A New Shrimp Genus (Crustacea: Decapoda) from the Deep Atlantic and an Unusual Cleaning Mechanism of Pelagic Decapods
Abstract
:1. Introduction
2. Methods
2.1. Morphological Analysis
2.2. Molecular Analyses
3. Results
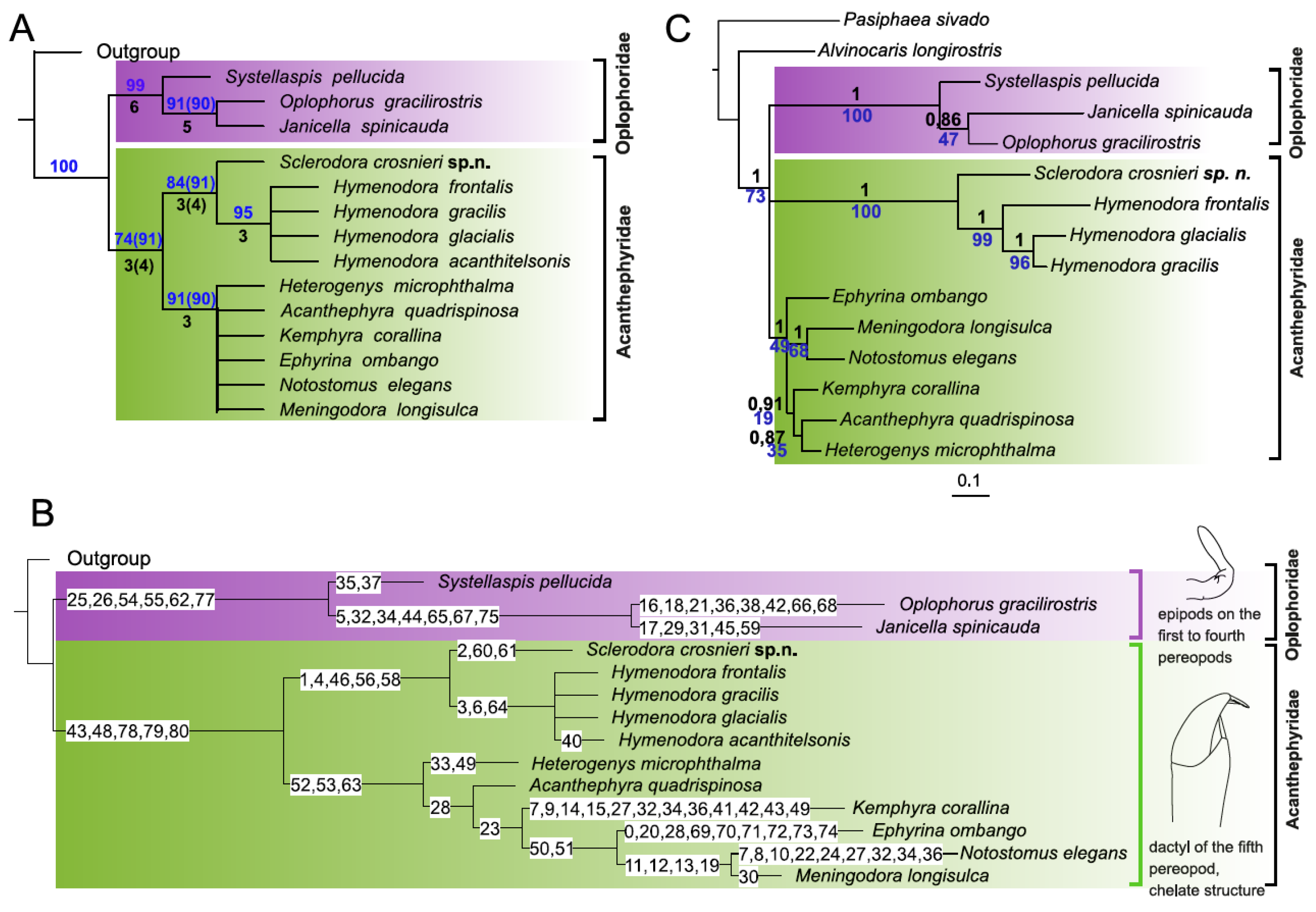
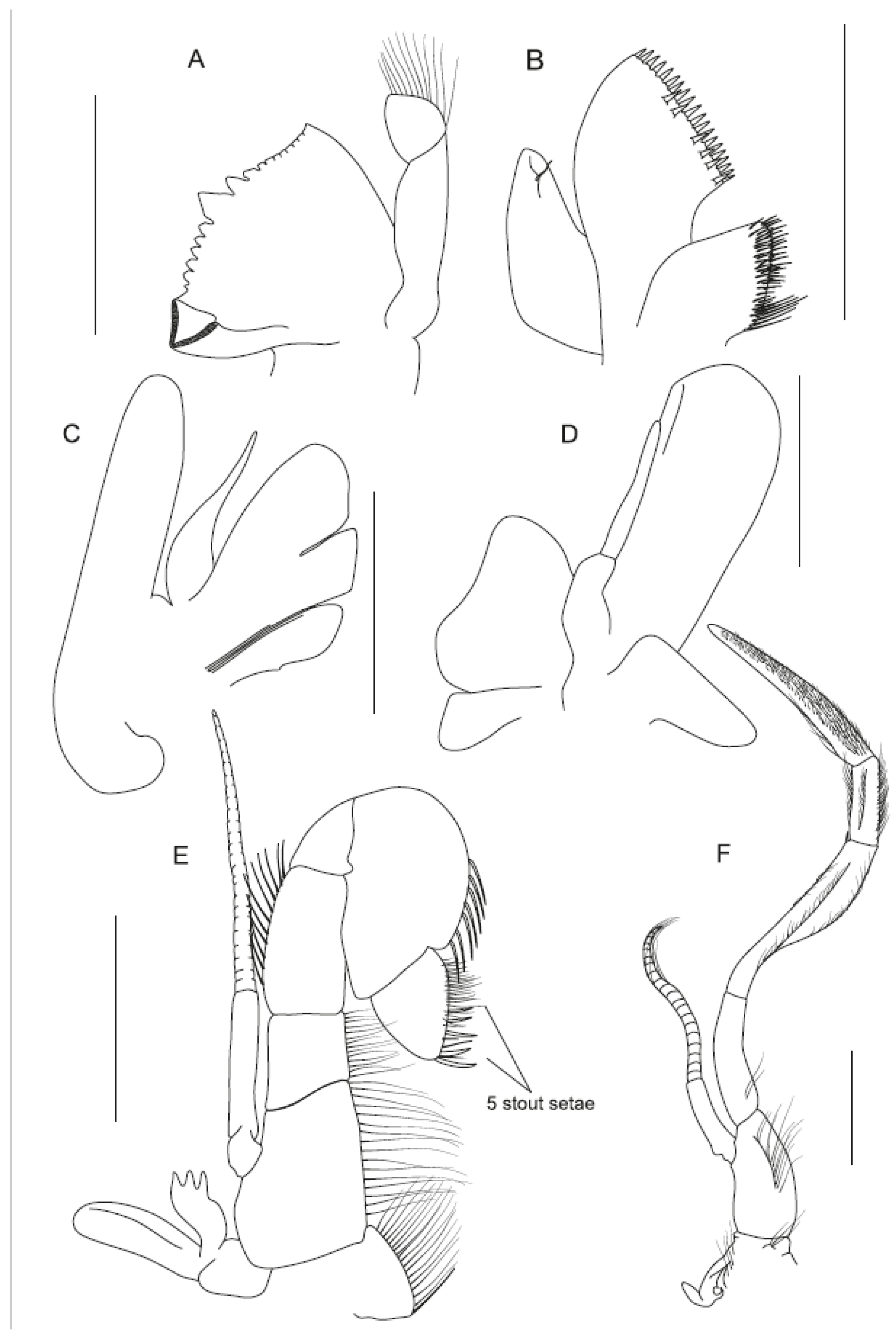
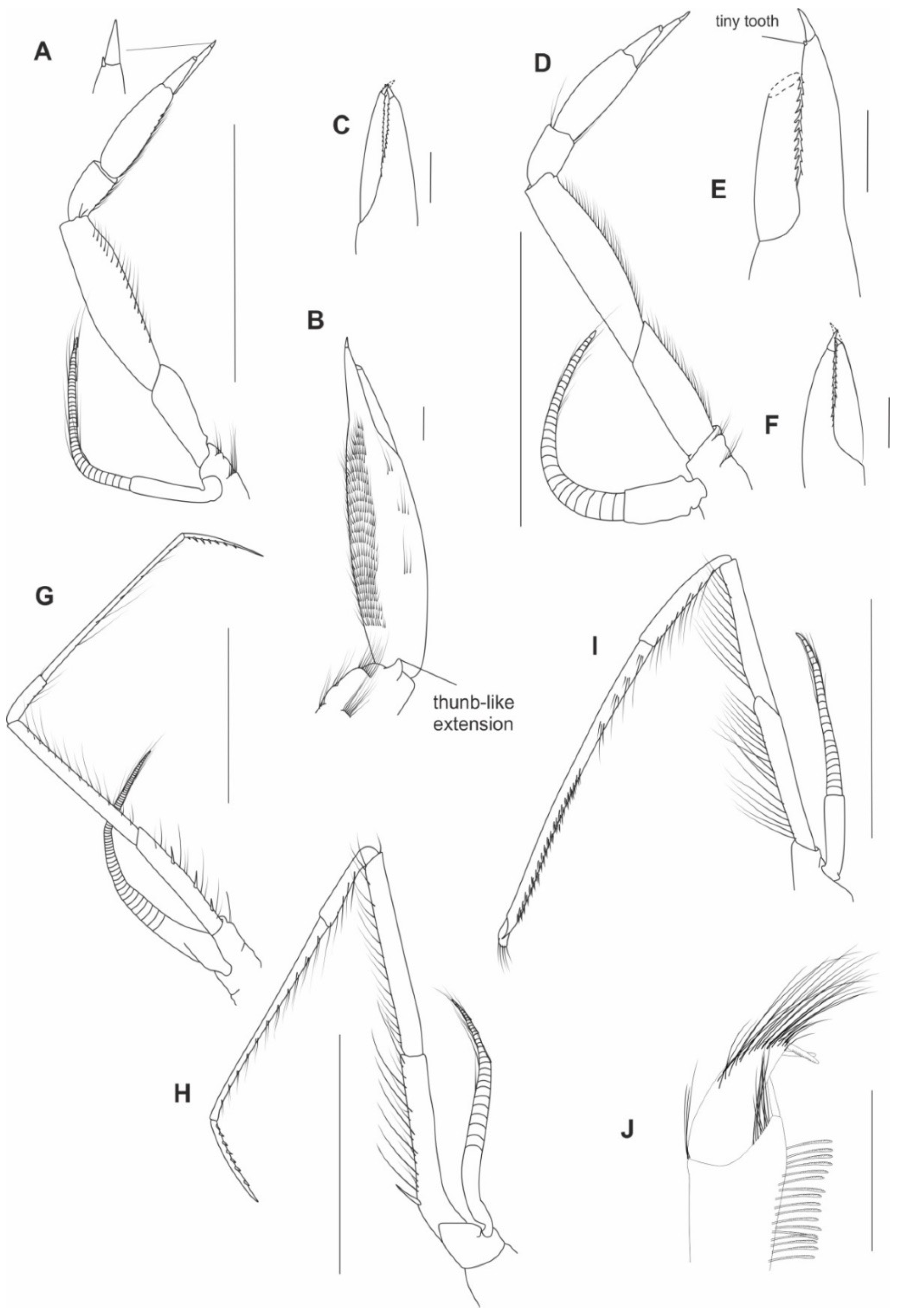
3.1. Morphological Analyses and Supporting Synapomorphies
3.2. Molecular Analyses
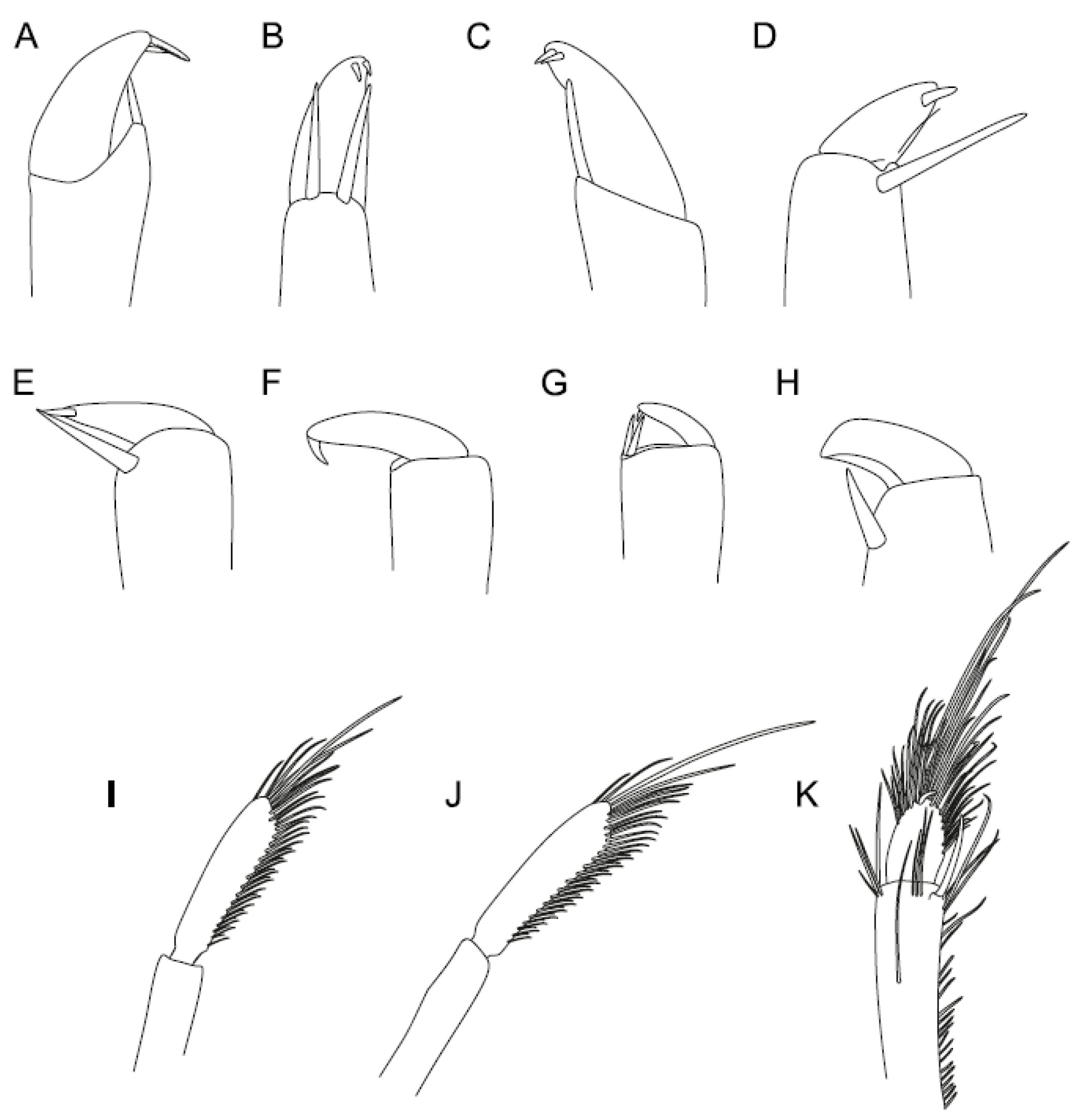
4. Discussion
4.1. Taxonomic Implication
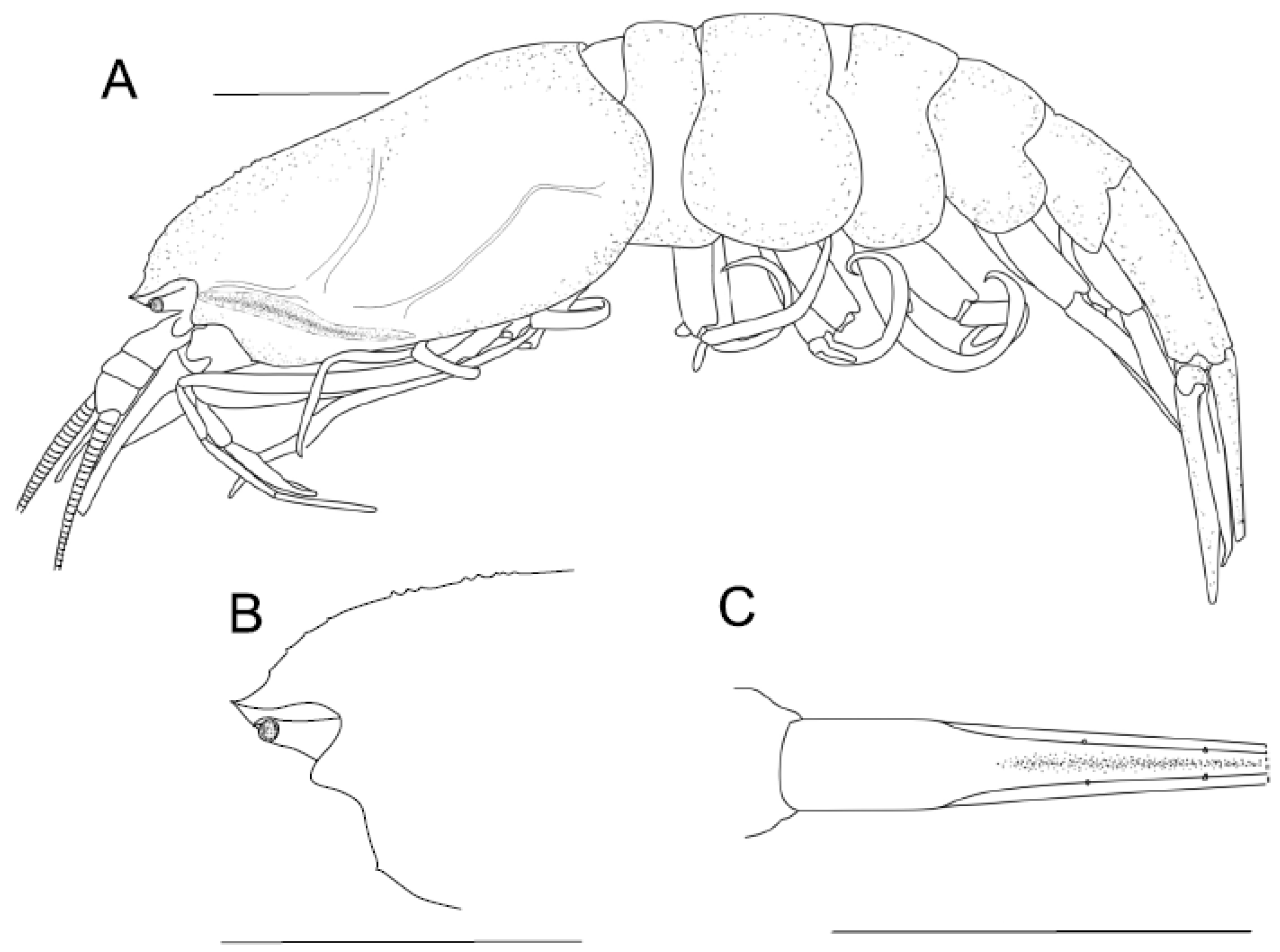
4.1.1. Sclerodora gen. nov.
4.1.2. Sclerodora crosnieri sp. nov.
4.1.3. Key to Genera of Oplophoroidea
| 1. Sixth abdominal somite with distinct dorsal carina………………………………………….2 |
| - Sixth abdominal somite dorsally smooth……………………………………………………….6 |
| 2. Hepatic spine present…………………………………………………....Kemphyra Chace, 1986 |
| - Hepatic spine absent……………………………………………………………………………....3 |
| 3. Third abdominal somite with long dorsal tooth overreaching fourth somite……………... ……………………………………………………………………………...Heterogenys Chace, 1986 |
| - Tooth on third abdominal somite, if present, not overreaching fourth somite…………….4 |
| 4. Carapace dorsally denticulate over nearly entire length; first abdominal somite dorsally carinate……………………………………………………...Notostomus A. Milne-Edwards, 1881 |
| - Carapace dorsally not denticulate on posterior half; first abdominal somite smooth…….5 |
| 5. A single continuous lateral carina on carapace (extending from near orbit to near posterior margin on carapace…………………………………………..Meningodora Smith, 1882 |
| - None or two continuous lateral carinae on carapace (one extending from near orbit, another extending from near branchiostegal spine) …………………………………………….. ……………………………………………………………...Acanthephyra A. Milne-Edwards, 1881 |
