Plant Broth- (Not Bovine-) Based Culture Media Provide the Most Compatible Vegan Nutrition for In Vitro Culturing and In Situ Probing of Plant Microbiota
Abstract
1. Introduction
2. Materials and Methods
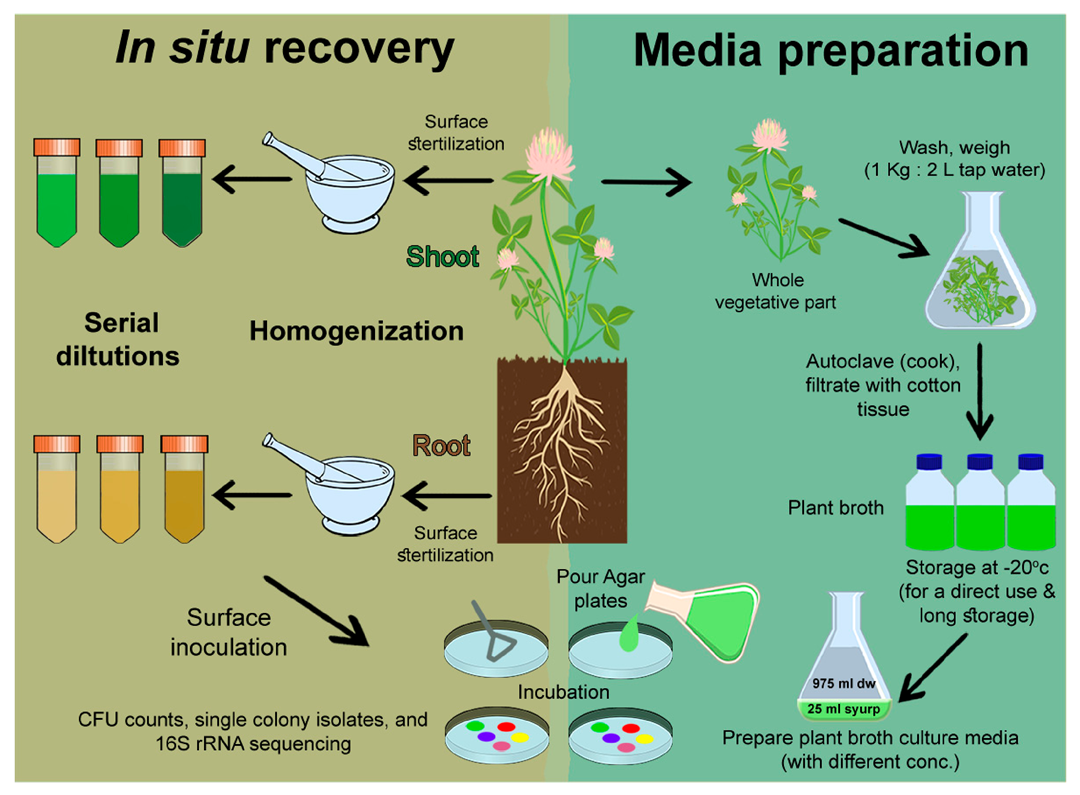
2.1. Tested Plant Materials
2.2. Plant Broth (PB)
2.3. Culture Media
2.4. In Vitro Growth of Rhizobacteria Isolates on Plant Broth-Based Culture Media
2.5. In Situ Cultivability of Endophytes of Plant Endo-Rhizosphere and Endo-Phyllosphere
2.6. In Situ Recovery and Cultivability of Wheat Endophytes on Homologous Wheat-Based Culture Media
2.7. Cultivability of Clover Endophytes on Culture Media Prepared from Homologous (Clover) and Heterologous (Wheat) Plant Broth
2.8. DNA Extraction and 16S rRNA Gene Sequencing of Bacterial Isolates
2.9. Protein Typing of Bacterial Isolates Using Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) Mass Spectrometry
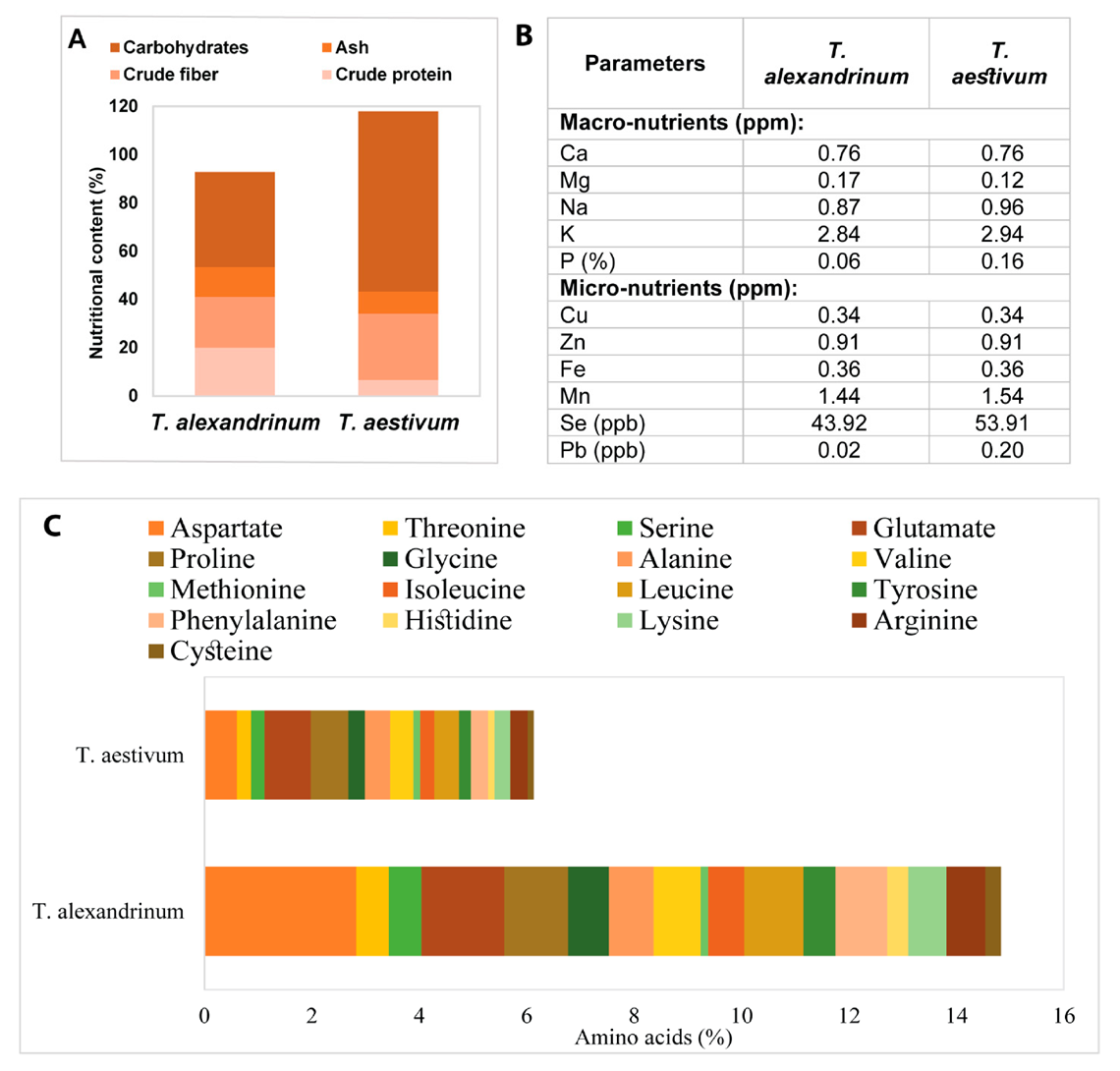
2.10. Chemical Analysis of the Dehydrated Plant Powders
2.11. Statistical and Phylogenetic Analyses
3. Results
3.1. In Vitro Growth of Rhizobacteria Isolates on Plant Broth-Based Culture Media
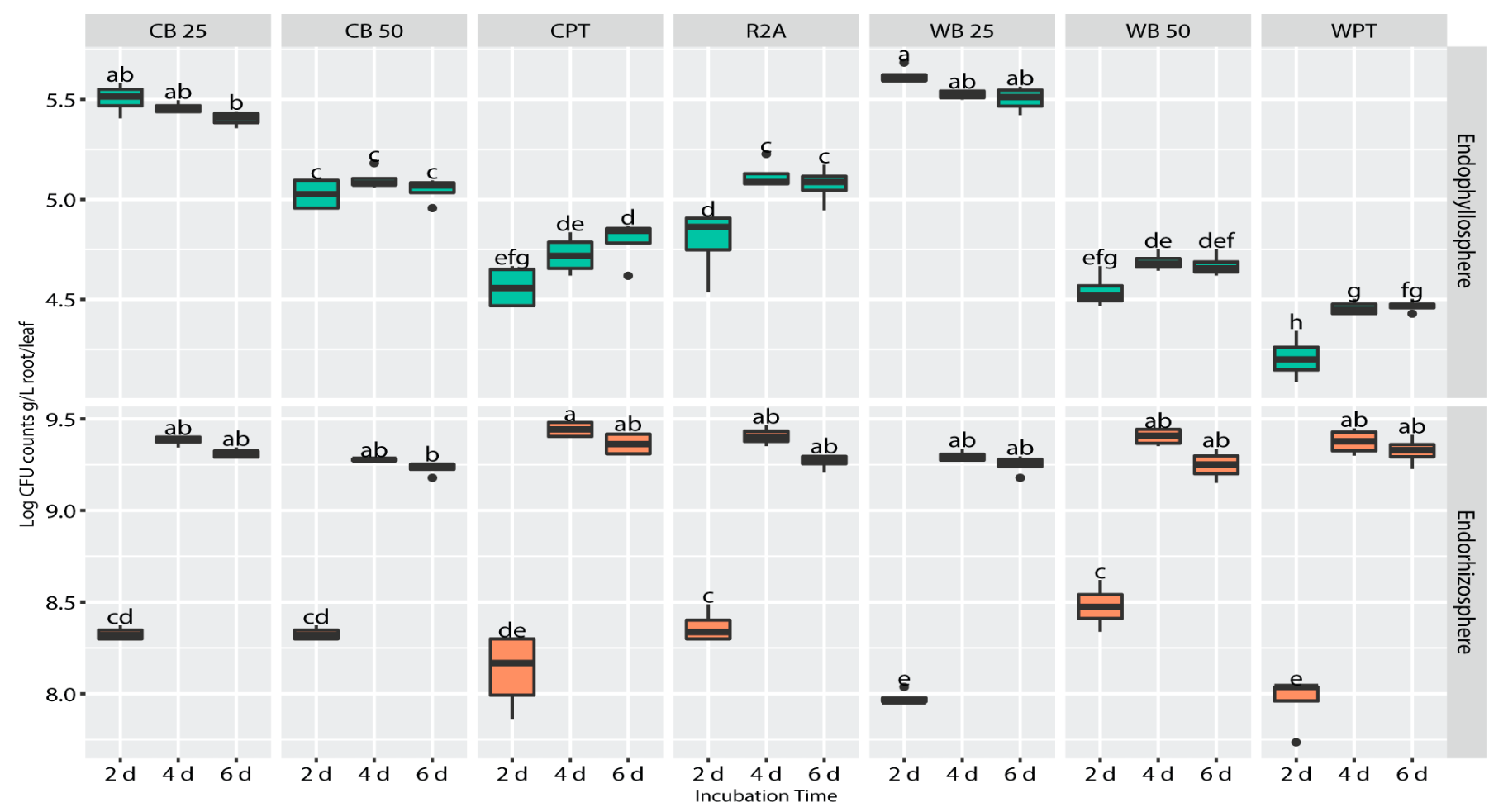
3.2. In Situ Recovery and Cultivability of Wheat Endophytes on Homologous Wheat Broth-Based Culture Media
3.3. Cultivability of Clover Endophytes on Culture Media Prepared from Homologous (Clover) and Heterologous (Wheat) Plant Broths
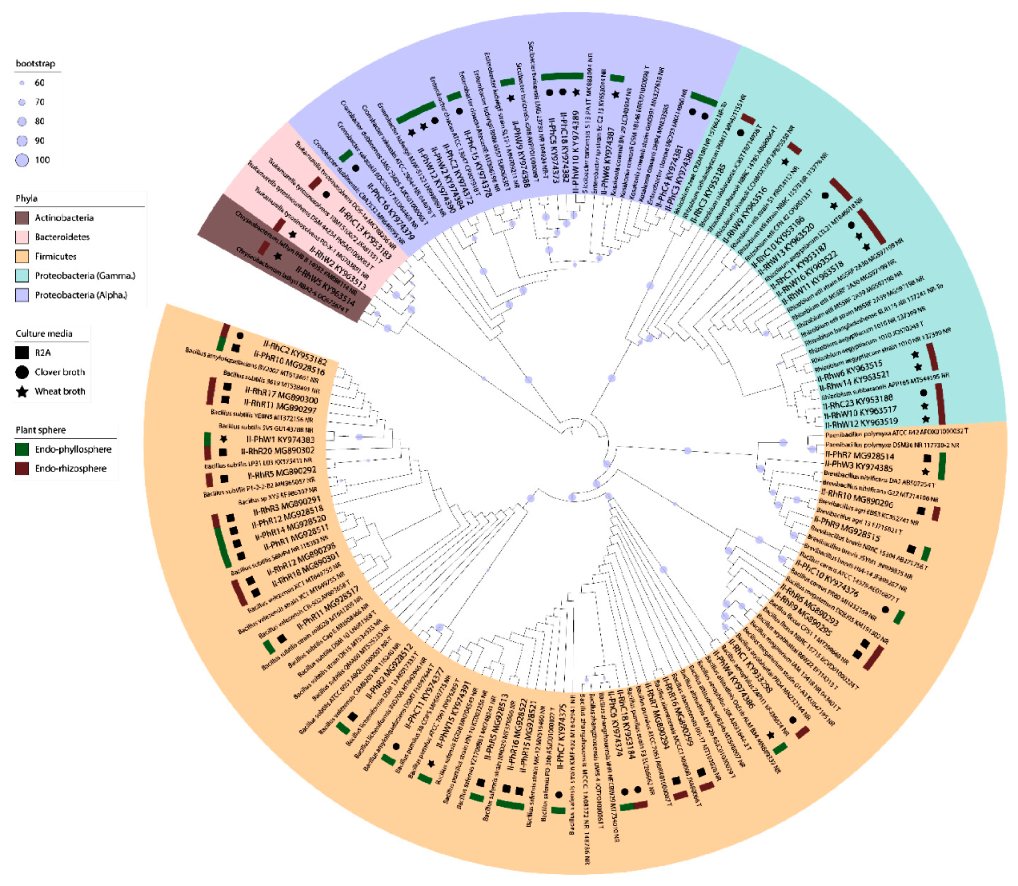
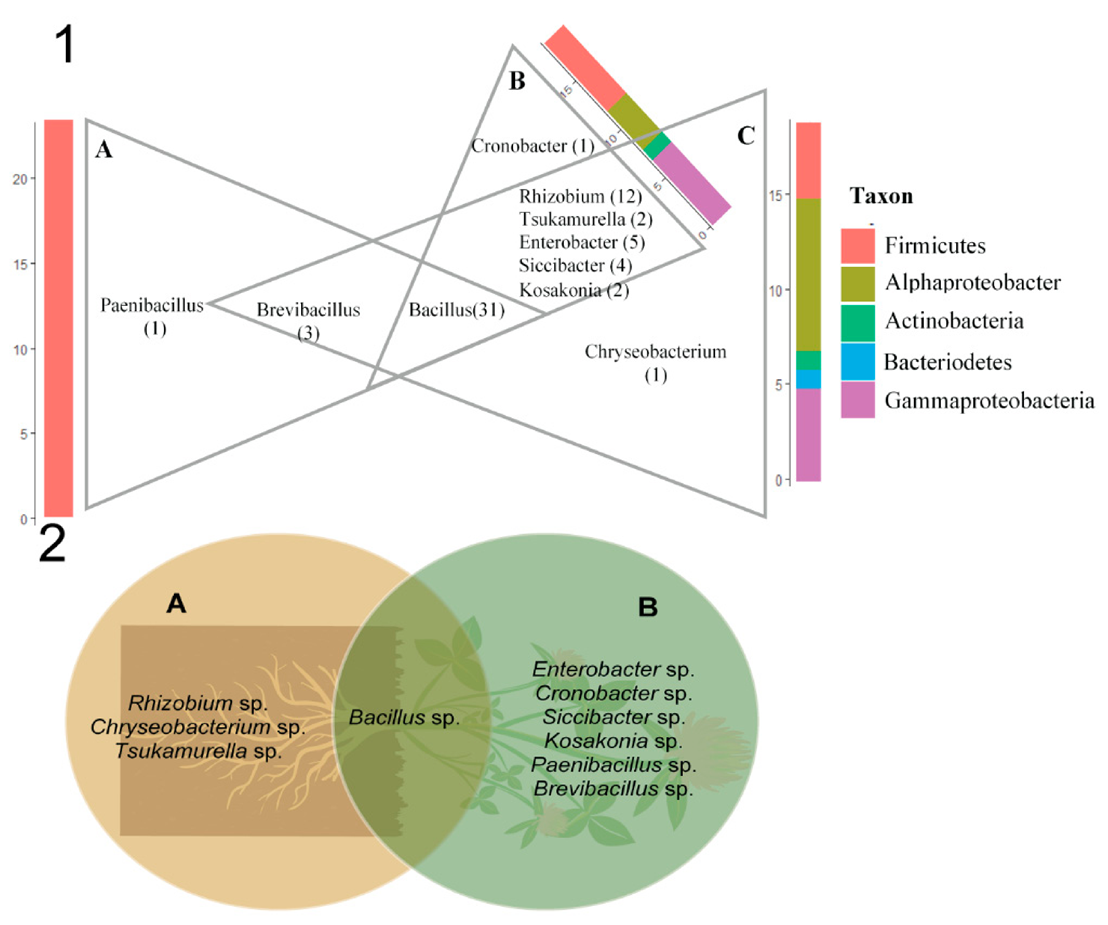
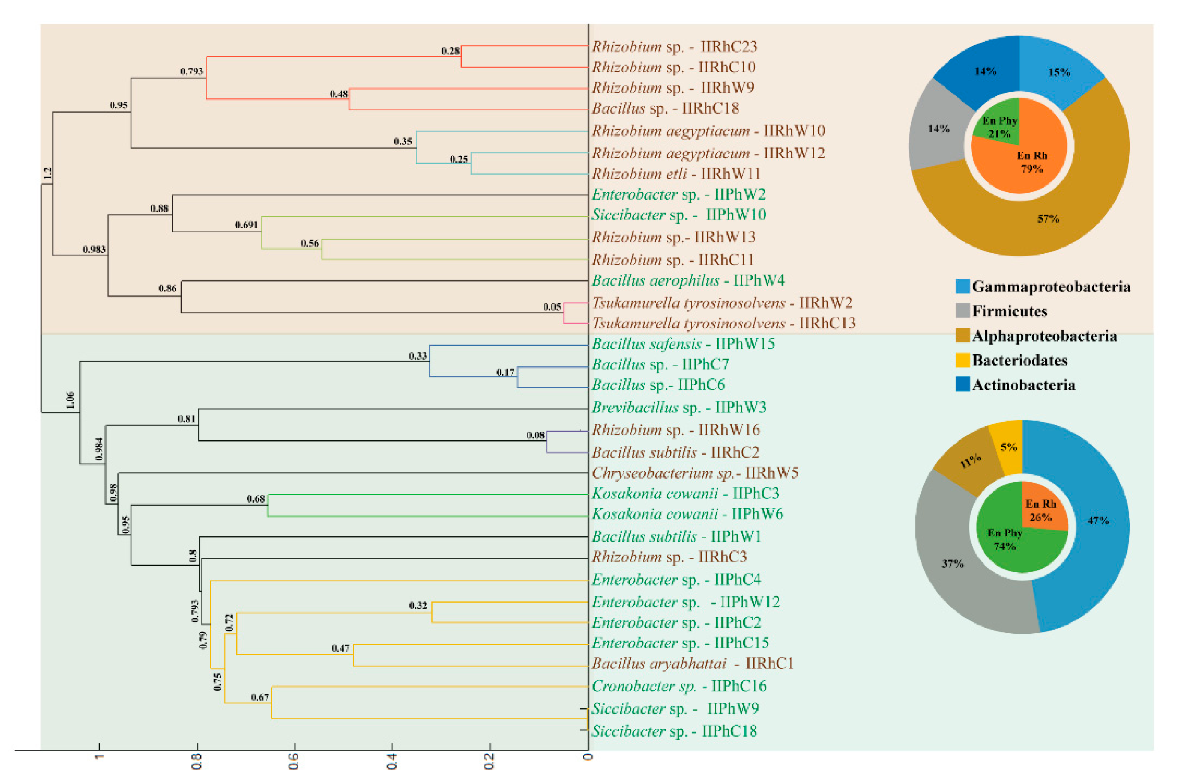
3.4. Diversity of Clover Endophytes Based on 16S rRNA Gene Sequences
