579–3590. [Google Scholar] [CrossRef]
- Panella, L.; Lewellen, R.T. Broadening the Genetic Base of Sugar Beet: Introgression from Wild Relatives. Euphytica 2007, 154, 383–400. [Google Scholar] [CrossRef]
- Wu, G.-Q.; Wang, C.-M.; Su, Y.-Y.; Zhang, J.-J.; Feng, R.-J.; Liang, N. Assessment of Drought Tolerance in Seedlings of Sugar Beet (Beta vulgaris L.) Cultivars Using Inorganic and Organic Solutes Accumulation Criteria. Soil Sci. Plant Nutr. 2014, 60, 565–576. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, C.; Ribeiro, I.C.; Reisinger, V.; Planchon, S.; Veloso, M.M.; Renaut, J.; Eichacker, L.; Ricardo, C.P. Salinity Effect on Germination, Seedling Growth and Cotyledon Membrane Complexes of a Portuguese Salt Marsh Wild Beet Ecotype. Theor. Exp. Plant Physiol. 2018, 30, 113–127. [Google Scholar] [CrossRef]
- Duminil, J.; Hardy, O.J.; Petit, R.J. Plant Traits Correlated with Generation Time Directly Affect Inbreeding Depression and Mating System and Indirectly Genetic Structure. BMC Evol. Biol. 2009, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sánchez, F.; Pérez-Barrales, R.; Ojeda, F.; Vargas, P.; Arroyo, J. The Strait of Gibraltar as a Melting Pot for Plant Biodiversity. Quat. Sci. Rev. 2008, 27, 2100–2117. [Google Scholar] [CrossRef]
- Fénart, S.; Arnaud, J.-F.; De Cauwer, I.; Cuguen, J. Nuclear and Cytoplasmic Genetic Diversity in Weed Beet and Sugar Beet Accessions Compared to Wild Relatives: New Insights into the Genetic Relationships within the Beta vulgaris Complex Species. Theor. Appl. Genet. 2008, 116, 1063–1077. [Google Scholar] [CrossRef]
- Richards, C.M.; Reeves, P.A.; Fenwick, A.L.; Panella, L. Genetic Structure and Gene Flow in Beta vulgaris subspecies maritima along the Atlantic Coast of France. Genet. Resour. Crop Evol. 2014, 61, 651–662. [Google Scholar] [CrossRef]
- Fievet, V.; Touzet, P.; Arnaud, J.-F.; Cuguen, J. Spatial Analysis of Nuclear and Cytoplasmic DNA Diversity in Wild Sea Beet (Beta vulgaris ssp. maritima) Populations: Do Marine Currents Shape the Genetic Structure? Mol. Ecol. 2007, 16, 1847–1864. [Google Scholar] [CrossRef]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape Genetics: Combining Landscape Ecology and Population Genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Smulders, M.J.; Esselink, G.D.; Everaert, I.; De Riek, J.; Vosman, B. Characterisation of Sugar Beet (Beta vulgaris L. ssp. vulgaris) Varieties Using Microsatellite Markers. BMC Genet. 2010, 11, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, P.G.; Snow, A.A.; Schug, M.D.; Booton, G.C.; Fuerst, P.A. What Molecules Can Tell Us About Populations: Choosing and Using a Molecular Marker. Ecology 1998, 79, 361–382. [Google Scholar] [CrossRef]
- Abbasi, Z.; Majidi, M.M.; Arzani, A.; Rajabi, A.; Mashayekhi, P.; Bocianowski, J. Association of SSR Markers and Morpho-Physiological Traits Associated with Salinity Tolerance in Sugar Beet (Beta vulgaris L.). Euphytica 2015, 205, 785–797. [Google Scholar] [CrossRef]
- Hawkes, J.G.; Maxted, N.; Ford-Lloyd, B.V. The Ex Situ Conservation of Plant Genetic Resources; Springer: Amsterdam, The Netherlands, 2000; ISBN 978-0-7923-6442-9. [Google Scholar]
