Molecular Species Delimitation of Larks (Aves: Alaudidae), and Integrative Taxonomy of the Genus Calandrella, with the Description of a Range-Restricted African Relic Taxon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphology
2.2. Phylogeny and Species Delimitation
2.3. Description of a New Taxon
3. Results
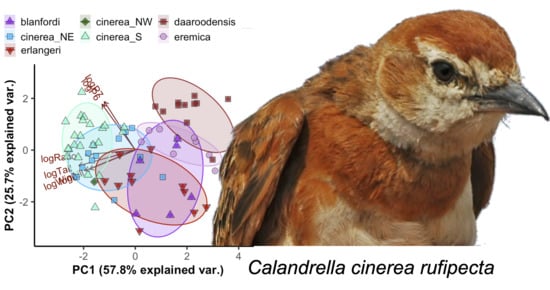
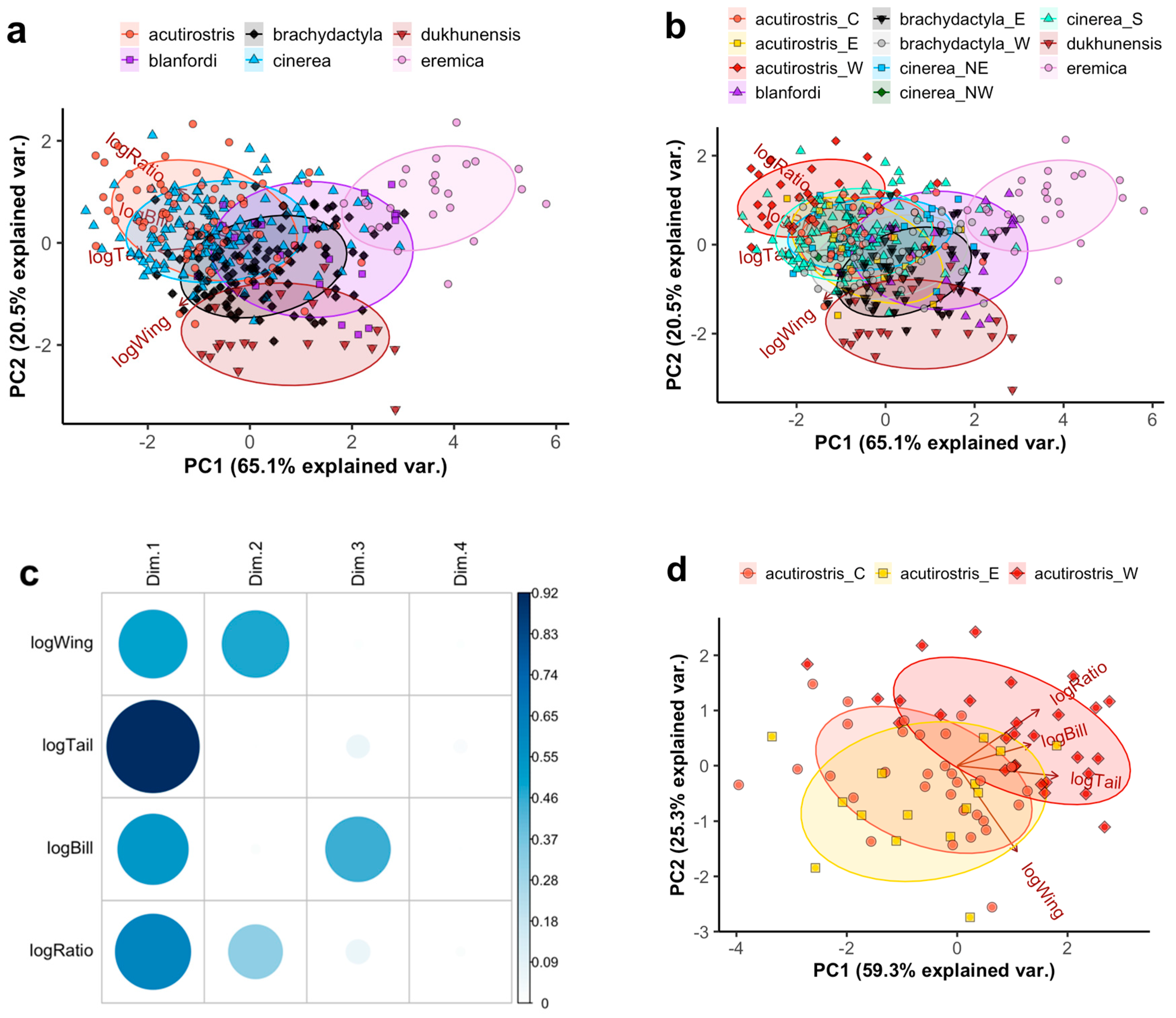
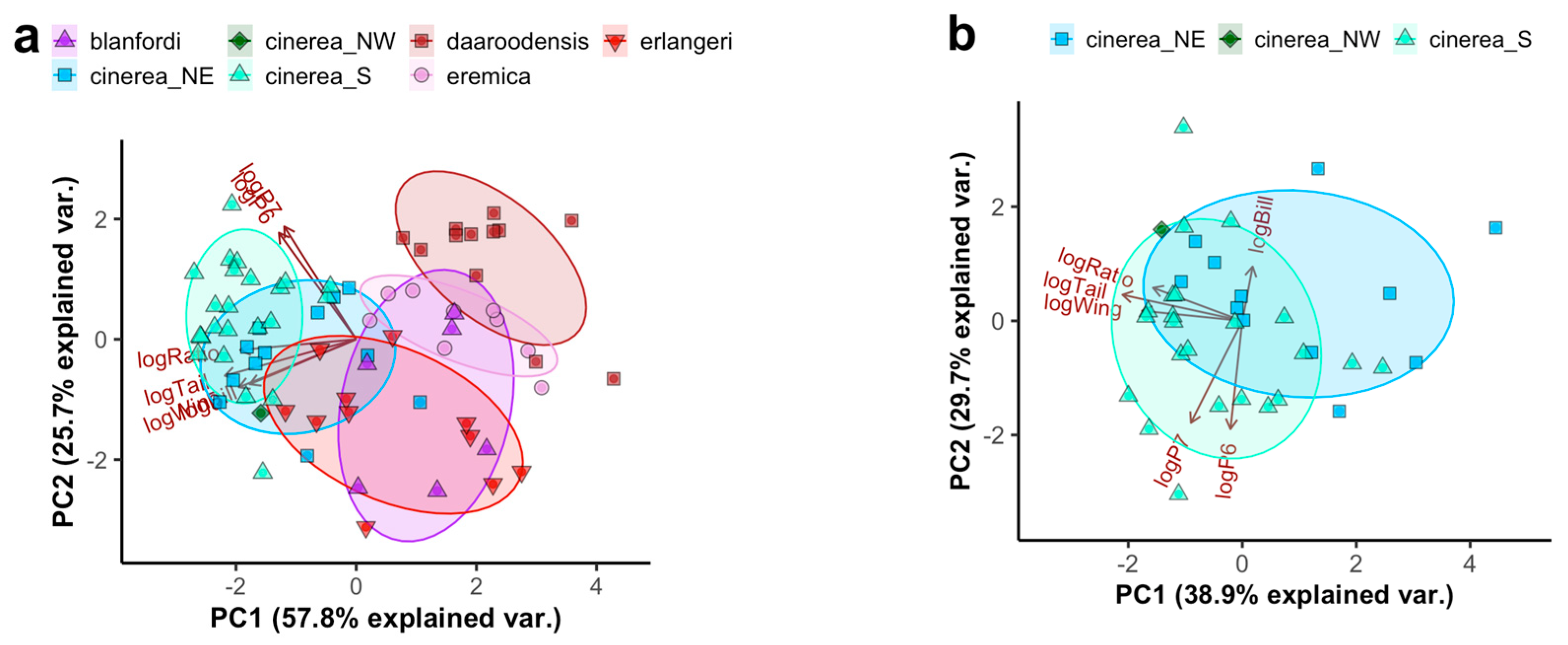
3.1. Morphology of Calandrella larks
3.1.1. Descriptive Statistics
3.1.2. Prediction of Taxon Membership
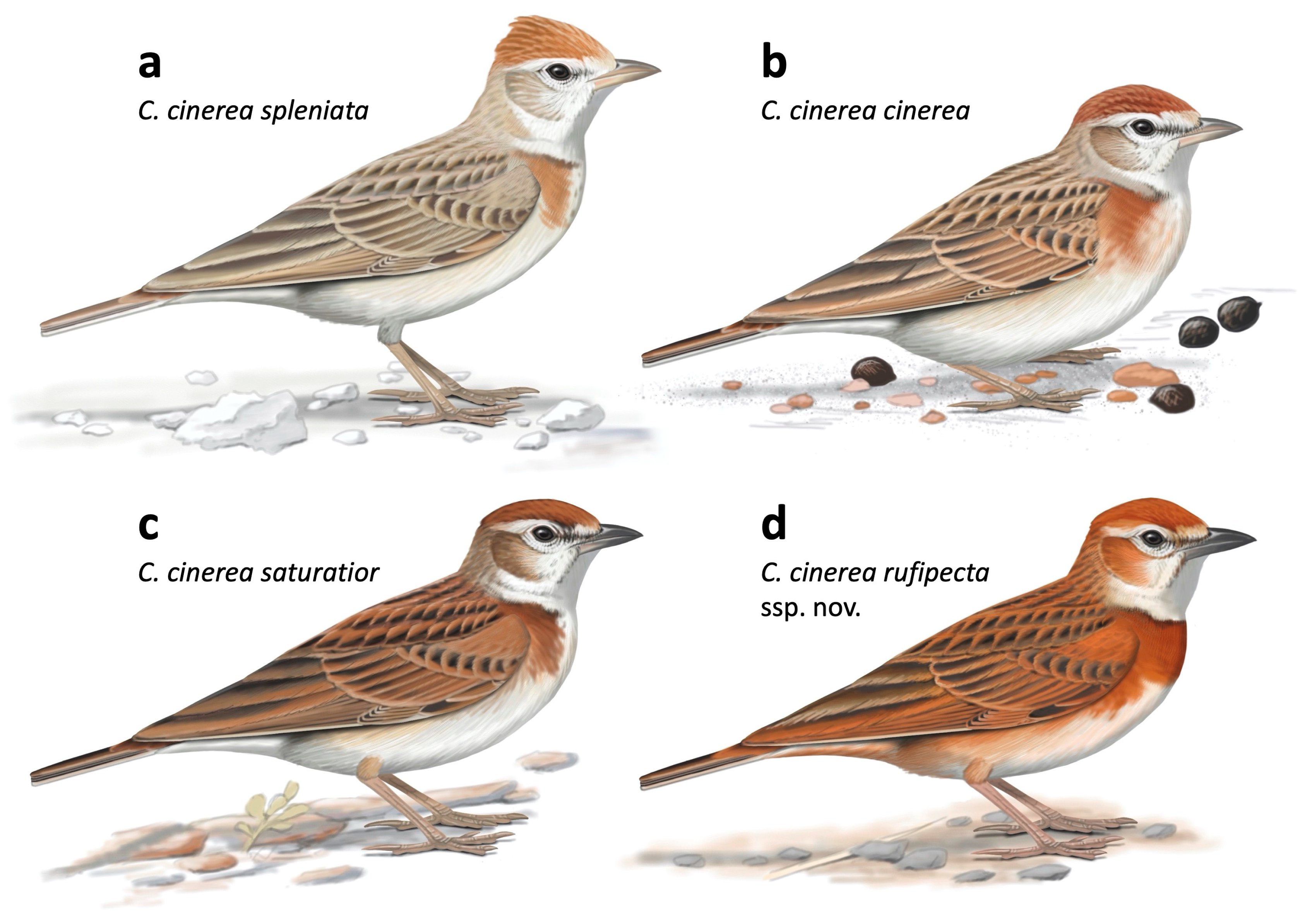
