Macroinvertebrate Taxonomic Richness in Minimally Disturbed Streams on the Southeastern USA Coastal Plain
Abstract
1. Introduction
2. Materials and Methods
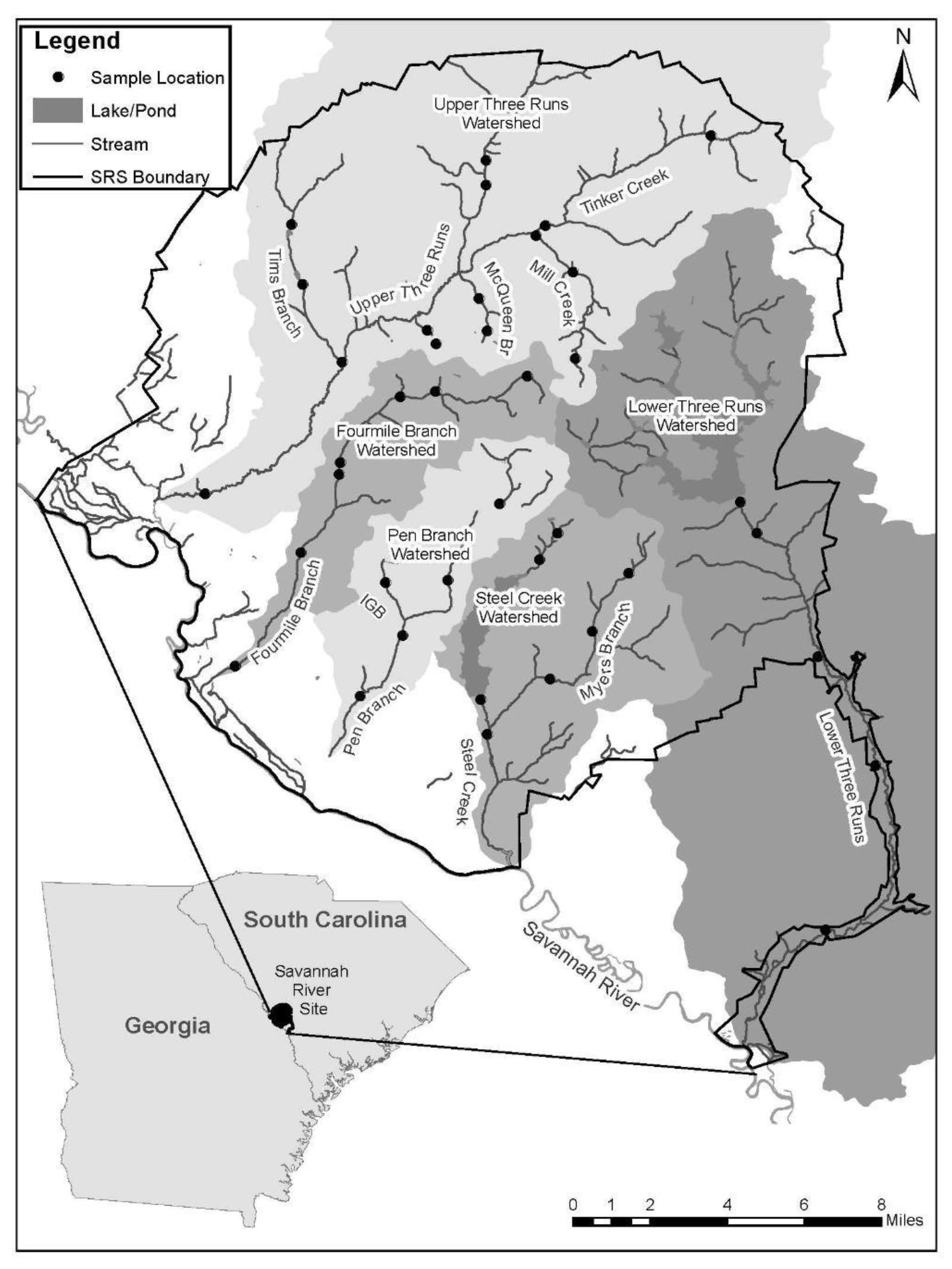
2.1. Study Area
2.2. Macroinvertebrate Sampling
2.3. Data Analysis
3. Results
3.1. Genera Richness
3.2. Factors Affecting Genera Richness
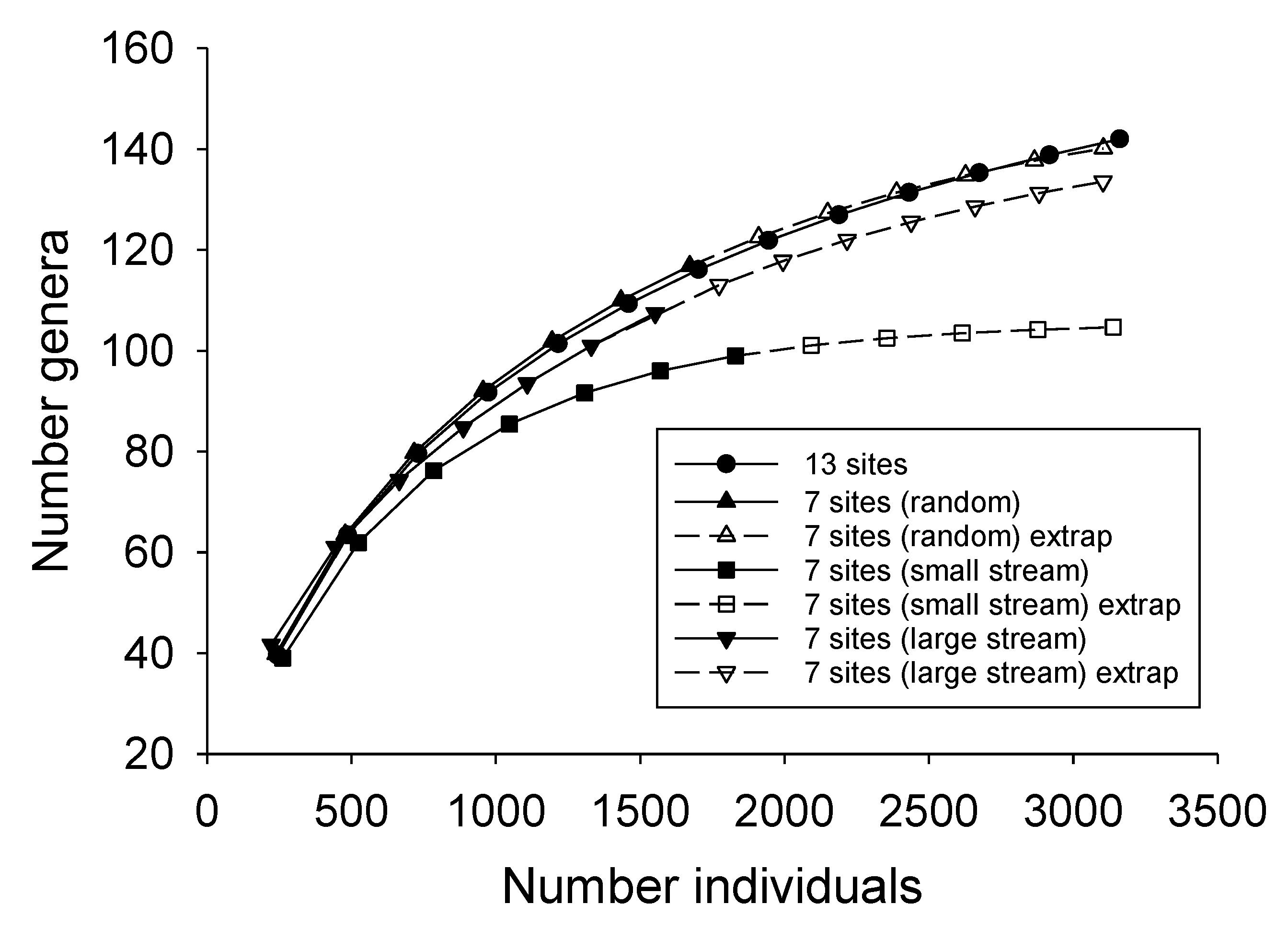
3.3. Effects of Sampling Effort and Site Selection on Estimates of Genera Richness
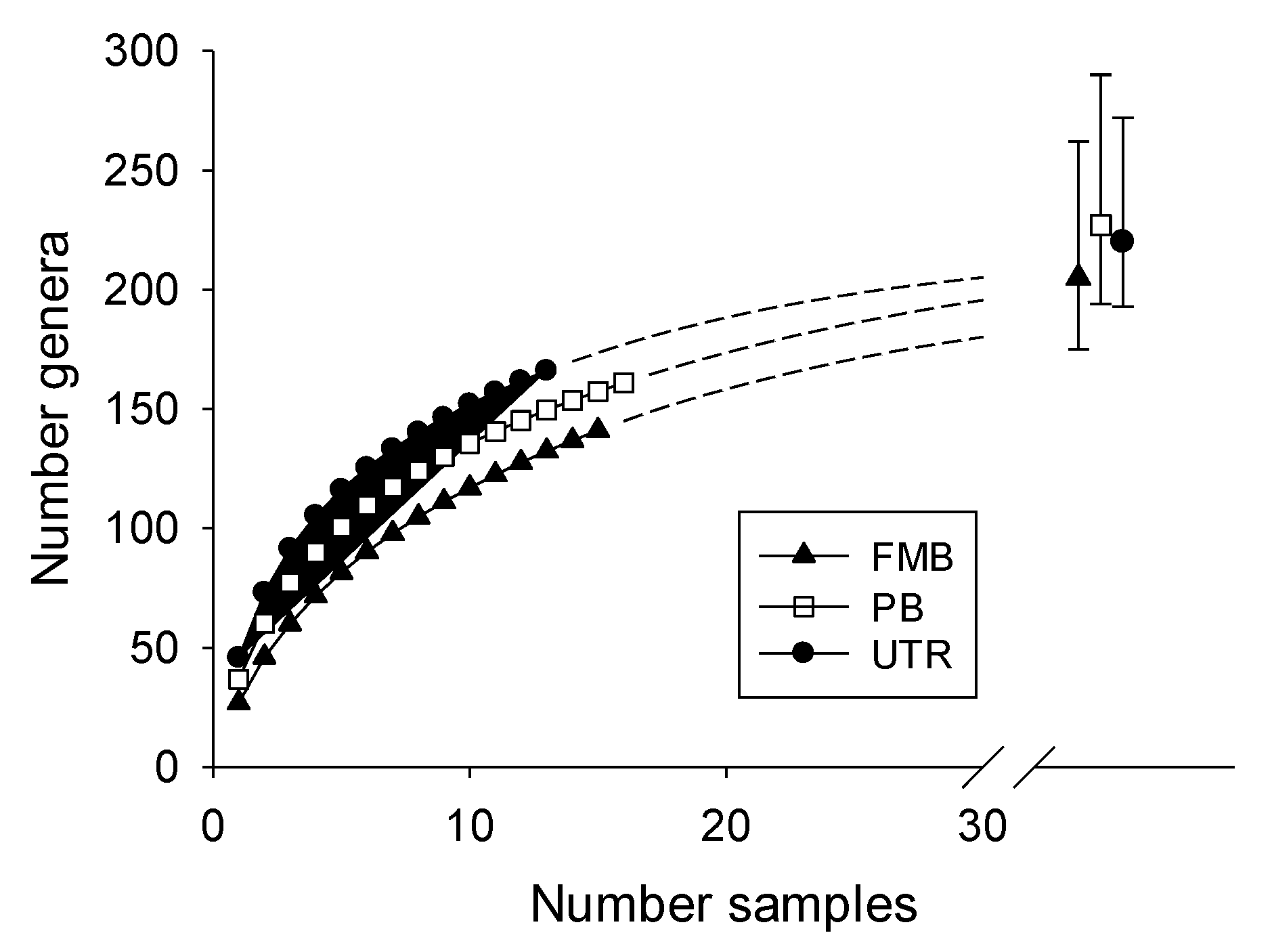
3.4. Genera Richness in Headwater and Lower Reach Streams
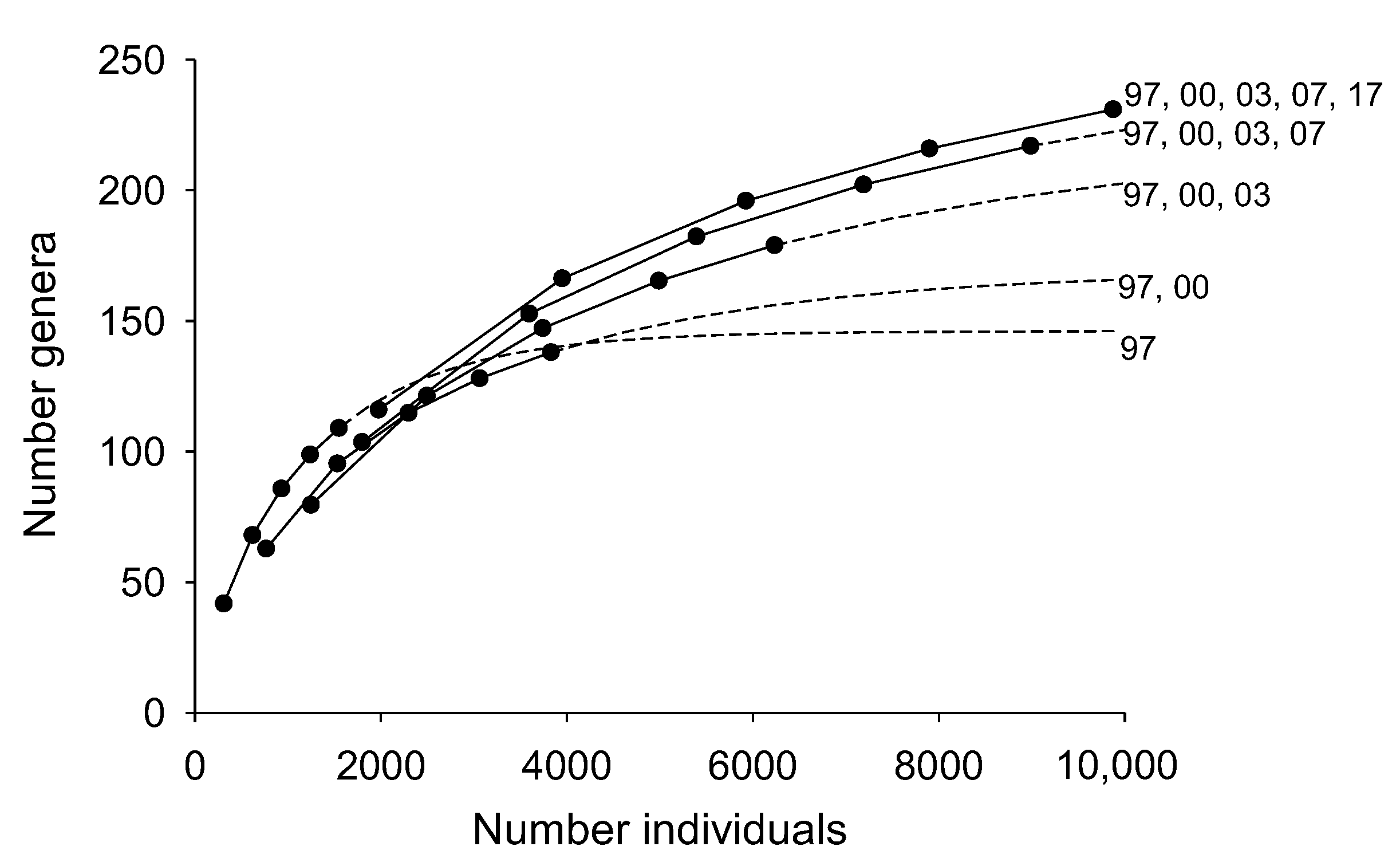
3.5. Effects of Sampling Duration on Estimates of Genera Richness
3.6. Comparison of UTR to Other Streams
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Roberto, C.G. Freshwater biodiversity: A review of local and global threats. Int. J. Environ. Stud. 2016, 73, 887–904. [Google Scholar]
- Vaughn, C.C. Biodiversity losses and ecosystem function in freshwaters: Emerging conclusions and research directions. BioScience 2016, 60, 25–35. [Google Scholar] [CrossRef]
- Pallottini, M.; Goretti, E.; Selvaggi, R.; Cappelletti, D.; Dedieu, N.; Céréghino, R. An efficient semiquantitative macroinvertebrate multimetric index for the assessment of water and sediment contamination in streams. Inland Waters 2017, 7, 314–322. [Google Scholar] [CrossRef]
- Pallottini, M.; Cappelletti, D.; Fabrizi, A.; Gaino, E.; Goretti, E.; Selvaggi, R.; Céréghino, R. Macroinvertebrate functional trait responses to chemical pollution in agricultural-industrial landscapes. River Res. Appl. 2017, 33, 505–513. [Google Scholar] [CrossRef]
- Carter, J.L.; Resh, V.H. Analytical Approaches Used in Stream Benthic Macroinvertebrate Biomonitoring Programs of State Agencies in the United States; U.S. Geological Survey: Reston, VA, USA, 2013; p. 50. [Google Scholar]