- Richards, C.M.; Brownson, M.; Mitchell, S.E.; Kresovich, S.; Panella, L. Polymorphic Microsatellite Markers for Inferring Diversity in Wild and Domesticated Sugar Beet (Beta vulgaris). Mol. Ecol. Notes 2004, 4, 243–245. [Google Scholar] [CrossRef]
- McGrath, J.M.; Trebbi, D.; Fenwick, A.; Panella, L.; Schulz, B.; Laurent, V.; Barnes, S.; Murray, S.C. An Open-Source First-Generation Molecular Genetic Map from a Sugarbeet × Table Beet Cross and Its Extension to Physical Mapping. Crop Sci. 2007, 47, S27–S44. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberto, F. StandArich_v1.00: An R Package to Estimate Population Allelic Richness Using Standardized Sample Size; University of Algarve: Faro, Portugal, 2006. [Google Scholar]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Del Carpio, P.D.; Basnet, R.K.; De Vos, R.C.H.; Maliepaard, C.; Visser, R.; Bonnema, G. The Patterns of Population Differentiation in a Brassica rapa Core Collection. Theor. Appl. Genet. 2011, 122, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software, Version 6. Available online: https://darwin.cirad.fr/ (accessed on 18 July 2020).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software Structure: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holtsford, T.P.; Ellstrand, N.C. Variation in Outcrossing Rate and Population Genetic Structure of Clarkia tembloriensis (Onagraceae). Theor. Appl. Genet. 1989, 78, 480–488. [Google Scholar] [CrossRef]
- Andrello, M.; Henry, K.; Devaux, P.; Desprez, B.; Manel, S. Taxonomic, Spatial and Adaptive Genetic Variation of Beta Section Beta. Theor. Appl. Genet. 2016, 129, 257–271. [Google Scholar] [CrossRef]
- Cao, Q.; Lu, B.-R.; Xia, H.; Rong, J.; Sala, F.; Spada, A.; Grassi, F. Genetic Diversity and Origin of Weedy Rice (Oryza sativa f. spontanea) Populations Found in North-Eastern China Revealed by Simple Sequence Repeat (SSR) Markers. Ann. Bot. 2006, 98, 1241–1252. [Google Scholar] [CrossRef] [Green Version]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and Speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Wang, Z.; Kang, X.; Zhang, J. Genetic Analysis of Admixture and Hybrid Patterns of Populus hopeiensis and P. tomentosa. Sci. Rep. 2019, 9, 4821. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zheng, Q.-J.; Qian, Z.-Q.; Yang, J.; Zhang, Y.-P.; Li, Z.-H.; Zhao, G.-F. Genetic Structure and Evolutionary History of Three Alpine Sclerophyllous Oaks in East Himalaya-Hengduan Mountains and Adjacent Regions. Front. Plant Sci. 2016, 7, 1688. [Google Scholar] [CrossRef] [Green Version]
- Mallet, J. Hybridization as an Invasion of the Genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef]
- Van Geyt, J.P.C.; Lange, W.; Oleo, M.; De Bock, T.S.M. Natural Variation within the Genus Beta and Its Possible Use for Breeding Sugar Beet: A Review. Euphytica 1990, 49, 57–76. [Google Scholar] [CrossRef]
- Petit, R.J.; Excoffier, L. Gene Flow and Species Delimitation. Trends Ecol. Evol. 2009, 24, 386–393. [Google Scholar] [CrossRef]