3.2. Description of a New Taxon Endemic to the Jos Plateau in Nigeria
3.3. Molecular Species Delimitation
3.3.1. Focal Group
- 1.
- Genus Calandrella
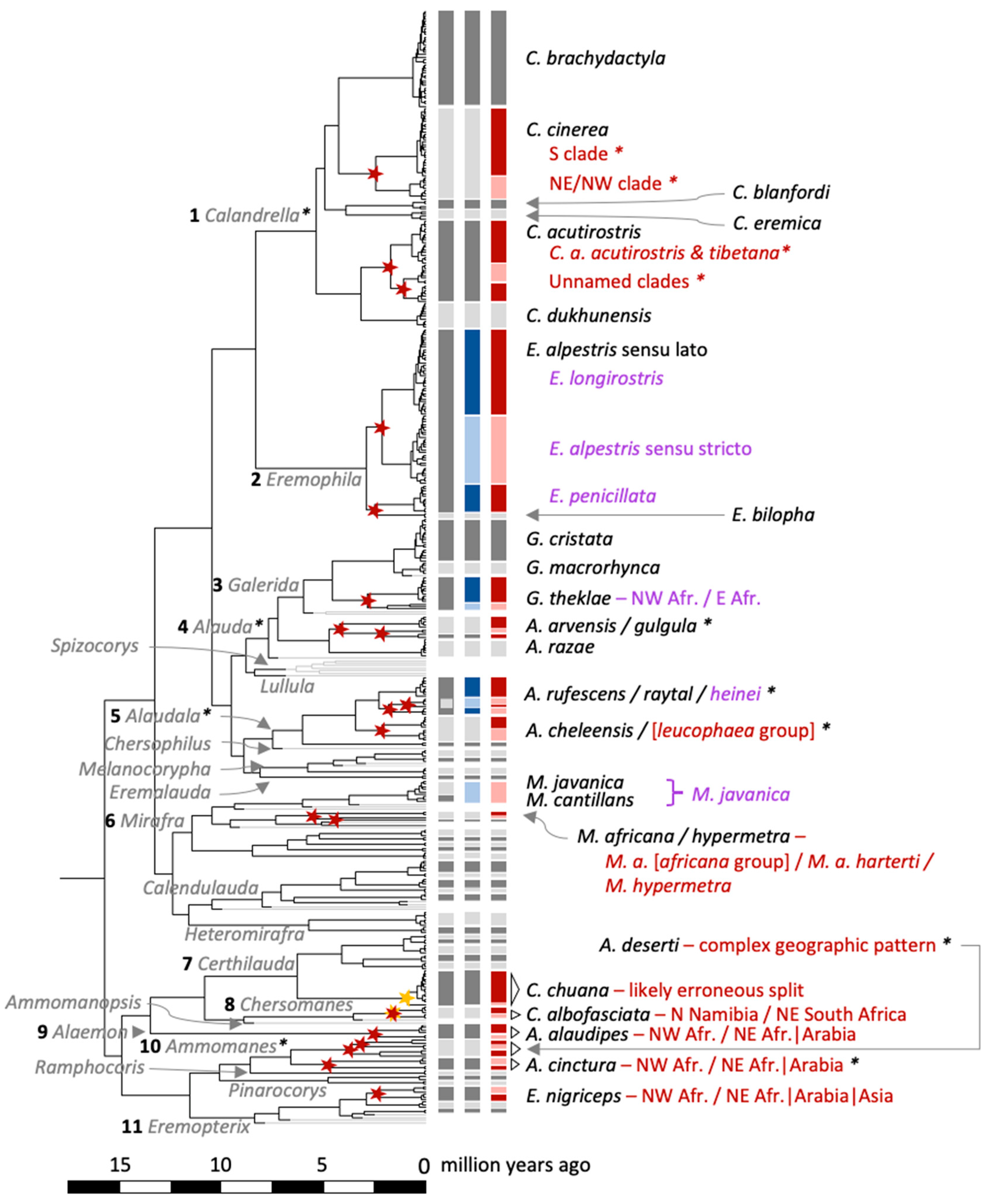
- C. brachydactyla, inferred as a single species by mPTP: Only two clear groups that split 0.71 (95% highest posterior density, HPD: 0.35–0.97) million years ago (MYA) (Figure S1), containing western birds (clade A1) and eastern birds (clade A2; Figure 5; Calandrella clade designations in accordance with Stervander, et al. [27]), but with no internal sorting within these clades according to currently recognized subspecies.