- Connor, E.F.; McCoy, E.D. The statistics and biology of the species-area relationship. Am. Nat. 1979, 113, 791–833. [Google Scholar] [CrossRef]
- Scheiner, S.M. Six types of species-area curves. Glob. Ecol. Biogeogr. 2003, 12, 441–447. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Colwell, R.K. Estimating species richness. In Biological Diversity: Frontiers in Measurement and Assessment; Oxford University Press: Oxford, UK, 2011; pp. 39–54. [Google Scholar]
- Carey, S.; Ostling, A.; Harte, J.; del Moral, R. Impact of curve construction and community dynamics on the species-time relationship. Ecology 2007, 88, 2145–2153. [Google Scholar] [CrossRef]
- Gotelli, N.; Colwell, R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001, 4, 379–391. [Google Scholar] [CrossRef]
- Rosenzweig, M.L. Preston’s ergodic conjecture: The accumulation of species in space and time. In Biodiversity Dynamics: Turnover of Populations, Taxa and Communities; McKinney, M.L., Drake, J., Eds.; Columbia University Press: New York, NY, USA, 1998; pp. 311–348. [Google Scholar]
- Poff, N.L. Landscape filters and species traits: Towards mechanistic understanding and prediction in stream ecology. J. N. Am. Benthol. Soc. 1997, 16, 391–409. [Google Scholar] [CrossRef]
- Vannote, R.L.; Minshall, G.W.; Cummins, K.W.; Sedell, J.R.; Cushing, C.E. The river continuum concept. Can. J. Fish. Aquat. Sci. 1980, 37, 130–137. [Google Scholar] [CrossRef]
- Clarke, A.; Mac Nally, R.; Bond, N.; Lake, P.S. Macroinvertebrate diversity in headwater streams: A review. Freshw. Biol. 2008, 53, 1707–1721. [Google Scholar] [CrossRef]
- Archaimbault, V.; Usseglio Polatera, P.; Garric, J.; Wasson, J.G.; Babut, M. Assessing pollution of toxic sediment in streams using bio-ecological traits of benthic macroinvertebrates. Freshw. Biol. 2010, 55, 1430–1446. [Google Scholar] [CrossRef]