| 6. Rostrum unarmed. Meri and ischia of pereopods greatly wide and compressed ………... ………………………………………………………………………………...Ephyrina Smith, 1885 |
| - Rostrum denticulate. Meri and ischia of pereopods not greatly wide and compressed…..7 |
| 7. Rostral teeth subuliform, spaced from each other. Cornea subequal or narrower than eyestalk……………………………………………………………………………………………….8 |
| - Rostral teeth subtriangular, extending from a common crest. Cornea wider than eyestalk ………………………………………………………………………………………………………....9 |
| 8. Dorsal subuliform teeth only on rostrum…………………………………………………….... ………………………………………………………………………....Hymenodora G.O. Sars, 1877 |
| - Dorsal subuliform teeth both on rostrum and anterior part of carapace …………………... ……………………………………………………………………………………Sclerodora gen.nov. |
| 9. Carapace strongly chitinized, subtriangular in cross-section. Abdomen with sixth somite not longer than fifth, third to fourth somites with strong dorsomedial spines (at least ½ of segment length)……………………………………………………………………………………10 |
| - Carapace moderately chitinized, suboval in cross-section. Abdomen with sixth somite nearly twice as long as fifth, third to fourth somites without strong dorsomedial spines ………………………………………………………………………..Systellaspis Spence Bate, 1888 |
| 10. Second abdominal somite with strong dorsomedial spine.…………Janicella Chace, 1986 |
| - Second abdominal somite without strong dorsomedial spine ……………………………..... ………………………………………………………………….......Oplophorus H. Milne Edwards |
4.2. A New Suggested Cleaning and Grooming Mechanism
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dawson, M.N. Species richness, habitable volume, and species densities in freshwater, the sea, and on land. Front. Biogeogr. 2012, 4. [Google Scholar] [CrossRef] [Green Version]