- C. cinerea, inferred as two species by mPTP: Split 2.55 (1.64–3.31) MYA between (1) a clade containing the cinerea group with all southern populations, the population in DR Congo and west of Lake Victoria (clade E), and (2) one clade comprising birds sampled in Kenya and Tanzania east of Lake Victoria (C. c. williamsi; clade D1) and Nigeria (described above as C. c. rufipecta ssp. nov.; clade D2).
- C. blanfordi, inferred as a single species by mPTP (clade F, including C. b. blanfordi and C. b. erlangeri).
- Rufous-crowned Lark C. eremica, inferred as a single species by mPTP (clade G, including C. e. eremica and C. e. daaroodensis).
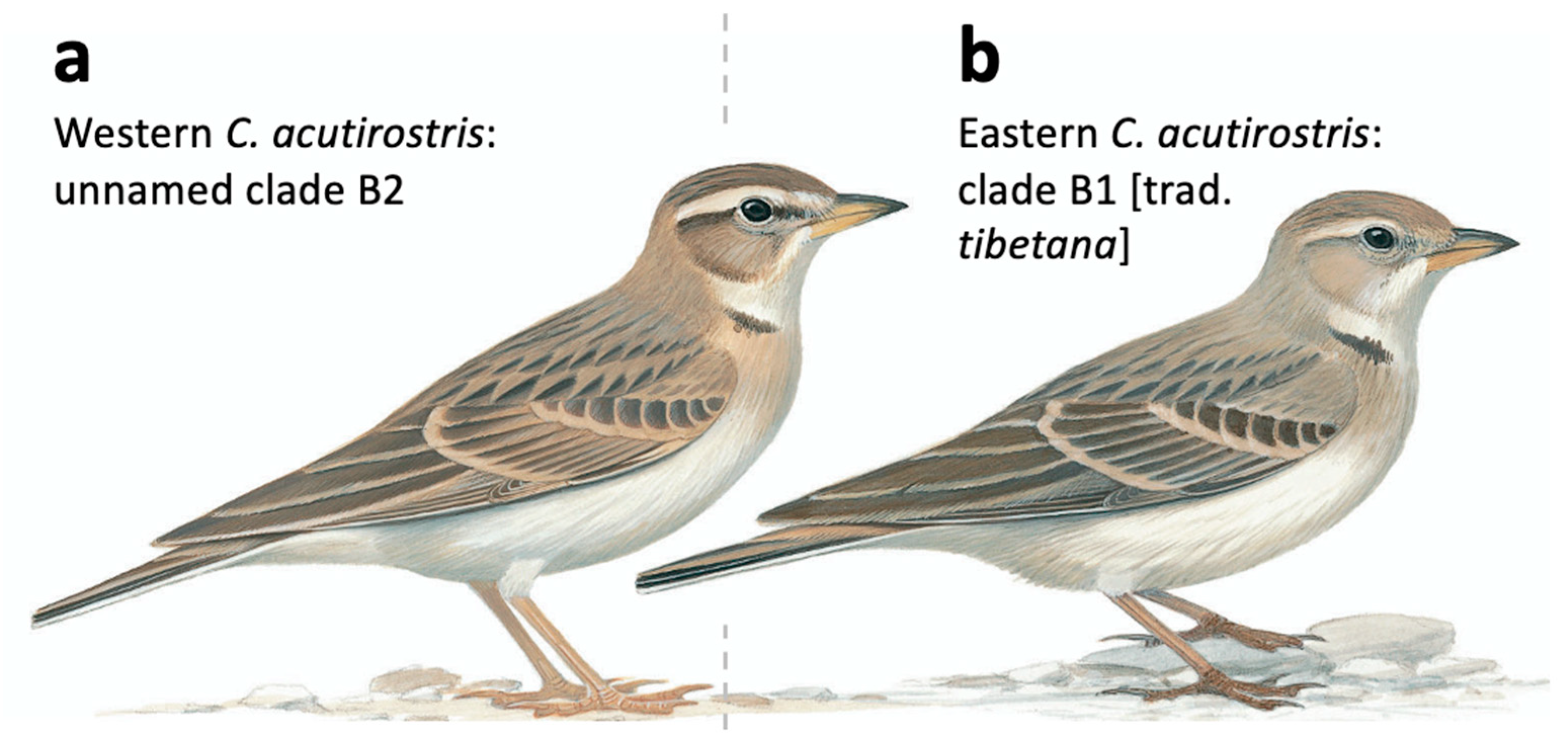
- C. acutirostris, three species delimited by mPTP: Split at 1.78 (1.11–2.35) MYA between (1) a clade containing eastern populations in Tibet (clade B1, corresponding to the population traditionally assigned as C. a. tibetana in eastern Tibet and Qinghai, China) and eastern Ladakh, India (clade B4, birds traditionally assigned to C. a. tibetana); and a clade that, in turn, is split at 1.16 (0.45–1.75) MYA between (2) birds sampled in Afghanistan (clade B2); and (3) birds sampled in Ladakh (clade B3).
- C. dukhunensis, inferred as a single species by mPTP (clade C), separated from C. acutirostris at 3.27 (2.29–4.18) MYA.
3.3.2. Other Larks
- 2.
- Horned Lark Eremophila alpestris and Temminck’s Lark E. bilopha, mPTP delimiting 4 species: Split at 2.25 (1.47–2.92) MYA between a clade containing (1) E. alpestris elwesi and E. a. deosaiensis (Tibet–Himalaya) and (2) a clade comprising multiple subspecies (e.g., alpestris, flava, brandti) across the North Palearctic and North America. A clade comprising (3) E. a. penicillata/albigula (Iran) and E. a. atlas (Morocco) split at 2.61 (1.75–3.41) MYA from (4) E. bilopha (Morocco and Saudi Arabia). The deepest split within Eremophila was at 3.00 (2.12–3.84) MYA.
- 3.
- Thekla’s Lark Galerida theklae, mPTP delimiting 2 species: Split 2.94 (1.99–3.77) MYA between (1) a clade of NW African birds sampled in Morocco (G. t. theklae) and Tunisia (G. t. superflua) and (2) a clade comprising E African birds sampled in Ethiopia (G. t. praetermissa and G. t. hueii) and Somalia (G. t. ellioti).
- 4.
- Eurasian Skylark Alauda arvensis and Oriental Skylark A. gulgula, mPTP delimiting 3 species: (1) western A. arvensis (sampled across Europe east to Kazakhstan) diverged at 4.33 (3.17–5.41) MYA from a clade comprising (2) A. gulgula (likely A. g. inconspicua) from Kazakhstan and India and (3) eastern birds (A. arvensis japonica from Japan and a bird sampled in Anhui Province in SE China, which split at 2.26 (1.43–3.02) MYA.
- 5.
- Genus Alaudala, mPTP delimiting 6 species in the rufescens/cheleensis/raytal complex that is traditionally divided in two or three species:
- Asian Short-toed Lark A. cheleensis: Split at 2.24 (1.36–2.94) MYA between (1) A. c. cheleensis (sampled in eastern Mongolia and Inner Mongolia, China) and (2) a clade containing A. c. leucophaea (Kazakhstan) and A. c. seebohmi (Xinjiang, China).
- Sand Lark A. raytal: Split at 1.01 (0.52–1.36) MYA between (3) a single sample of A. r. adamsi (Punjab, India) and (4) a clade containing A. r. raytal (Haryana and Delhi, India).
- Lesser Short-toed Lark A. rufescens: A clade comprising (5) A. r. heinei (Kazakhstan) and A. r. aharoni (Turkey) split at 1.90 (1.23–2.44) MYA from A. raytal, whose common ancestor split at 2.39 (1.64–3.05) MYA from (6) a clade comprising A. rufescens rufescens and A. r. polatzeki (Canary Islands), A. r. minor (Morocco and Saudi Arabia), and A. r. apetzii (Spain).
- 6.