- Mondy, C.P.; Villeneuve, B.; Archaimbault, V.; Usseglio-Polatera, P. A new macroinvertebrate-based multimetric index (I2M2) to evaluate ecological quality of French wadeable streams fulfilling the WFD demands: A taxonomical and trait approach. Ecol. Indic. 2012, 18, 452–467. [Google Scholar] [CrossRef]
- HydroGeoLogic. Prepared for the legacy resource management program, strategic environmental research and development program, and environmental security technology certification program. In Proceedings of the Southeast Region Threatened, Endangered, and At-Risk Species Workshop, Cocoa Beach, FL, USA, 1 March 2007. [Google Scholar]
- Voelz, N.J.; McArthur, J.V. An exploration of factors influencing lotic insect species richness. Biodivers. Conserv. 2000, 9, 1543–1570. [Google Scholar] [CrossRef]
- Schmidt, J.P. Sandhills. In New Georgia Encyclopedia; University of Georgia Press: Athens, GA, USA, 2004. [Google Scholar]
- Sabater, F.; Meyer, J.L.; Edwards, R.T. Longitudinal patterns of dissolved organic carbon concentration and suspended bacterial density along a blackwater river. Biogeochemistry 1993, 21, 73–93. [Google Scholar] [CrossRef]
- Benke, A.C.; Meyer, J.L. Structure and function of a blackwater river in the southeastern USA. Proc. Int. Assoc. Theor. 1988, 23, 1209–1218. [Google Scholar]
- Paller, M.H.; Dyer, S.A.; Coughlin, D.P. Exposure of Ecological Receptors to Metals in the Savannah River Site Integrator Operable Units, Analysis of Refined Data from Multiple Environmental Media; WSRC-STI-2008-00365; Savannah River National Laboratory: Aiken, SC, USA, 2008. [Google Scholar]
- Paller, M.H. Estimating fish species richness across multiple watersheds. Diversity 2018, 10, 42. [Google Scholar] [CrossRef]