3.5. MALDI-TOF MS Analysis of Clover Endophytes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alivisatos, A.P.; Blaser, M.J.; Brodie, E.L.; Chun, M.; Dangl, J.L.; Donohue, T.J.; Dorrestein, P.C.; Gilbert, J.A.; Green, J.L.; Jansson, J.K.; et al. A unified initiative to harness Earth’s microbiomes. Science 2015, 350, 507–508. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Meyer, F.; Antonopoulos, D.; Balaji, P.; Brown, C.T. Meeting Report: The Terabase Metagenomics Workshop and the Vision of an Earth Microbiome. Stand. Genom. Sci. 2010, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Fraune, S.; Bosch, T.C. Why bacteria matter in animal development and evolution. BioEssays 2010, 7, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E. Research priorities for harnessing plant microbiomes in sustainable agriculture Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Musilova, L.; Ridl, J.; Polivkova, M.; Macek, T.; Uhlik, O. Effects of secondary plant metabolites on microbial populations: Changes in community structure and metabolic activity in contaminated environments. Int. J. Mol. Sci. 2016, 17, 1205. [Google Scholar] [CrossRef]
- Becker, M.; Patz, S.; Becker, Y.; Berger, B.; Drungowski, M.; Bunk, B.; Overmann, J.; Spröer, C.; Reetz, J.; Tchakounte, G.V.T.; et al. Comparative genomics reveal a flagellar system, a type VI secretion system and plant growth-promoting gene clusters unique to the endophytic bacterium Kosakonia radicincitans. Front. Microbiol. 2018, 9, 1–22. [Google Scholar] [CrossRef]
- Huang, X.-F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities 1. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-oter, R.; Mchardy, A.C.; Schulze-lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 392–403. [Google Scholar] [CrossRef]