- Genus Mirafra
- Rufous-naped Lark M. africana, mPTP delimiting 2 species: Split at 5.69 (3.87–7.36) MYA between M. a. transvaalensis (South Africa) and M. a. harterti (Kenya; the latter grouping with low support with Red-winged Lark M. hypermetra).
- Singing Bush Lark M. cantillans and Horsfield’s Bush Lark M. javanica were inferred by mPTP to be conspecific, with a most recent common ancestor at 0.97 (0.54–1.27) MYA.
- 7.
- Short-clawed Lark Certhilauda chuana, mPTP delimiting 2 species: Split at 0.98 (0.40–1.48) MYA between (1) a clade comprising a dozen samples from the disjunct western and eastern South African populations, and (2) a single bird from the eastern South African population.
- 8.
- Spike-heeled Lark Chersomanes albofasciata, mPTP delimiting 2 species: Split at 1.62 (0.72–2.53) MYA between one clade comprising (1) C. a. boweni (Namibia) and (2) another clade comprising samples from the North West and Limpopo provinces of South Africa, presumably all of C. a. alticola.
- 9.
- Greater Hoopoe-Lark Alaemon alaudipes, mPTP delimiting 2 species: Split at 2.65 (1.56–3.85) MYA between (1) Northwest African A. a. alaudipes (sampled in Morocco) and A. a. boavistae (Cape Verde) and (2) Northeast African/Arabian A. a. desertorum (Egypt and Saudi Arabia).
- 10.
- Genus Ammomanes
- Bar-tailed Lark Ammomanes cinctura, mPTP delimiting 2 species: Split at 4.86 (3.33–6.25) MYA between (1) Northwest African A. c. arenicolor (sampled in Morocco) and A. c. cinctura (Cape Verde) and (2) Northeast African/Arabian A. c. arenicolor (sampled in Saudi Arabia).
- Desert Lark Ammomanes deserti, mPTP delimiting 3 species: Split at 3.28 (2.26–4.22) MYA between (1) Northwest African A. d. payni (sampled in Morocco) and (2) Northeast African/Arabian A. d. isabellina (sampled in Saudi Arabia) and A. d. annae (Jordan). This clade split at 3.80 (2.73–4.75) MYA from a clade comprising (3) A. d. deserti (sampled in Israel) and A. d. phoenicuroides (sampled in Pakistan).
- 11.
- Black-crowned Sparrow-Lark Eremopterix nigriceps, mPTP delimiting 2 species: Split at 2.45 (1.50–3.30) MYA between (1) Northwest African E. n. albifrons (sampled in Mauritania) and E. n. nigriceps (Cape Verde) and (2) Northeast African/Arabian/Asian E. n. melanauchen (in Saudi Arabia) and E. n. affinis (in Pakistan).
4. Discussion
4.1. Divergence within African and African–Arabian Lineages
4.1.1. Fragmented Sub-Saharan Species Distributions and Relictual Lineages
4.1.2. Fragmented NW African vs. E African/Arabian Populations
4.1.3. Biogeography of Fragmented African Populations
4.2. Taxonomy of Calandrella Larks
4.2.1. The Calandrella cinerea Complex
4.2.2. Calandrella dukhunensis and Calandrella brachydactyla
4.2.3. The Calandrella acutirostris Complex
4.3. Reliability of Molecular Species Delimitation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Accessibility
References
- Dobzhansky, T. A Critique of the Species Concept in Biology. Philos. Sci. 1935, 2, 344–355. [Google Scholar] [CrossRef]
- Mishler, B.D.; Brandon, R.N. Individuality, Pluralism, and the Phylogenetic Species Concept. Biol. Philos. 1987, 2, 397–414. [Google Scholar] [CrossRef]
- Hausdorf, B. Progress toward a General Species Concept. Evolution 2011, 65, 923–931. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz, K. The General Lineage Concept of Species, Species Criteria, and the Process of Speciation: A Conceptual Unification and Terminological Recommendations. In Endless Forms: Species and Speciation; Howard, D.J., Berlocher, S.H., Eds.; Oxford University Press: Oxford, UK, 1998; pp. 57–75. [Google Scholar]
- de Queiroz, K. Species Concepts and Species Delimitation. Syst. Biol. 2007, 56, 879–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agapow, P.M.; Bininda-Emonds, O.R.P.; Crandall, K.A.; Gittleman, J.L.; Mace, G.M.; Marshall, J.C.; Purvis, A. The Impact of Species Concept on Biodiversity Studies. Q. Rev. Biol. 2004, 79, 161–179. [Google Scholar] [CrossRef] [Green Version]
- Groves, C.P.; Cotterill, F.P.D.; Gippoliti, S.; Robovský, J.; Roos, C.; Taylor, P.J.; Zinner, D. Species Definitions and Conservation: A Review and Case Studies from African Mammals. Conserv. Genet. 2017, 18, 1247–1256. [Google Scholar] [CrossRef]
- Barrowclough, G.F.; Cracraft, J.; Klicka, J.; Zink, R.M. How Many Kinds of Birds Are There and Why Does It Matter? PLoS ONE 2016, 11, e0166307. [Google Scholar] [CrossRef]
- Isaac, N.J.; Mallet, J.; Mace, G.M. Taxonomic Inflation: Its Influence on Macroecology and Conservation. Trends Ecol. Evol. 2004, 19, 464–469. [Google Scholar] [CrossRef] [Green Version]
- Haffer, J. The History of Species Concepts and Species Limits in Ornithology. Bull. Br. Ornithol. Club 1992, 112, 107–158. [Google Scholar]
- Sangster, G. Increasing Numbers of Bird Species Result from Taxonomic Progress, Not Taxonomic Inflation. Proc. R. Soc. Biol. Sci. Ser. B 2009, 276, 3185–3191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poelstra, J.W.; Vijay, N.; Bossu, C.M.; Lantz, H.; Ryll, B.; Muller, I.; Baglione, V.; Unneberg, P.; Wikelski, M.; Grabherr, M.G.; et al. The Genomic Landscape Underlying Phenotypic Integrity in the Face of Gene Flow in Crows. Science 2014, 344, 1410–1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toews, D.P.L.; Taylor, S.A.; Vallender, R.; Brelsford, A.; Butcher, B.G.; Messer, P.W.; Lovette, I.J. Plumage Genes and Little Else Distinguish the Genomes of Hybridizing Warblers. Curr. Biol. 2016, 26, 2313–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irwin, D.E.; Alström, P.; Olsson, U.; Benowitz-Fredericks, Z.M. Cryptic Species in the Genus Phylloscopus (Old World Leaf Warblers). Ibis 2008, 143, 233–247. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-Genome Analyses Resolve Early Branches in the Tree of Life of Modern Birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Tuinen, M.; Butvill, D.B.; Kirsch, J.A.W.; Hedges, S.B. Convergence and Divergence in the Evolution of Aquatic Birds. Proc. R. Soc. Biol. Sci. Ser. B 2001, 268, 1345–1350. [Google Scholar] [CrossRef]
- Alström, P.; Hooper, D.M.; Liu, Y.; Olsson, U.; Mohan, D.; Gelang, M.; Le Manh, H.; Zhao, J.; Lei, F.; Price, T.D. Discovery of a Relict Lineage and Monotypic Family of Passerine Birds. Biol. Lett. 2014, 10, 20131067. [Google Scholar] [CrossRef] [Green Version]
- Garcia-R, J.C.; Lemmon, E.M.; Lemmon, A.R.; French, N. Phylogenomic Reconstruction Sheds Light on New Relationships and Timescale of Rails (Aves: Rallidae) Evolution. Diversity 2020, 12, 70. [Google Scholar] [CrossRef] [Green Version]
- Alström, P.; Jønsson, K.A.; Fjeldså, J.; Ödeen, A.; Ericson, P.G.P.; Irestedt, M. Dramatic Niche Shifts and Morphological Change in Two Insular Bird Species. R. Soc. Open Sci. 2015, 2. [Google Scholar] [CrossRef] [Green Version]
- Alström, P.; Olsson, U.; Lei, F. A Review of the Recent Advances in the Systematics of the Avian Superfamily Sylvioidea. Chin. Birds 2013, 4, 99–131. [Google Scholar] [CrossRef] [Green Version]
- IOC World Bird List (v. 10.2). 2020. Available online: https://www.worldbirdnames.org/new/ioc-lists/crossref (accessed on 25 July 2020).