- SCDHEC (South Carolina Department of Health and Environmental Control). Standard Operating and Quality Control Procedures for Macroinvertebrate Sampling; Technical Report No. 004-98; Bureau of Water, Division of Water Monitoring, Assessment and Protection, Aquatic Biology Section: Columbia, SC, USA, 1998.
- Hester, F.E.; Dendy, J.S. A multiple-plate sampler for aquatic macroinvertebrates. Trans. Am. Fish. Soc. 1962, 91, 420–421. [Google Scholar] [CrossRef]
- Plafkin, J.L.; Barbour, M.T.; Porter, K.D.; Gross, S.K.; Hughes, R.M. Rapid Bioassessment Protocols for Use in Streams and Rivers: Benthic Macroinvertebrates and Fish; EPA/444/4-89-0001; U.S. Environmental Protection Agency: Washington, DC, USA, 1989.
- Barbour, M.T.; Gerritsen, J.; Snyder, B.D.; Stribling, J.B. Rapid Bioassessment Protocols for Use in Streams and Wadeable Rivers: Periphyton, Benthic Macroinvertebrates and Fish, 2nd ed.; EPA-841-B-99-002; U.S. Environmental Protection Agency, Office of Water: Washington, DC, USA, 1999.
- Brigham, A.R.; Brigham, W.U.; Gnilka, A. The Aquatic Insects and Oligochaetes of North and South Carolina; Brigham, A.R., Gnilka, A., Eds.; Midwest Enterprises: Mahomet, IL, USA, 1982; p. 837. [Google Scholar]
- Epler, J.H. Identification Manual for Larval Chironomidae (Diptera) of North and South Carolina; North Carolina Department of Environment and Natural Resources: Raleigh, NC, USA, 2001; p. 526.
- Fitzpatrick, J.F. How to Know the Freshwater Crustacea; Wm. C. Brown Company Publishers: Dubuque, IA, USA, 1983; p. 227. [Google Scholar]
- Merritt, R.W.; Cummins, K.W.; Berg, M.B. An Introduction to the Aquatic Insects of North America, 4th ed.; Kendall/Hunt Publishing Co.: Dubuque, IA, USA, 2008; p. 1158. [Google Scholar]