- Abbasi, M.K. Isolation and characterization of rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Koch, R. Zur Untersuchung von pathogenen Organismen (1881). Entdeck. Doppelhelix 2018, 45–111. [Google Scholar] [CrossRef][Green Version]
- Basu, S.; Bose, C.; Ojha, N.; Das, N.; Das, J.; Pal, M.; Khurana, S. Evolution of bacterial and fungal growth media. Bioinformation 2015, 11, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; ASM Press: Washington, DC, USA; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2010; ISBN 9781439804087. [Google Scholar]
- Austin, B. The value of cultures to modern microbiology. Antonie Van Leeuwenhoek 2017, 110, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, M.S.; Hamza, M.A.; Youssef, H.H.; Patz, S.; Becker, M.; Elsawey, H.; Nemr, R.; Daanaa, H.A.; Mourad, E.F.; Morsi, A.T.; et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant- based culture media—A review. J. Adv. Res. 2019, 19, 15–27. [Google Scholar] [CrossRef]
- Ferrari, B.C.; Binnerup, S.J.; Gillings, M. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 2005, 71, 8714–8720. [Google Scholar] [CrossRef]
- Bollmann, A.; Lewis, K.; Epstein, S.S. Incubation of Environmental Samples in a Diffusion Chamber Increases the Diversity of Recovered Isolates. Appl. Environ. Microbiol. 2007, 73, 6386–6390. [Google Scholar] [CrossRef]
- Aoi, Y.; Kinoshita, T.; Hata, T.; Ohta, H.; Obokata, H.; Tsuneda, S. Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol. 2009, 75, 3826–3833. [Google Scholar] [CrossRef]
- Nour, E.H.; Hamza, M.A.; Fayez, M.; Monib, M.; Ruppel, S.; Hegazi, N.A. The crude plant juices of desert plants as appropriate culture media for the cultivation of rhizospheric microorganisms. J. Adv. Res. 2012, 3, 35–43. [Google Scholar] [CrossRef]