- Costello, M.J.; Chaudhary, C. Marine biodiversity, biogeography, deep-sea gradients, and conservation. Curr. Biol. 2017, 27, 511–527. [Google Scholar] [CrossRef] [PubMed]
- WoRMS Editorial Board. World Register of Marine Species. 2021. Available online: http://www.marinespecies.org (accessed on 4 October 2021). [CrossRef]
- Bauer, R.T. Decapod crustacean grooming: Functional morphology, adaptive value, and phylogenetic significance. In Functional Morphology of Feeding and Grooming in Crustacea; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Bauer, R.T. Antifouling adaptations of caridcan shrimp (Decapoda: Caridea): Gill cleaning mechanisms and grooming of brooded embryos. Zool. J. Linn. Soc. 1979, 65, 281–303. [Google Scholar] [CrossRef]
- Bauer, R.T. Grooming behavior and morphology in the decapod Crustacea. J. Crust. Biol. 1981, 1, 153–173. [Google Scholar] [CrossRef]
- Wong, J.M.; Pérez-Moreno, J.L.; Chan, T.Y.; Frank, T.M.; Bracken-Grissom, H.D. Phylogenetic and transcriptomic analyses reveal the evolution of bioluminescence and light detection in marine deep-sea shrimps of the family Oplophoridae (Crustacea: Decapoda). Mol. Phylogenet. Evol. 2015, 83, 278–292. [Google Scholar] [CrossRef]
- Chace, F.A. The Caridean Shrimps (Crustacea: Decapoda) of the Albatross Philippine Expedition, 1907–1910, Part 4: Families Oplophoridae and Nematocarcinidae; Smithsonian Institution Press: Washington, DC, USA, 1986; p. 81. [Google Scholar]
- Lunina, A.; Vereshchaka, A. The role of the male copulatory organs in the colonization of the pelagic by shrimp-like eucarids. Deep Sea Res. Part II Top. Stud. Oceanogr. 2017, 137, 327–334. [Google Scholar] [CrossRef]
- Lunina, A.A.; Kulagin, D.N.; Vereshchaka, A.L. Oplophoridae (Decapoda: Crustacea): Phylogeny, taxonomy and evolution studied by a combination of morphological and molecular methods. Zool. J. Linn. Soc. 2019, 186, 213–232. [Google Scholar] [CrossRef]