- Morse, J.C.; McCafferty, W.P.; Stark, B.P.; Jacobus, L.M. Larvae of the Southeastern USA Mayfly, Stonefly, and Caddisfly Species; Clemson Public Service and Agriculture: Clemson, SC, USA, 2017; p. 482. [Google Scholar]
- Needham, J.G.; Westfall, M.J.; May, M.L. Dragonflies of North America; Scientific Publishers: Gainesville, FL, USA, 2000; p. 939. [Google Scholar]
- Pennak, R.W. Freshwater Invertebrates of the United States, 2nd ed.; John Wiley and Sons: New York, NY, USA, 1978; p. 803. [Google Scholar]
- Smith, D.G. Pennak’s Freshwater Invertebrates of the United States: Porifera to Crustacea, 4th ed.; John Wiley and Sons: Indianapolis, IN, USA, 2001; p. 638. [Google Scholar]
- Stewart, K.W.; Stark, B.P. Nymphs of North American Stonefly Genera (Plecoptera); University of North Texas Press: Texas City, TX, USA, 1988; p. 460. [Google Scholar]
- Thorp, J.H.; Rogers, D.C. Thorp and Covich’s Freshwater Invertebrates: Keys to Nearctic Fauna, 4th ed.; Academic Press-Elsevier: Cambridge, MA, USA, 2016; Volume 2, p. 740. [Google Scholar]
- Westfall, M.J.; May, M.L. Damselflies of North America; Scientific Publishers: Gainesville, FL, USA, 1996; p. 649. [Google Scholar]
- SYSTAT Software, Inc. SigmaPlot 13.0; SYSTAT Software, Inc.: San Jose, CA, USA, 2007. [Google Scholar]
- McCune, B.; Mefford, M.J. PC-ORD Multivariate Analysis of Ecological Data; Version 6; MJM Software: Gleneden Beach, OR, USA, 2011. [Google Scholar]
- Colwell, R.K.; Mao, C.X.; Chang, J. Interpolating, extrapolating, and comparing incidence-based species accumulation curves. Ecology 2004, 85, 2717–2727. [Google Scholar] [CrossRef]
- Colwell, R.K. EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples, Version 9; User’s Guide and Application. 2013. Available online: http://viceroy.eeb.uconn.edu/estimates/EstimateSPages/EstSUsersGuide/EstimateSUsersGuide.htm (accessed on 9 September 2020).