- Eevers, N.; Gielen, M.; Sánchez-López, A.; Jaspers, S.; White, J.C.; Vangronsveld, J.; Weyens, N. Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Microb. Biotechnol. 2015, 8, 707–715. [Google Scholar] [CrossRef]
- Murphy, B.R.; Batke, S.P.; Doohan, F.M.; Hodkinson, T.R. Media Manipulations and the Culture of Beneficial Fungal Root Endophytes. Int. J. Biol. 2015, 7, 94–102. [Google Scholar] [CrossRef]
- Sarhan, M.S.; Mourad, E.F.; Hamza, M.A.; Youssef, H.H.; Scherwinski, A.C.; El-Tahan, M.; Fayez, M.; Ruppel, S.; Hegazi, N.A. Plant powder teabags: A novel and practical approach to resolve culturability and diversity of rhizobacteria. Physiol. Plant. 2016, 157, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Youssef, H.H.; Hamza, M.A.; Fayez, M.; Mourad, E.F.; Saleh, M.Y.; Sarhan, M.S.; Suker, R.M.; Eltahlawy, A.A.; Nemr, R.A.; El-Tahan, M.; et al. Plant-based culture media: Efficiently support culturing rhizobacteria and correctly mirror their in-situ diversity. J. Adv. Res. 2016, 7, 305–316. [Google Scholar] [CrossRef]
- Saleh, M.Y.; Sarhan, M.S.; Mourad, E.F.; Hamza, M.A.; Abbas, M.T.; Othman, A.A.; Youssef, H.H.; Morsi, A.T.; Youssef, G.H.; El-Tahan, M.; et al. A novel plant-based-sea water culture media for in vitro cultivation and in situ recovery of the halophyte microbiome. J. Adv. Res. 2017, 8, 577–590. [Google Scholar] [CrossRef]
- Sarhan, M.S.; Patz, S.; Hamza, M.A.; Youssef, H.H.; Mourad, E.F.; Fayez, M.; Murphy, B.; Ruppel, S.; Hegazi, N.A. G3 PhyloChip Analysis Confirms the Promise of Plant-Based Culture Media for Unlocking the Composition and Diversity of the Maize Root Microbiome and for Recovering Unculturable Candidate Divisions/Phyla. Microbes Environ. 2018, 33, 317–325. [Google Scholar] [CrossRef]