- Lunina, A.A.; Kulagin, D.N.; Vereshchaka, A.L. Phylogenetic revision of the shrimp genera Ephyrina, Meningodora and Notostomus (Acanthephyridae: Caridea). Zool. J. Linn. Soc. 2020, zlaa161. [Google Scholar] [CrossRef]
- Li, C.P.; De Grave, S.; Chan, T.Y.; Lei, H.C.; Chu, K.H. Molecular systematics of caridean shrimps based on five nuclear genes: Implications for superfamily classification. Zool. Anz. 2011, 250, 270–279. [Google Scholar] [CrossRef]
- Nixon, K. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics 1999, 15, 407–414. [Google Scholar] [CrossRef]
- Goloboff, P.; Farris, S.; Nixon, K. TNT: Tree Analysis Using New Technology. 2000. Available online: http://www.lillo.org.ar/phylogeny/tnt (accessed on 24 October 2021).
- Schubart, C.D.; Huber, M.G.J. Genetic comparisons of German populations of the stone crayfish, Austropotamobius torrentium (Crustacea: Astacidae). Bull. Français Pêche Piscic. 2006, 380–381, 1019–1028. [Google Scholar] [CrossRef] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Schubart, C.D. Mitochondrial DNA and decapod phylogenies: The importance of pseudogenes and primer optimization. Crustacean 2009, 18, 47–65. [Google Scholar]
- Schubart, C.D.; Cuesta, J.A.; Felder, D.L. Glyptograpsidae, a new brachyuran family from Central America: Larval and adult morphology, and a molecular phylogeny of the Grapsoidea. J. Crust. Biol. 2002, 22, 28–44. [Google Scholar] [CrossRef] [Green Version]
- Reuschel, S.; Schubart, C.D. Phylogeny and geographic differentiation of Atlanto–Mediterranean species of the genus Xantho (Crustacea: Brachyura: Xanthidae) based on genetic and morphometric analyses. Mar. Biol. 2006, 148, 853–866. [Google Scholar] [CrossRef]