- Boothroyd, I.K.G.; Stark, J.D. Use of invertebrates in Monitoring. In New Zealand Stream Invertebrates: Ecology and Implications for Management; Collier, K., Winterbourn, M.J., Eds.; New Zealand Limnological Society: Hamilton, New Zealand, 2000; pp. 344–373. [Google Scholar]
- Collier, K.J.; Wilcock, R.J.; Meredith, A.S. Influence of substrate type and physico-chemical conditions on macroinvertebrate faunas and biotic indices of some lowland Waikato, New Zealand, streams. N. Z. J. Mar. Freshw. Res. 1998, 32, 1–19. [Google Scholar] [CrossRef]
- Morse, J.C.; Chapin, J.W.; Herlong, D.D.; Harvey, R.S. Aquatic insects of Upper Three Runs, Savannah River Plant, South Carolina, Part I: Orders other than Diptera. J. Ga. Entomol. Soc. 1980, 15, 73–101. [Google Scholar]
- Morse, J.C.; Chapin, J.W.; Herlong, D.D.; Harvey, R.S. Aquatic insects of Upper Three Runs, Savannah River Plant, South Carolina, Part I: Diptera. J. Ga. Entomol. Soc. 1983, 18, 303–316. [Google Scholar]
- Weatherley, N.S.; Ormerod, S.J. The Constancy of invertebrate assemblages in soft-water streams: Implications for the prediction and detection of environmental change. J. Appl. Ecol. 1990, 27, 952–964. [Google Scholar] [CrossRef]
- Huttunen, K.-L.; Mykrä, H.; Muotka, T. Temporal variability in taxonomic completeness of stream macroinvertebrate assemblages. Freshw. Sci. 2012, 31, 423–441. [Google Scholar] [CrossRef]
- Rosillon, D. The influence of abiotic factors and density-dependent mechanisms on between-year variations in a stream invertebrate community. Hydrobiologia 1989, 179, 25–38. [Google Scholar] [CrossRef]
- Gutiérrez-Fonseca, P.E.; Ramírez, A.; Pringle, C.M. Large-scale climatic phenomena drive fluctuations in macroinvertebrate assemblages in lowland tropical streams, Costa Rica: The importance of ENSO events in determining long-term (15y) patterns. PLoS ONE 2016, 13, e0191781. [Google Scholar] [CrossRef]
- Minshall, G.W.; Petersen, R.C., Jr.; Nimz, C.F. Species richness in streams of different size from the same drainage basin. Am. Nat. 1985, 125, 16–38. [Google Scholar] [CrossRef]
- Melo, A.S.; Froehlich, C.G. Macroinvertebrates in neotropical streams: Richness patterns along a catchment and assemblage structure between 2 seasons. J. N. Am. Benthol. Soc. 2001, 20, 1–16. [Google Scholar] [CrossRef][Green Version]
- Paller, M.H.; Specht, W.L.; Dyer, S.A. Effects of stream size on taxa richness and other commonly used benthic bioassessment metrics. Hydrobiologia 2006, 568, 309–316. [Google Scholar] [CrossRef]
- Floyd, M.A.; Morse, J.C.; McArthur, J.V. Aquatic insects of Upper Three Runs Creek, Savannah River Site, South Carolina. Part IV: Caddisflies (Trichoptera) of the lower reaches. J. Ga. Entomol. Soc. 1993, 28, 85–95. [Google Scholar] [CrossRef]
- Henriques-Oliveira, A.L.; Nessimian, J.L. Aquatic macroinvertebrate diversity and composition in streams along an altitudinal gradient in Southeastern Brazil. Biota Neotrop. 2010, 10, 115–128. [Google Scholar] [CrossRef]
- Prommi, T.; Payakka, A. Aquatic insect biodiversity and water quality parameters of streams in northern Thailand. Sains Malays. 2015, 44, 707–717. [Google Scholar] [CrossRef]