- Mourad, E.F.; Sarhan, M.S.; Daanaa, H.-S.A.; Abdou, M.; Morsi, A.T.; Abdelfadeel, M.R.; Elsawey, H.; Nemr, R.; El-Tahan, M.; Hamza, M.A.; et al. Plant Materials are Sustainable Substrates Supporting New Technologies of Plant-Only-Based Culture Media for in vitro Culturing of the Plant Microbiota. Microbes Environ. 2018, 33, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.N.; Akter, A.; Humayan, S.; Mohanto, L.C.; Begum, S.; Ahmed, M.M. Edible Growth Medium: A New Window for Probiotic Research. Adv. Microbiol. 2020, 10, 39–51. [Google Scholar] [CrossRef]
- Hong, S.; Bunge, J.; Leslin, C.; Jeon, S.; Epstein, S.S. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 2009, 3, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lei, L.; Duan, Y.; Zhang, K.Q.; Yang, J. Culture-independent methods for studying environmental microorganisms: Methods, application, and perspective. Appl. Microbiol. Biotechnol. 2012, 93, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Adewale, M.; Kheng, J.; Ping, S.; Su, A.; Ting, Y. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. GDATA 2015, 6, 159–163. [Google Scholar] [CrossRef]
- Knight, R.; Navas, J.; Quinn, R.A.; Sanders, J.G.; Zhu, Q. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Fricker, A.M.; Podlesny, D.; Fricke, W.F. What is new and relevant for sequencing-based microbiome research? A mini-review. J. Adv. Res. 2019, 19, 105–112. [Google Scholar] [CrossRef]