- Apakupakul, K.; Siddall, M.E.; Burreson, E.M. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Mol. Phylogenetics Evol. 1999, 12, 350–359. [Google Scholar] [CrossRef] [Green Version]
- Colgan, D.J.; McLauchlan, A.; Wilson, G.D.F.; Livingston, S.P.; Edgecombe, G.D.; Macaranas, J.; Gray, M.R. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust. J. Zool. 1998, 46, 419–437. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0 beta: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
| Species | Coordinates | Other Information | Museum, Number |
|---|---|---|---|
| Acanthephyra quadrispinosa | 29°39′ S, 44°16′ E | Expedition ATIMO VATAE. SUD MADAGASCAR, Sud Pointe Barrow. Chaultier “Nosy Be 11”, Stn. CP 3596, 986–911 m. 12.05.2010. | MNHN-IU-2010-4285 |
| Acanthephyra acutifrons | 14°43′ N, 45° 02′ W | “Professor Logatchev” 39 cruise St 215 RT, RTAK | IO RAN 39L 215 RT №1 |
| Ephyrina ombango | 10°23,17′ N, 46°45,34′ W | DEMERABY, CP07, chalutage 4850 m. 20.09.80 | MNHN-IU-2018-1579 |
| Ephyrina ombango | 9°18′ S, 11°10′ E | “Ombango”, C14, St.325,midwater traul, 0–725 m, 02.03.1961, 23h00–23h15 | MNHN-IU-2014-11098 |
| Heterogenis microphtalma | No data | Collection de S.A.S.le Prince de Monaco, Station 7/3. №12h, 16–19.8.96. Chal 4360 m | MNHN-IU-2018-1578 |
| Hymenodora acanthitelsonis | 45°18′ N, 125°43′ W–45°17′ N, 125°49′ W | Pacific Ocean, Unated States, Oregon, W of Pacific City. Yaqina BMT.189, 18.03.1970. | USNM 137500 |
| Hymenodora glacialis | 02°03′ S, 118°45′ E | Indonesie, CORINDON -Makassar. St CH286, 1710–1730 m | Na 10655 |
| Hymenodora glacialis | 73°28′ N, 10°07′ W | Mer de Norvege, Campagne NORBI, N.O. “Jean Charcot”, Stn CP16, 2937 m, 07.08.1975 | MNHN-IU-2008-16833 |