- de Juana, E.; Suárez, F.; Ryan, P.G.; Alström, P.; Donald, P.F. Family Alaudidae (Larks). In Handbook of the Birds of the World; del Hoyo, J., Elliott, A., Christie, D.A., Eds.; Lynx Edicions: Barcelona, Spain, 2004; Volume 9, pp. 496–601. [Google Scholar]
- Alström, P.; Barnes, K.N.; Olsson, U.; Barker, F.K.; Bloomer, P.; Khan, A.A.; Qureshi, M.A.; Guillaumet, A.; Crochet, P.A.; Ryan, P.G. Multilocus Phylogeny of the Avian Family Alaudidae (Larks) Reveals Complex Morphological Evolution, Non-Monophyletic Genera and Hidden Species Diversity. Mol. Phylogenet. Evol. 2013, 69, 1043–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghorbani, F.; Aliabadian, M.; Zhang, R.; Irestedt, M.; Hao, Y.; Sundev, G.; Lei, F.; Ma, M.; Olsson, U.; Alström, P. Densely Sampled Phylogenetic Analyses of the Lesser Short-toed Lark (Alaudala rufescens)—Sand Lark (A. raytal) species Complex (Aves, Passeriformes) Reveal Cryptic Diversity. Zool. Scr. 2020, 49, 427–439. [Google Scholar] [CrossRef]
- Stervander, M.; Alström, P.; Olsson, U.; Ottosson, U.; Hansson, B.; Bensch, S. Multiple Instances of Paraphyletic Species and Cryptic Taxa Revealed by Mitochondrial and Nuclear RAD Data for Calandrella Larks (Aves: Alaudidae). Mol. Phylogenet. Evol. 2016, 102, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Alström, P.; Sundev, G. Mongolian Short-toed Lark Calandrella dukhunensis, an Overlooked East Asian Species. J. Ornithol. 2020. [Google Scholar] [CrossRef]
- Donald, P.F.; Alström, P.; Engelbrecht, D. Possible Mechanisms of Substrate Colour-Matching in Larks (Alaudidae) and Their Taxonomic Implications. Ibis 2017, 159, 699–702. [Google Scholar] [CrossRef] [Green Version]
- Guillaumet, A.; Crochet, P.A.; Pons, J.M. Climate-Driven Diversification in Two Widespread Galerida Larks. BMC Evol. Biol. 2008, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- Guillaumet, A.; Crochet, P.A.; Godelle, B. Phenotypic Variation in Galerida Larks in Morocco: The Role of History and Natural Selection. Mol. Ecol. 2005, 14, 3809–3821. [Google Scholar] [CrossRef]
- Guillaumet, A.; Pons, J.-M.; Godelle, B.; Crochet, P.-A. History of the Crested Lark in the Mediterranean Region as Revealed by MtDNA Sequences and Morphology. Mol. Phylogenet. Evol. 2006, 39, 645–656. [Google Scholar] [CrossRef]
- Ryan, P.G.; Bloomer, P. The Long-billed Lark Complex: A Species Mosaic in Southwestern Africa. Auk 1999, 116, 194–208. [Google Scholar] [CrossRef]
- Ryan, P.G.; Hood, I.; Bloomer, P.; Komen, J.; Crowe, T.M. Barlow’s Lark: A New Species in the Karoo Lark Certhilauda albescens Complex of Southwest Africa. Ibis 1998, 140, 605–619. [Google Scholar] [CrossRef]
- Svensson, L. Identification Guide to European Passerines, 4th ed.; British Trust for Ornithology: Stockholm, Sweden, 1992. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Ghorbani, F.; Aliabadian, M.; Olsson, U.; Donald, P.F.; Khan, A.A.; Alström, P. Mitochondrial Phylogeography of the Genus Eremophila Confirms Underestimated Species Diversity in the Palearctic. J. Ornithol. 2020, 161, 297–312. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. Jmodeltest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, M.; Kishino, H.; Yano, T.A. Dating of the Human Ape Splitting by a Molecular Clock of Mitochondrial-DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Schwarz, G. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. Beast 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Weir, J.T.; Schluter, D. Calibrating the Avian Molecular Clock. Mol. Ecol. 2008, 17, 2321–2328. [Google Scholar] [CrossRef]
- Alström, P.; Ericson, P.G.; Olsson, U.; Sundberg, P. Phylogeny and Classification of the Avian Superfamily Sylvioidea. Mol. Phylogenet. Evol. 2006, 38, 381–397. [Google Scholar] [CrossRef]
- Moyle, R.G.; Oliveros, C.H.; Andersen, M.J.; Hosner, P.A.; Benz, B.W.; Manthey, J.D.; Travers, S.L.; Brown, R.M.; Faircloth, B.C. Tectonic Collision and Uplift of Wallacea Triggered the Global Songbird Radiation. Nat. Commun. 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ericson, P.G.P.; Johansson, U.S. Phylogeny of Passerida (Aves: Passeriformes) Based on Nuclear and Mitochondrial Sequence Data. Mol. Phylogenet. Evol. 2003, 29, 126–138. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heibl, C. Phyloch: Interfaces and Graphic Tools for Phylogenetic Data in R. Available online: http://www.christophheibl.de/Rpackages.html (accessed on 30 April 2013).
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Paradis, E. Analysis of Phylogenetics and Evolution with R. In Use R! 2nd ed.; Springer: New York, NY, USA, 2012; p. 386. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [Green Version]
- Heled, J.; Bouckaert, R.R. Looking for Trees in the Forest: Summary Tree from Posterior Samples. BMC Evol. Biol. 2013, 13, 221. [Google Scholar] [CrossRef] [Green Version]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-Rate Poisson Tree Processes for Single-Locus Species Delimitation under Maximum Likelihood and Markov Chain Monte Carlo. Bioinformatics 2017. [Google Scholar] [CrossRef] [Green Version]
- Sharland, R.E. Two Interesting Plateau Birds: The Red-capped Lark Calandrella cinerea and the Three-banded Plover Afroxyechus tricollaris. Bull. Niger. Ornithol. Soc. 1964, 1, 4. [Google Scholar]
- Sharland, R.E. Bird-Ringing in West Africa, 1959. Niger. Field 1960, 25, 125–127. [Google Scholar] [CrossRef]
- International Commission on Zoological Nomenclature. Amendment of Articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to Expand and Refine Methods of Publication. ZooKeys 2012. [Google Scholar] [CrossRef]
- Hulme, M.F.; Cresswell, W. Density and Behaviour of Whinchats Saxicola rubetra on African Farmland Suggest That Winter Habitat Conditions Do Not Limit European Breeding Populations. Ibis 2012, 154, 680–692. [Google Scholar] [CrossRef]
- Donald, P.F.; Alström, P. Larks of the World. Bloomsbury Publishing: London, UK, forthcoming.
- Payne, R.B. A New Species of Firefinch Lagonosticta from Northern Nigeria and Its Association with the Jos Plateau Indigobird Vidua maryae. Ibis 2008, 140, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Ryan, P.G. Red-capped Lark (Calandrella cinerea). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- BirdLife International. Species Factsheet: Mirafra africana. Available online: http://datazone.birdlife.org/species/factsheet/rufous-naped-lark-mirafra-africana (accessed on 4 May 2020).