- Rius, M.; Darling, J.A. How Important Is Intraspecific Genetic Admixture to the Success of Colonising Populations? Trends Ecol. Evol. 2014, 29, 233–242. [Google Scholar] [CrossRef]
- Keller, S.R.; Taylor, D.R. Genomic Admixture Increases Fitness during a Biological Invasion. J. Evol. Biol. 2010, 23, 1720–1731. [Google Scholar] [CrossRef]
- Roussel, V.; Koenig, J.; Beckert, M.; Balfourier, F. Molecular Diversity in French Bread Wheat Accessions Related to Temporal Trends and Breeding Programmes. Theor. Appl. Genet. 2004, 108, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Semagn, K.; Magorokosho, C.; Ogugo, V.; Makumbi, D.; Warburton, M.L. Genetic Relationships and Structure among Open-Pollinated Maize Varieties Adapted to Eastern and Southern Africa Using Microsatellite Markers. Mol. Breed. 2014, 34, 1423–1435. [Google Scholar] [CrossRef]
- Oliveira, H.R.; Tomás, D.; Silva, M.; Lopes, S.; Viegas, W.; Veloso, M.M. Genetic Diversity and Population Structure in Vicia faba L. Landraces and Wild Related Species Assessed by Nuclear SSRs. PLoS ONE 2016, 11, e0154801. [Google Scholar] [CrossRef] [Green Version]
- Meirmans, P.G.; Hedrick, P.W. Assessing Population Structure: FST and Related Measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi, S.A.; Prasanna, B.M. Analysis of Genetic Diversity in Crop Plants—Salient Statistical Tools and Considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, F.; Vidigal, P.; Barros, A.B.; Monteiro, A.; Oliveira, H.R.; Viegas, W. Genetic Distinctiveness of Rye In Situ Accessions from Portugal Unveils a New Hotspot of Unexplored Genetic Resources. Front. Plant Sci. 2016, 7, 1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saccomani, M.; Stevanato, P.; Trebbi, D.; McGrath, J.M.; Biancardi, E. Molecular and Morpho-Physiological Characterization of Sea, Ruderal and Cultivated Beets. Euphytica 2009, 169, 19–29. [Google Scholar] [CrossRef]
- Ennos, R.A. Estimating the Relative Rates of Pollen and Seed Migration among Plant Populations. Heredity 1994, 72, 250–259. [Google Scholar] [CrossRef] [Green Version]
- Petit, R.J.; Mousadik, A.E.; Pons, O. Identifying Populations for Conservation on the Basis of Genetic Markers. Conserv. Biol. 1998, 12, 844–855. [Google Scholar] [CrossRef]

| Locus | Na | Ne | Ho | He | PIC | FST |
|---|---|---|---|---|---|---|
| SB04 | 14 | 3.03 | 0.590 | 0.651 | 0.731 | 0.202 |
| SB06 | 10 | 3.55 | 0.610 | 0.681 | 0.808 | 0.233 |
| SB07 | 23 | 5.43 | 0.610 | 0.769 | 0.898 | 0.216 |
| SB13 | 7 | 2.19 | 0.440 | 0.536 | 0.559 | 0.191 |
| SB15 | 21 | 3.52 | 0.635 | 0.667 | 0.812 | 0.225 |
| BQ588629 | 25 | 3.90 | 0.669 | 0.694 | 0.832 | 0.254 |
| Population | N | Na | Ne | Npa | Ar | Ho | He | F |
|---|---|---|---|---|---|---|---|---|
| FIN | 30 | 6.500 | 3.036 | 3 | 6.667 | 0.547 | 0.613 | 0.108 |
| VCA | 28 | 4.833 | 2.052 | 0 | 4.833 | 0.500 | 0.507 | 0.014 |
| AVE | 23 | 6.000 | 3.091 | 2 | 6.167 | 0.658 | 0.658 | 0.000 |
| RIO | 28 | 6.167 | 2.928 | 0 | 6.167 | 0.649 | 0.652 | 0.005 |
| VMT | 31 | 7.667 | 4.227 | 4 | 7.667 | 0.634 | 0.739 | 0.142 |