| Hymenodora gracilis | 37°39′ S, 77°26′ E | Ile Amsterdam, Campagne Jasus (MD 50), N.O. “Marion Dufresne”, Stn CP193, 2800–3075 m. 27.06.1986 | MNHN-IU-2008-16839 |
| Hymenodora frontalis | 15° N, 45° W | ROV “Vityaz”, 59 th cruise, st. 7497, № 271. 18.06.1976, 1500–2500 m | |
| Janicella spinicauda | 1°28′ S, 48°06′ E | ROV “Vityaz”, 17th cruise, St. 2604,13.11.88, 670–690 m | ZMUK |
| Janicella spinicauda | 8°44′ S, 43°54′ E | Dana Expedition, St. 3939-1, 23.12.1929, 500 meter wire | ZMUK |
| Kemphyra corallina | 37°54′ S, 77°22′ E | Iles St Paul et Amsterdam, “Marion Dufresne” Cne MD Jasus Stn CP 56. 2280–2310 m. 14.07.1986. 20h02–22h31 | MNHN-IU-2018-1581 |
| Kemphyra corallina | 33°59′ S, 43°55′ E | Indian Ocean: Walters shoal, Plaine Sud. N.O. “Marion Dufresne”, Campagne MD208(Walters Shoal). Stn CP49156 1865–2058 m, 12.05.2017 | MNHN-IU-2016-9402 |
| Meningodora longiscula | 9°55′ N, 142°00′ E | Nouvelle-Caledonie, Campagne Caride V. Stn 15, 1000 m. 12.09.1969 | MNHN-IU-2011-5635 |
| Notostomus elegans | 37 cruise RV Logatchev, St 156 TS | IO RAN | |
| Oplophorus gracilirostris | 25°11′ N, 122°35′ E | Dana Expedition, St. 3722-3, 300 m wire | ZMUK |
| Oplophorus gracilirostris | 20°08′ N, 82°59′ W | Dana Expedition, St. 1218, 800 m wire | ZMUK |
| Oplophorus gracilirostris | 12°30′ S, 48°16′ E | ROV “Vityaz”, 17th cruise, St. 2597, 12.11.88, 360–555 m wire. | ZMUK |
| Oplophorus gracilirostris | 22°06′ N, 84°58′ W | Dana Expedition, St. 1223, 500 m wire | ZMUK |
| Pasiphaea sivado | 35°47′ N, 05°17′ W | Detroit de Gibraltar, N.O. “Cryos”, BALGIM, St. CP150, 280–300 m, 18.06.1984 | MNHN-IU-2018-1611 |
| Sclerodora crosnierisp.nov. | 16° N, 46° W | 39th Cruise of R/V “Professor Logatchev”, March 2018 | ZMUK |
| Systellaspis pellucida | 12°30′ S, 48°16′ E | Indian Ocean. North end of Madagascar. ROV “Vityaz”, 17th cruise, St. 2597, 360–555 m | IO RAN |