- Ryan, P.G. Red-winged Lark (Mirafra hypermetra). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Fuchs, J.; Alström, P.; Yosef, R.; Olsson, U. Miocene Diversification of an Open-Habitat Predatorial Passerine Radiation, the Shrikes (Aves: Passeriformes: Laniidae). Zool. Scr. 2019, 48, 571–588. [Google Scholar] [CrossRef]
- Fuchs, J.; Douno, M.; Bowie, R.C.K.; Fjeldså, J. Taxonomic Revision of the Square-tailed Drongo Species Complex (Passeriformes: Dicruridae) with Description of a New Species from Western Africa. Zootaxa 2018, 4438, 105–127. [Google Scholar] [CrossRef] [Green Version]
- Olsson, U.; Yosef, R.; Alström, P. Assessment of Species Limits in African ‘Brown Buntings’ (Emberiza, Passeriformes) Based on Mitochondrial and Nuclear Sequence Data. Ibis 2013, 155, 534–543. [Google Scholar] [CrossRef]
- Bertola, L.D.; Jongbloed, H.; van der Gaag, K.J.; de Knijff, P.; Yamaguchi, N.; Hooghiemstra, H.; Bauer, H.; Henschel, P.; White, P.A.; Driscoll, C.A.; et al. Phylogeographic Patterns in Africa and High Resolution Delineation of Genetic Clades in the Lion (Panthera leo). Sci. Rep. 2016, 6, 30807. [Google Scholar] [CrossRef] [Green Version]
- Lorenzen, E.D.; Heller, R.; Siegismund, H.R. Comparative Phylogeography of African Savannah Ungulates. Mol. Ecol. 2012, 21, 3656–3670. [Google Scholar] [CrossRef]
- Flagstad, O.; Syvertsen, P.O.; Stenseth, N.C.; Jakobsen, K.S. Environmental Change and Rates of Evolution: The Phylogeographic Pattern within the Hartebeest Complex as Related to Climatic Variation. Proc. R. Soc. Biol. Sci. Ser. B 2001, 268, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Moodley, Y.; Bruford, M.W. Molecular Biogeography: Towards an Integrated Framework for Conserving Pan-African Biodiversity. PLoS ONE 2007, 2, e454. [Google Scholar] [CrossRef] [Green Version]
- Muwanika, V.B.; Nyakaana, S.; Siegismund, H.R.; Arctander, P. Phylogeography and Population Structure of the Common Warthog (Phacochoerus africanus) Inferred from Variation in Mitochondrial DNA Sequences and Microsatellite Loci. Heredity 2003, 91, 361–372. [Google Scholar] [CrossRef]
- Rohland, N.; Pollack, J.L.; Nagel, D.; Beauval, C.; Airvaux, J.; Paabo, S.; Hofreiter, M. The Population History of Extant and Extinct Hyenas. Mol. Biol. Evol. 2005, 22, 2435–2443. [Google Scholar] [CrossRef] [PubMed]
- Droissart, V.; Dauby, G.; Hardy, O.J.; Deblauwe, V.; Harris, D.J.; Janssens, S.; Mackinder, B.; Blach-Overgaard, A.; Sonke, B.; Sosef, M.S.M.; et al. Beyond Trees: Biogeographical Regionalization of Tropical Africa. J. Biogeogr. 2018, 45, 1153–1167. [Google Scholar] [CrossRef]
- Ryan, P.G. Flappet Lark (Mirafra rufocinnamomea). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Ryan, P.G. White-tailed Lark (Mirafra albicauda). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- BirdLife International. Species Factsheet: Mirafra javanica. Available online: http://datazone.birdlife.org/species/factsheet/rufous-naped-lark-mirafra-javanica (accessed on 4 May 2020).
- Tieleman, B.I.; Williams, J.B.; Bloomer, P. Adaptation of Metabolism and Evaporative Water Loss Along an Aridity Gradient. Proc. R. Soc. B Biol. Sci. 2003, 270, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, P.G.; Christie, D. Kordofan Lark (Mirafra cordofanica). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Ryan, P.G. Rusty Lark (Mirafra rufa). Birds of the World. In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Ryan, P.G. Chestnut-backed Sparrow-Lark (Eremopterix leucotis). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- de Juana, E.; Suárez, F. Greater Hoopoe-Lark (Alaemon alaudipes). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- de Juana, E.; Suárez, F. Bar-Tailed Lark (Ammomanes cinctura). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- de Juana, E.; Suárez, F. Desert Lark (Ammomanes deserti). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- de Juana, E.; Suárez, F. Temminck’s Lark (Eremophila bilopha). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- de Juana, E.; Suárez, F. Thick-Billed Lark (Ramphocoris clotbey). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Ryan, P.G. Black-Crowned Sparrow-Lark (Eremopterix nigriceps). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Valente, L.; Illera, J.C.; Havenstein, K.; Pallien, T.; Etienne, R.S.; Tiedemann, R. Equilibrium Bird Species Diversity in Atlantic Islands. Curr. Biol. 2017, 27, 1660–1666. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, J.; Pons, J.-M.; Bowie, R.C.K. Biogeography and Diversification Dynamics of the African Woodpeckers. Mol. Phylogenet. Evol. 2017, 108, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Spottiswoode, C.N.; Olsson, U.; Mills, M.S.L.; Cohen, C.; Francis, J.E.; Toye, N.; Hoddinott, D.; Dagne, A.; Wood, C.; Donald, P.F.; et al. Rediscovery of a Long-Lost Lark Reveals the Conspecificity of Endangered Heteromirafra Populations in the Horn of Africa. J. Ornithol. 2013, 154, 813–825. [Google Scholar] [CrossRef]
- Larrasoaña, J.C.; Roberts, A.P.; Rohling, E.J. Dynamics of Green Sahara Periods and Their Role in Hominin Evolution. PLoS ONE 2013, 8, e76514. [Google Scholar] [CrossRef] [Green Version]
- Larrasoaña, J.C.; Roberts, A.P.; Rohling, E.J.; Winklhofer, M.; Wehausen, R. Three Million Years of Monsoon Variability over the Northern Sahara. Clim. Dyn. 2003, 21, 689–698. [Google Scholar] [CrossRef]
- deMenocal, P.B. African Climate Change and Faunal Evolution during the Pliocene–Pleistocene. Earth Planet. Sci. Lett. 2004, 220, 3–24. [Google Scholar] [CrossRef]
- Trauth, M.H.; Larrasoaña, J.C.; Mudelsee, M. Trends, Rhythms and Events in Plio–Pleistocene African Climate. Quat. Sci. Rev. 2009, 28, 399–411. [Google Scholar] [CrossRef]
- Linder, H.P. East African Cenozoic Vegetation History. Evol. Anthr. 2017, 26, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Potts, R.; Faith, J.T. Alternating High and Low Climate Variability: The Context of Natural Selection and Speciation in Plio–Pleistocene Hominin Evolution. J. Hum. Evol. 2015, 87, 5–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Päckert, M.; Martens, J.; Hering, J.; Kvist, L.; Illera, J.C. Return Flight to the Canary Island—The Key Role of Peripheral Populations of Afrocanarian Blue Tits (Aves: Cyanistes teneriffae) in Multi-Gene Reconstructions of Colonization Pathways. Mol. Phylogenet. Evol. 2013, 67, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Stervander, M.; Illera, J.C.; Kvist, L.; Barbosa, P.; Keehnen, N.P.; Pruisscher, P.; Bensch, S.; Hansson, B. Disentangling the Complex Evolutionary History of the Western Palearctic Blue Tits (Cyanistes spp.)—Phylogenomic Analyses Suggest Radiation by Multiple Colonization Events and Subsequent Isolation. Mol. Ecol. 2015, 24, 2477–2494. [Google Scholar] [CrossRef]
- Irestedt, M.; Gelang, M.; Sangster, G.; Olsson, U.; Ericson, P.G.P.; Alström, P. Neumann’s Warbler Hemitesia neumanni (Sylvioidea): The Sole African Member of a Palaeotropic Miocene Avifauna. Ibis 2011, 153, 78–86. [Google Scholar] [CrossRef]