| OEI | 35 | 9.000 | 4.749 | 1 | 9.167 | 0.709 | 0.738 | 0.039 |
| CMP | 30 | 10.000 | 5.316 | 3 | 10.167 | 0.783 | 0.786 | 0.004 |
| TAV | 15 | 7.666 | 5.316 | 4 | 7.667 | 0.782 | 0.762 | 0.054 |
| FUS | 34 | 8.667 | 5.123 | 1 | 8.667 | 0.750 | 0.774 | 0.031 |
| ML | 30 | 8.333 | 5.022 | 2 | 8.333 | 0.650 | 0.789 | 0.176 |
| PM | 10 | 6.833 | 5.111 | 0 | 7.000 | 0.783 | 0.785 | 0.105 |
| ALM | 27 | 8.833 | 5.079 | 5 | 8.333 | 0.630 | 0.778 | 0.190 |
| AZO | 11 | 4.333 | 2.688 | 0 | 4.333 | 0.561 | 0.615 | 0.088 |
| MAD | 9 | 4.500 | 3.394 | 0 | 4.500 | 0.630 | 0.710 | 0.113 |
| TAVX | 30 | 2.500 | 0.899 | 1 | 2.500 | 0.044 | 0.074 | 0.287 |
| PMX | 19 | 2.333 | 1.092 | 0 | 2.333 | 0.088 | 0.084 | −0.041 |
| Source of Variation | df | Sum of Squares | Variance Components | Variation (%) |
|---|---|---|---|---|
| All populations | ||||
| Among populations | 15 | 397.135 | 0.505 | 21 |
| Among individuals | 374 | 771.772 | 0.161 | 7 |
| Within individuals | 390 | 679.000 | 1.741 | 72 |
| Northern populations | ||||
| Among populations | 2 | 61.238 | 0.533 | 23 |
| Among individuals | 78 | 146.824 | 0.102 | 4 |
| Within individuals | 81 | 136.000 | 1.679 | 73 |
| Southern populations | ||||
| Among populations | 6 | 41.988 | 0.088 | 4 |
| Among individuals | 174 | 442.261 | 0.207 | 8 |
| Within individuals | 181 | 385.000 | 2.127 | 88 |
| Algarve populations | ||||
| Among populations | 5 | 140.259 | 0.588 | 27 |
| Among individuals | 132 | 234.969 | 0.169 | 8 |
| Within individuals | 138 | 199.000 | 1.442 | 65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veloso, M.M.; Simões-Costa, M.C.; Guimarães, J.B.; Ribeiro, C.M.; Evaristo, I.; Espírito-Santo, D.; Pinto-Ricardo, C.; Paulo, O.S.; Duarte, M.C. Genetic Diversity and Population Structure of Wild Beets (Beta spp.) from the Western Iberian Peninsula and the Azores and Madeira Islands. Diversity 2021, 13, 593. https://doi.org/10.3390/d13110593
Veloso MM, Simões-Costa MC, Guimarães JB, Ribeiro CM, Evaristo I, Espírito-Santo D, Pinto-Ricardo C, Paulo OS, Duarte MC. Genetic Diversity and Population Structure of Wild Beets (Beta spp.) from the Western Iberian Peninsula and the Azores and Madeira Islands. Diversity. 2021; 13(11):593. https://doi.org/10.3390/d13110593
Chicago/Turabian StyleVeloso, Maria Manuela, Maria Cristina Simões-Costa, Joana Bagoin Guimarães, Carla Marques Ribeiro, Isabel Evaristo, Dalila Espírito-Santo, Cândido Pinto-Ricardo, Octávio S. Paulo, and Maria Cristina Duarte. 2021. "Genetic Diversity and Population Structure of Wild Beets (Beta spp.) from the Western Iberian Peninsula and the Azores and Madeira Islands" Diversity 13, no. 11: 593. https://doi.org/10.3390/d13110593
APA StyleVeloso, M. M., Simões-Costa, M. C., Guimarães, J. B., Ribeiro, C. M., Evaristo, I., Espírito-Santo, D., Pinto-Ricardo, C., Paulo, O. S., & Duarte, M. C. (2021). Genetic Diversity and Population Structure of Wild Beets (Beta spp.) from the Western Iberian Peninsula and the Azores and Madeira Islands. Diversity, 13(11), 593. https://doi.org/10.3390/d13110593