| Systellaspis pellucida | 25°11′ N, 122°35′ E | North Western Pacific Ocean. S.E. and E. of Formosa. Dana Expedition 3722(2) 29.05.1929, 600 mw | ZMUK |
| Systellaspis pellucida | 25°11′ N, 122°35′ E | North Western Pacific Ocean. S.E. and E. of Formosa. Dana Expedition 3722(1) 29.05.1929, 1000 mw | ZMUK |
| Taxon | GenBank Accession Numbers | Source | |||
|---|---|---|---|---|---|
| COI | 16S | 18S | H3 | ||
| Outgroup taxa | |||||
| Pasiphaeoidea Dana, 1852 | |||||
| Pasiphaeidae Dana, 1852 | |||||
| Pasiphaea sivado (Risso, 1816) | KP759487 | KP725629 | KP725826 | MF279416 | Aznar-Cormano et al., 2015; Liao et al., 2017 |
| Bresilioidea Calman, 1896 | |||||
| Alvinocarididae Christoffersen, 1986 | |||||
| Alvinocaris longirostris Kikuchi & Ohta, 1995 | KP215329 | KP215285 | KP215300 | KP215342 | Aznar-Cormano et al., 2015 |
| Ingroup taxa | |||||
| Oplophoroidea Dana, 1852 | |||||
| Oplophoridae Dana, 1852 | |||||
| Janicella spinicauda (A. Milne-Edwards, 1883) | MH572546 | KP075932 | MH100869 | MH107256 | Wilkins and Bracken-Grissom, 2020 (GenBank); Wong et al., 2015; Lunina et al., 2019 |
| Oplophorus gracilirostris A. Milne-Edwards, 1881 | KP076150 | KP075920 | KP075847 | KP076072 | Wong et al., 2015 |
| Systellaspis pellucida (Filhol, 1884) | JQ306184 | KP075925 | JF346250 | KP076077 | Matzen da Silva et al., 2011; Wong et al., 2015; Li et al., 2011 |
| Acanthephyridae Spence Bate, 1888 | |||||
| Acanthephyra quadrispinosa Kemp, 1939 | KP759363 | KP725479 | KP725677 | KP726051 | Aznar-Cormano et al., 2015 |
| Ephyrina ombango Crosnier & Forest, 1973 | MW043004 | MW043448 | MW043463 | MW052289 | Lunina et al., 2020 |
| Heterogenys microphthalma (Smith, 1885) | KP076183 | KP075898 | KP075787 | KP076124 | Wong et al., 2015 |