- Rheindt, F.E.; Edwards, S.V. Genetic Introgression: An Integral but Neglected Component of Speciation in Birds. Auk 2011, 128, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Toews, D.P.L.; Brelsford, A. The Biogeography of Mitochondrial and Nuclear Discordance in Animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Ottenburghs, J. Ghost Introgression: Spooky Gene Flow in the Distant Past. Bioessays 2020. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Tang, L.; Cheng, Y.; Hao, Y.; Xiong, Y.; Song, G.; Qu, Y.; Rheindt, F.E.; Alström, P.; Jia, C.; et al. “Ghost Introgression” as a Cause of Deep Mitochondrial Divergence in a Bird Species Complex. Mol. Biol. Evol. 2019, 36, 2375–2386. [Google Scholar] [CrossRef]
- Hogner, S.; Laskemoen, T.; Lifjeld, J.T.; Porkert, J.; Kleven, O.; Albayrak, T.; Kabasakal, B.; Johnsen, A. Deep Sympatric Mitochondrial Divergence without Reproductive Isolation in the Common Redstart Phoenicurus phoenicurus. Ecol. Evol 2012, 2, 2974–2988. [Google Scholar] [CrossRef]
- Alström, P. Hume’s Lark (Calandrella acutirostris). In Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Zink, R.M.; Barrowclough, G.F. Mitochondrial DNA under Siege in Avian Phylogeography. Mol. Ecol. 2008, 17, 2107–2121. [Google Scholar] [CrossRef] [PubMed]
- Funk, D.J.; Omland, K.E. Species-Level Paraphyly and Polyphyly: Frequency, Causes, and Consequences, with Insights from Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 397–423. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.B.; Alström, P.; Ödeen, A.; Leaché, A.D. Discordance between Genomic Divergence and Phenotypic Variation in a Rapidly Evolving Avian Genus (Motacilla). Mol. Phylogenet. Evol. 2018, 120, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayrat, B. Towards Integrative Taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Padial, J.M.; Miralles, A.; De la Riva, I.; Vences, M. The Integrative Future of Taxonomy. Front. Zool. 2010, 7. [Google Scholar] [CrossRef]
- Sangster, G. Integrative Taxonomy of Birds: The Nature and Delimitation of Species. In Bird Species: How They Arise, Modify and Vanish; Tietze, D.T., Ed.; Springer Open: Cham, Switzerland, 2018; pp. 9–37. [Google Scholar]
- Alström, P.; Rasmussen, P.C.; Olsson, U.; Sundberg, P. Species Delimitation Based on Multiple Criteria: The Spotted Bush Warbler Bradypterus thoracicus Complex (Aves: Megaluridae). Zool. J. Linn. Soc. 2008, 154, 291–307. [Google Scholar] [CrossRef] [Green Version]
- Alström, P.; Rasmussen, P.C.; Sangster, G.; Dalvi, S.; Round, P.D.; Zhang, R.; Yao, C.T.; Irestedt, M.; Le Manh, H.; Lei, F.; et al. Multiple Species within the Striated Prinia Prinia crinigera–Brown Prinia P. polychroa Complex Revealed through an Integrative Taxonomic Approach. Ibis 2019, 162, 936–967. [Google Scholar] [CrossRef]
- Alström, P.; Rasmussen, P.C.; Zhao, C.; Xu, J.; Dalvi, S.; Cai, T.; Guan, Y.; Zhang, R.; Kalyakin, M.V.; Lei, F.; et al. Integrative Taxonomy of the Plain-backed Thrush (Zoothera mollissima) Complex (Aves, Turdidae) Reveals Cryptic Species, Including a New Species. Avian Res. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Alström, P.; Xia, C.; Rasmussen, P.C.; Olsson, U.; Dai, B.; Zhao, J.; Leader, P.J.; Carey, G.J.; Dong, L.; Cai, T.; et al. Integrative Taxonomy of the Russet Bush Warbler Locustella mandelli Complex Reveals a New Species from Central China. Avian Res. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Chen, G.; Huang, Q.; Jia, C.; Carey, G.; Leader, P.; Li, Y.; Zou, F.; Yang, X.; Olsson, U.; et al. Species Delimitation of the White-tailed Rubythroat Calliope pectoralis Complex (Aves, Muscicapidae) Using an Integrative Taxonomic Approach. J. Avian Biol. 2016, 47, 899–910. [Google Scholar] [CrossRef]
- Sangster, G.; Rodríguez-Godoy, F.; Roselaar, C.S.; Robb, M.S.; Luksenburg, J.A. Integrative Taxonomy Reveals Europe’s Rarest Songbird Species, the Gran Canaria Blue Chaffinch Fringilla polatzeki. J. Avian Biol. 2016, 47, 159–166. [Google Scholar] [CrossRef]
- Gjershaug, J.O.; Diserud, O.H.; Kleven, O.; Rasmussen, P.C.; Espmark, Y. Integrative Taxonomy of the Changeable Hawk-Eagle Nisaetus cirrhatus Complex (Accipitriformes: Accipitridae) in India. Zootaxa 2020, 4789, 554–574. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, M.X.; Deraad, D.A.; Tsai, W.L.E.; Zarza, E.; Zellmer, A.J.; Maley, J.M.; McCormack, J.E. Cloudy with a Chance of Speciation: Integrative Taxonomy Reveals Extraordinary Divergence within a Mesoamerican Cloud Forest Bird. Biol. J. Linn. Soc. 2019, 126, 1–15. [Google Scholar] [CrossRef]
- Younger, J.L.; Strozier, L.; Maddox, J.D.; Nyári, Á.S.; Bonfitto, M.T.; Raherilalao, M.J.; Goodman, S.M.; Reddy, S. Hidden Diversity of Forest Birds in Madagascar Revealed Using Integrative Taxonomy. Mol. Phylogenet. Evol. 2018, 124, 16–26. [Google Scholar] [CrossRef]
- Alström, P.; van Linschooten, J.; Donald, P.F.; Sundev, G.; Mohammadi, Z.; Ghorbani, F.; Shafaeipour, A.; van den Berg, A.; Robb, M.; Aliabadian, M.; et al. Multiple Species Delimitation Approaches Applied to the Avian Lark Genus Alaudala. Mol. Phylogenet. Evol. 2020. submitted. [Google Scholar] [CrossRef]
- Marki, P.Z.; Fjeldså, J.; Irestedt, M.; Jønsson, K.A. Molecular Phylogenetics and Species Limits in a Cryptically Coloured Radiation of Australo-Papuan Passerine Birds (Pachycephalidae: Colluricincla). Mol. Phylogenet. Evol. 2018, 124, 100–105. [Google Scholar] [CrossRef]
- Drovetski, S.V.; Raković, M.; Semenov, G.; Fadeev, I.V.; Red’kin, Y.A. Limited Phylogeographic Signal in Sex-Linked and Autosomal Loci Despite Geographically, Ecologically, and Phenotypically Concordant Structure of MtDNA Variation in the Holarctic Avian Genus Eremophila. PLoS ONE 2014, 9, e87570. [Google Scholar] [CrossRef] [Green Version]
- del Hoyo, J.; Collar, N.J.; Christie, D.A.; Elliott, A.; Fishpool, L.D.C.; Boesman, P.; Kirwan, G.M. Passerines. In HBW and Birdlife International Illustrated Checklist of the Birds of the World; Lynx Edicions and BirdLife International: Barcelona, Spain; Cambridge, UK, 2016; Volume 2. [Google Scholar]
- Ryan, P.G. Spike-heeled Lark (Chersomanes albofasciata). In Birds of the World; Billerman, S.M., Keeney, B.K., Rodewald, P.G., Schulenberg, T.S., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Barnes, K.N. The Phylogenetics and Evolution of Africa’s Larks (Alaudidae). Ph.D. Thesis, University of Cape Town, Cape Town, South Africa, 2007. [Google Scholar]
- Dierickx, E.G.; Shultz, A.J.; de Brooke, M.L.; Alström, P.; Liu, Y. Phylogeography of the Raso Lark, a single-island endemic, and its widespread, continental relatives. Ibis 2020. under review. [Google Scholar]
- Campbell, R.W.; Van Damme, L.M.; Johnson, S.R.; Donald, P.F.; Garcia, E. Eurasian Skylark (Alauda arvensis). In Birds of the World; Billerman, S.M., Ed.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Qian, C.; Wang, Y.; Guo, Z.; Yang, J.; Kan, X. Complete Mitochondrial Genome of Skylark, Alauda arvensis (Aves: Passeriformes): The First Representative of the Family Alaudidae with Two Extensive Heteroplasmic Control Regions. Mitochondrial DNA 2013, 24, 246–248. [Google Scholar] [CrossRef]
- Zink, R.M.; Pavlova, A.; Drovetski, S.; Rohwer, S. Mitochondrial Phylogeographies of Five Widespread Eurasian Bird Species. J. Ornithol. 2008, 149, 399–413. [Google Scholar] [CrossRef]
- Li, Y.; Chen, G.; Liu, S.; Dierickx, E.G.; Wang, P.; Birks, S.; Yao, C.; Shiraki, S.; Lei, F.; Brooke, M.d.L.; et al. Complex divergence patterns across the speciation continuum of Alauda skylarks reject a ‘ring species’ origin. Manuscript in preparation.