- Maneechan, W.; Prommi, T.O. Diversity and distribution of aquatic insects in streams of the Mae Klong watershed, western Thailand. Psyche 2015, 2, 1–7. [Google Scholar] [CrossRef]
- Marshall, J.C.; Steward, A.L. Taxonomic resolution and quantification of freshwater macroinvertebrate samples from an Australian dryland river: The benefits and costs of using species abundance data. Hydrobiologia 2006, 572, 171–194. [Google Scholar] [CrossRef]
- Growns, J.E.; Growns, I.O. Predicting species richness for Australasian freshwater macroinvertebrates: Do we want to know? Mem. Mus. Vic. 1997, 56, 483–490. [Google Scholar] [CrossRef]
| Drainage | Streams within Drainage | Number Samples * | Number Sites | Number Years Sampled | Observed Number Genera | Estimated Complete Number of Genera ** | % Difference between Observed and Estimated |
|---|---|---|---|---|---|---|---|
| FMB | FMB | 31 (15, 16) * | 7 | 5 | 184 | 217 (199–257) | 17.9 |
| LTR | LTR | 15 (6, 9) | 5 | 4 | 135 | 150 (139–180) | 11.1 |
| PB | PB | 29 (16, 13) | 4 | 5 | 177 | 201 (188–232) | 13.6 |
| IGB | 9 (5, 4) | 1 | 5 | 97 | 111(102–134) | 14.4 | |
| PB Drainage Total | 38 (21, 17) | 5 | 5 | 195 | 212 (201–238) | 8.7 | |
| SC | MB | 12 (7, 5) | 3 | 5 | 139 | 169 (151–212) | 21.6 |
| SC | 17 (9, 8) | 4 | 5 | 149 | 158 (152–178) | 6.0 | |
| SC Drainage Total | 29 (16, 13) | 7 | 5 | 172 | 189 (177–221) | 9.9 | |
| UTR | CB | 12 (7, 5) | 2 | 5 | 97 | 108 (101–133) | 11.3 |
| MC | 11 (7, 4) | 3 | 5 | 137 | 159 (146–191) | 16.1 | |
| MQ | 9 (5, 4) | 2 | 4 | 109 | 130 (117–162) | 19.3 | |
| TB | 9 (5, 4) | 1 | 5 | 120 | 141 (129–173) | 17.5 | |
| TC | 8 (5, 3) | 3 | 3 | 120 | 147 (130–194) | 22.5 | |
| UTR | 23 (13, 10) | 4 | 5 | 193 | 212 (201–238) | 9.8 | |
| UTR Drainage Total | 72 (42, 30) | 15 | 5 | 259 | 280 (268–307) | 8.1 |
| Taxonomic Groups | FMB | LTR | PB | SC | UTR | HD (All Drainages) | MHSP (All Drainages) | Total Numbers | Total Genera | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Num | Gen | Num | Gen | Num | Gen | Num | Gen | Num | Gen | Num | Gen | Num | Gen | |||
| Turbellaria | 4 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 4 | 1 | 10 | 1 | 0 | 0 | 10 | 1 |
| Nematoda | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 1 | 0 | 0 | 2 | 1 |
| Annelida-Hirudinea | 6 | 2 | 1 | 1 | 21 | 1 | 0 | 0 | 35 | 2 | 4 | 3 | 59 | 4 | 63 | 6 |
| Annelida-Lumbriculidae | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 |
| Annelida-Naididae | 179 | 5 | 9 | 1 | 226 | 3 | 165 | 3 | 80 | 3 | 406 | 5 | 253 | 3 | 659 | 5 |
| Annelida-Tubificidae | 12 | 1 | 11 | 2 | 24 | 2 | 71 | 1 | 40 | 2 | 28 | 1 | 130 | 2 | 158 | 2 |
| Mollusca-Bivalvia | 105 | 2 | 42 | 1 | 135 | 3 | 72 | 2 | 63 | 3 | 9 | 2 | 408 | 3 | 417 | 3 |
| Mollusca-Gastropoda | 119 | 12 | 123 | 6 | 181 | 10 | 157 | 10 | 183 | 11 | 177 | 11 | 586 | 13 | 762 | 16 |
| Arachnida-Acari | 282 | 1 | 190 | 1 | 49 | 1 | 229 | 1 | 79 | 1 | 332 | 1 | 497 | 1 | 829 | 1 |
| Crustacea-Cladocera | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 5 | 1 | 0 | 0 | 5 | 1 |
| Crustaceae-Amphipoda | 95 | 2 | 26 | 2 | 91 | 2 | 29 | 2 | 38 | 2 | 118 | 2 | 161 | 2 | 279 | 2 |