- Hegazi, N.A.; Sarhan, M.S.; Fayez, M.; Patz, S.; Murphy, B.R.; Ruppel, S. Plant-fed versus chemicals-fed rhizobacteria of Lucerne: Plant-only teabags culture media not only increase culturability of rhizobacteria but also recover a previously uncultured Lysobacter sp., Novosphingobium sp. and Pedobacter sp. PLoS ONE 2017, 12, e0180424. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef]
- Jensen, V. The dilution plate count technique for the enumeration of bacteria and fungi in soil. Zbl Bakteriol Parasitenkde 1962, 116, 13–32. [Google Scholar]
- Hegazi, N.A.; Hamza, M.A.; Osman, A.; Ali, S.; Sedik, M.Z. Modified combined carbon N-deficient medium for isolation, enumeration and biomass production of diazotrophs. Nitrogen Fixat. Non-Legumes 1998, 1, 247–253. [Google Scholar] [CrossRef]
- Youssef, H.H.; Fayez, M.; Monib, M.; Hegazi, N. Gluconacetobacter diazotrophicus: A natural endophytic diazotroph of Nile Delta sugarcane capable of establishing an endophytic association with wheat. Biol. Fertil. Soils 2004, 39, 391–397. [Google Scholar] [CrossRef]
- de Oliveira Costa, L.E.; de Queiroz, M.V.; Borges, A.C.; de Moraes, C.A.; de Araújo, E.F. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 2012, 43, 1562–1575. [Google Scholar] [CrossRef]
- Jackson, C.R.; Randolph, K.C.; Osborn, S.L.; Tyler, H.L. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013, 13, 274. [Google Scholar] [CrossRef]
- Mühling, M.; Woolven-Allen, J.; Murrell, J.C.; Joint, I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2008, 2, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Lewis, K.; Orjala, J.; Mo, S.; Ortenberg, R.; O’Connor, P.; Zhao, C.; Vouros, P.; Kaeberlein, T.; Epstein, S.S. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 2008, 74, 4889–4897. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef]