- van den Berg, A.B. The Sound Approach. In Morocco: Sharing the Birds—A Sound Approach Guide to Birds of the Maghreb; The Sound Approach: Poole, UK, 2020. [Google Scholar]
- BirdLife International. Species Factsheet: Calandrella cinerea. Available online: http://datazone.birdlife.org/species/factsheet/red-capped-lark-calandrella-cinerea (accessed on 4 May 2020).


| Taxa | Dependent Variable | F | d.f. 1 | Adjusted r2 | p |
|---|---|---|---|---|---|
| All | Wing length | 45.51 | 15, 353 | 0.66 | <0.001 |
| All | Bill length | 18.86 | 15, 353 | 0.44 | <0.001 |
| All | Tail length (adjusted) | 42.46 | 15, 353 | 0.64 | <0.001 |
| All | Tail/wing ratio | 26.96 | 15, 353 | 0.53 | <0.001 |
| African | Wing length | 33.30 | 8, 65 | 0.80 | <0.001 |
| African | Bill length | 18.96 | 8, 65 | 0.70 | <0.001 |
| African | Tail length | 32.61 | 8, 65 | 0.80 | <0.001 |
| African | Tail/wing ratio | 08.70 | 8, 65 | 0.52 | <0.001 |
| African | Distance to wing tip: P2 | 0.86 | 8, 65 | 0.10 | 0.554 |
| African | Distance to wing tip: P3 | 0.94 | 8, 65 | 0.10 | 0.494 |
| African | Distance to wing tip: P4 | 3.71 | 8, 65 | 0.31 | 0.001 |
| African | Distance to wing tip: P5 | 4.70 | 8, 65 | 0.37 | <0.001 |
| African | Distance to wing tip: P6 | 10.78 | 8, 65 | 0.57 | <0.001 |
| African | Distance to wing tip: P7 | 14.97 | 8, 65 | 0.65 | <0.001 |
| Wing Length (mm) | Tail Length (adjusted; mm) | Bill length (mm) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Calandrella Taxon | N | Mean (±SD) | Range | Mean (±SD) | (n) 1 | Range | Mean (±SD) | (n) 1 | Range |
| C. cinerea | 156 | 93.0 ± 3.68 | 84.5–101.5 | 60.8 ± 3.59 | (−15) | 52.0–69.5 | 14.3 ± 0.85 | (−6) | 11.6–16.3 |
| cinerea_S (southern sspp.) | 123 | 93.2 ± 3.73 | 84.5–101.5 | 61.1 ± 3.48 | (−10) | 52.0–69.5 | 14.2 ± 0.82 | (−5) | 12.0–16.2 |
| cinerea_NE (williamsi) | 32 | 92.2 ± 3.44 | 85.0–99.0 | 59.6 ± 3.86 | (−5) | 52.5–67.0 | 14.6 ± 0.92 | (−1) | 11.6–16.3 |
| cinerea_NW (Nigerian pop.) | 1 | 97 | 65 | 13.7 | |||||
| C. blanfordi | 22 | 90.3 ± 3.96 | 82.5–97.0 | 55.9 ± 3.75 | 50.0–62.5 | 13.4 ± 0.58 | 12.3–14.3 | ||
| C. b. blanfordi | 7 | 88.1 ± 4.16 | 82.5–93.0 | 54.3 ± 3.19 | 50.5–59.5 | 13.3 ± 0.67 | 12.3–14.3 | ||
| C. b. erlangeri | 15 | 91.3 ± 3.56 | 84.0–97.0 | 56.7 ± 3.84 | 50.0–62.5 | 13.4 ± 0.55 | 12.3–14.1 | ||
| C. eremica | 26 | 82.3 ± 3.32 | 76.5–88.5 | 50.3 ± 2.95 | (−1) | 44.5–57.5 | 12.5 ± 0.55 | 11.0–13.6 | |
| C. e. daaroodensis | 15 | 80.7 ± 2.76 | 76.5–86.0 | 49.1 ± 2.42 | 44.5–53.0 | 12.4 ± 0.66 | 11.0–13.6 | ||
| C. e. eremica | 11 | 84.5 ± 2.74 | 79.5–88.5 | 52.1 ± 2.88 | (−1) | 48.5–57.5 | 12.7 ± 0.30 | 12.2–13.2 | |
| C. brachydactyla | 97 | 93.5 ± 3.80 | 84.0–101.5 | 58.6 ± 3.50 | 44.5–66.0 | 13.7 ± 0.70 | 12.5–16.0 | ||
| brachydactyla_E | 54 | 93.8 ± 3.81 | 85.5–101.5 | 58.2 ± 3.51 | 44.5–63.0 | 13.5 ± 0.75 | 12.5–16.0 | ||
| brachydactyla_W | 43 | 93.2 ± 3.80 | 84.0–99.5 | 59.0 ± 3.48 | 51.0–66.0 | 13.9 ± 0.58 | 12.9–15.0 | ||
| C. dukhunensis | 20 | 97.2 ± 3.77 | 91.0–102.5 | 57.9 ± 4.04 | 51.0–63.0 | 13.5 ± 0.55 | 12.5–14.7 | ||
| C. acutirostris sensu lato | 79 | 92.7 ± 3.24 | 85.0–100.5 | 61.6 ± 3.37 | (−5) | 54.0–68.5 | 14.3 ± 0.81 | 12.7–16.5 | |
| acutirostris_C (trad. C. a. acutirostris) | 31 | 92.9 ± 3.23 | 86.0–100.5 | 60.4 ± 2.90 | 54.0–64.5 | 14.1 ± 0.71 | 12.7–15.4 | ||
| acutirostris_E (trad. C. a. tibetana) | 18 | 93.2 ± 2.81 | 87.5–100.5 | 60.7 ± 3.27 | (−3) | 54.5–67.5 | 13.8 ± 0.48 | 13.0–14.6 | |
| acutirostris_W (unnamed clade) | 30 | 92.1 ± 3.48 | 85.0–99.0 | 63.3 ± 3.26 | (−2) | 54.5–68.5 | 14.9 ± 0.70 | 13.4–16.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stervander, M.; Hansson, B.; Olsson, U.; Hulme, M.F.; Ottosson, U.; Alström, P. Molecular Species Delimitation of Larks (Aves: Alaudidae), and Integrative Taxonomy of the Genus Calandrella, with the Description of a Range-Restricted African Relic Taxon. Diversity 2020, 12, 428. https://doi.org/10.3390/d12110428
Stervander M, Hansson B, Olsson U, Hulme MF, Ottosson U, Alström P. Molecular Species Delimitation of Larks (Aves: Alaudidae), and Integrative Taxonomy of the Genus Calandrella, with the Description of a Range-Restricted African Relic Taxon. Diversity. 2020; 12(11):428. https://doi.org/10.3390/d12110428
Chicago/Turabian StyleStervander, Martin, Bengt Hansson, Urban Olsson, Mark F. Hulme, Ulf Ottosson, and Per Alström. 2020. "Molecular Species Delimitation of Larks (Aves: Alaudidae), and Integrative Taxonomy of the Genus Calandrella, with the Description of a Range-Restricted African Relic Taxon" Diversity 12, no. 11: 428. https://doi.org/10.3390/d12110428
APA StyleStervander, M., Hansson, B., Olsson, U., Hulme, M. F., Ottosson, U., & Alström, P. (2020). Molecular Species Delimitation of Larks (Aves: Alaudidae), and Integrative Taxonomy of the Genus Calandrella, with the Description of a Range-Restricted African Relic Taxon. Diversity, 12(11), 428. https://doi.org/10.3390/d12110428