| Crustaceae-Decapoda | 113 | 3 | 56 | 3 | 231 | 4 | 184 | 3 | 210 | 4 | 100 | 3 | 694 | 4 | 794 | 4 |
| Crustaceae-Isopoda | 3 | 1 | 20 | 1 | 55 | 1 | 18 | 1 | 23 | 1 | 17 | 1 | 102 | 1 | 119 | 1 |
| Insecta-Collembola | 1 | 1 | 0 | 0 | 4 | 1 | 8 | 1 | 5 | 1 | 18 | 1 | 0 | 0 | 18 | 1 |
| Insecta-Coleoptera | 230 | 15 | 103 | 8 | 226 | 19 | 232 | 16 | 1152 | 33 | 500 | 22 | 1443 | 35 | 1943 | 40 |
| Insecta-Diptera-Chironomini | 1284 | 13 | 619 | 10 | 1202 | 14 | 864 | 15 | 2110 | 21 | 3857 | 18 | 2222 | 22 | 6079 | 25 |
| Insecta-Diptera-Orthocladiinae | 881 | 16 | 496 | 14 | 1075 | 23 | 1306 | 16 | 1553 | 25 | 2937 | 24 | 2374 | 25 | 5311 | 28 |
| Insecta-Diptera-Tanypodinae | 246 | 11 | 189 | 11 | 247 | 12 | 189 | 9 | 647 | 15 | 675 | 12 | 843 | 15 | 1518 | 17 |
| Insecta-Diptera-Tanytarsini | 1113 | 4 | 1001 | 5 | 1154 | 5 | 996 | 6 | 1843 | 7 | 3997 | 6 | 2110 | 7 | 6107 | 8 |
| Insecta-Diptera-other | 471 | 14 | 44 | 8 | 307 | 13 | 370 | 11 | 970 | 22 | 752 | 19 | 1410 | 24 | 2162 | 28 |
| Insecta-Ephemeroptera | 581 | 16 | 407 | 16 | 1399 | 21 | 759 | 17 | 1520 | 23 | 2262 | 24 | 2404 | 24 | 4666 | 25 |
| Insecta-Heteroptera | 50 | 8 | 29 | 8 | 53 | 7 | 38 | 6 | 102 | 12 | 4 | 1 | 268 | 15 | 272 | 15 |
| Insecta-Lepidoptera | 236 | 1 | 18 | 2 | 83 | 2 | 94 | 1 | 723 | 1 | 59 | 1 | 1095 | 3 | 1154 | 3 |
| Insecta-Megaloptera | 23 | 3 | 43 | 4 | 40 | 3 | 12 | 3 | 121 | 4 | 102 | 4 | 137 | 4 | 239 | 4 |
| Insecta-Odonata | 334 | 22 | 197 | 10 | 606 | 18 | 636 | 17 | 1366 | 24 | 229 | 13 | 2910 | 27 | 3139 | 29 |
| Insecta-Plecoptera | 119 | 10 | 90 | 8 | 338 | 11 | 324 | 10 | 779 | 14 | 901 | 14 | 748 | 13 | 1649 | 16 |
| Insecta-Trichoptera | 622 | 18 | 365 | 12 | 830 | 19 | 845 | 19 | 2995 | 26 | 2563 | 24 | 3094 | 29 | 5658 | 29 |
| Totals | 7113 | 184 | 4080 | 135 | 8577 | 195 | 7601 | 172 | 16,643 | 259 | 20,065 | 216 | 23,948 | 276 | 44,013 | 312 |
| Totals (insects only) | 6191 | 152 | 3601 | 116 | 7564 | 168 | 6673 | 147 | 15,886 | 228 | 18,856 | 183 | 21,058 | 243 | 39,915 | 268 |
| Num families | 82 | 68 | 85 | 79 | 101 | 86 | 106 | 114 | ||||||||
| Num Insect families | 61 | 53 | 66 | 61 | 79 | 63 | 83 | 87 | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paller, M.H.; Blas, S.A.; Kelley, R.W. Macroinvertebrate Taxonomic Richness in Minimally Disturbed Streams on the Southeastern USA Coastal Plain. Diversity 2020, 12, 459. https://doi.org/10.3390/d12120459
Paller MH, Blas SA, Kelley RW. Macroinvertebrate Taxonomic Richness in Minimally Disturbed Streams on the Southeastern USA Coastal Plain. Diversity. 2020; 12(12):459. https://doi.org/10.3390/d12120459
Chicago/Turabian StylePaller, Michael H., Susan A. Blas, and Robert W. Kelley. 2020. "Macroinvertebrate Taxonomic Richness in Minimally Disturbed Streams on the Southeastern USA Coastal Plain" Diversity 12, no. 12: 459. https://doi.org/10.3390/d12120459
APA StylePaller, M. H., Blas, S. A., & Kelley, R. W. (2020). Macroinvertebrate Taxonomic Richness in Minimally Disturbed Streams on the Southeastern USA Coastal Plain. Diversity, 12(12), 459. https://doi.org/10.3390/d12120459