| Kemphyra corallina (A.Milne-Edwards, 1883) | MW043006 | MW043450 | MW043465 | MW052291 | Lunina et al., 2020 |
| Meningodora longisulca Kikuchi, 1985 | MW043007 | MW043451 | MW043466 | MW052292 | Lunina et al., 2020 |
| Notostomus elegans A. Milne-Edwards, 1881 | MW043011 | MW043455 | MW043470 | MW052296 | Lunina et al., 2020 |
| Hymenodora frontalis Rathbun, 1902 | DQ882080 | N | N | N | Costa et al., 2007 |
| Hymenodora glacialis (Buchholz, 1874) | FJ602519 | GQ131896 | GQ131915 | N | Bucklin et al., 2010; Chan et al., 2010 |
| Hymenodora gracilis Smith, 1886 | MH572613 | MH542891 | KP075827 | KP076134 | Wilkins and Bracken-Grissom, 2020 (GenBank); Wong et al., 2015 |
| Sclerodora crosnieri gen. nov., sp. nov. | OK382996 | OK382953 | OK382952 | OK424597 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vereshchaka, A.; Kulagin, D.; Lunina, A. A New Shrimp Genus (Crustacea: Decapoda) from the Deep Atlantic and an Unusual Cleaning Mechanism of Pelagic Decapods. Diversity 2021, 13, 536. https://doi.org/10.3390/d13110536
Vereshchaka A, Kulagin D, Lunina A. A New Shrimp Genus (Crustacea: Decapoda) from the Deep Atlantic and an Unusual Cleaning Mechanism of Pelagic Decapods. Diversity. 2021; 13(11):536. https://doi.org/10.3390/d13110536
Chicago/Turabian StyleVereshchaka, Alexander, Dmitry Kulagin, and Anastasiia Lunina. 2021. "A New Shrimp Genus (Crustacea: Decapoda) from the Deep Atlantic and an Unusual Cleaning Mechanism of Pelagic Decapods" Diversity 13, no. 11: 536. https://doi.org/10.3390/d13110536
APA StyleVereshchaka, A., Kulagin, D., & Lunina, A. (2021). A New Shrimp Genus (Crustacea: Decapoda) from the Deep Atlantic and an Unusual Cleaning Mechanism of Pelagic Decapods. Diversity, 13(11), 536. https://doi.org/10.3390/d13110536