- Neumann, G.; Römheld, V. The release of root exudates as affected by the plant physiological status. In The Rhizosphere: Biochemic and Organic Substances at the Soil-Plant Interface; Pinton, R., Varani, Z., Nannipieri, P., Eds.; Marrcel Dekker, Inc.: New York, NY, USA, 2000; pp. 41–98. ISBN 0849338557. [Google Scholar]
- Tesfaye, M.; Dufault, N.S.; Dornbusch, M.R.; Allan, D.L.; Vance, C.P.; Samac, D.A. Influence of enhanced malate dehydrogenase expression by alfalfa on diversity of rhizobacteria and soil nutrient availability. Soil Biol. Biochem. 2003, 35, 1103–1113. [Google Scholar] [CrossRef]
- Hawkes, C.V.; DeAngelis, K.M.; Firestone, M.K. Root Interactions with Soil Microbial Communities and Processes. Rhizosphere 2007, 1–29. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Piromyou, P.; Songwattana, P.; Greetatorn, T.; Okubo, T.; Kakizaki, K.C.; Prakamhang, J.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N.; Minamisawa, K. The Type III Secretion System (T3SS) is a Determinant for Rice-Endophyte Colonization by Non-Photosynthetic Bradyrhizobium. Microbes Environ. 2015, 30, 291–300. [Google Scholar] [CrossRef]
- Straub, D.; Rothballer, M.; Hartmann, A.; Ludewig, U. The genome of the endophytic bacterium H. frisingense GSF30Tidentifies diverse strategies in the Herbaspirillum genus to interact with plants. Front. Microbiol. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Sheibani-Tezerji, R.; Rattei, T.; Sessitsch, A.; Trognitz, F.; Mitter, B. Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. MBio 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Compant, S.; Reiter, B.; Nowak, J.; Sessitsch, A.; Clément, C.; Barka, E.A. Endophytic Colonization of Vitis vinifera L. by Plant Growth- Promoting Bacterium Burkholderia sp. Strain PsJN. Appl. Environ. Microbiol. 2005, 71, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Terashima, M.; Yyano, A.; Sato, M.; Kitagawa, W.; Kawasaki, K.; Kamagata, Y. Improved Isolation of Uncultured Anaerobic Bacteria using Medium Prepared with Separate Sterilization of Agar and Phosphate. Microbes Environ. 2020, 35, 2–5. [Google Scholar] [CrossRef]
- Watve, M.; Shejval, V.; Sonawane, C.; Rahalkar, M. The’K’selected oligophilic bacteria: A key to uncultured diversity? Curr. Sci. 2000, 17, 1535–1542. [Google Scholar]
- Sun, J.; Guo, J.; Yang, Q.; Huang, J. Diluted conventional media improve the microbial cultivability from aquarium seawater. J. Microbiol. 2019, 57, 759–768. [Google Scholar] [CrossRef]
- Pereira, P.; Ibáñez, F.; Rosenblueth, M.; Etcheverry, M.; Martínez-Romero, E. Analysis of the Bacterial Diversity Associated with the Roots of Maize (Zea mays L.) through Culture-Dependent and Culture-Independent Methods. ISRN Ecol. 2011, 2011, 1–10. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Baar, B.L.M. Van Characterisation of bacteria by matrix-assisted laser desorption/ionisation and electrospray mass spectrometry. FEMS Microbiol. Rev. 2000, 24, 193–219. [Google Scholar] [CrossRef]
- Lay, J.O. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 2001, 20, 172–194. [Google Scholar] [CrossRef]
- Uhlik, O.; Strejcek, M.; Junkova, P.; Sanda, M.; Hroudova, M.; Vlcek, C.; Mackova, M.; Macek, T. Matrix-assisted laser desorption ionization (MALDI)-time of flight mass spectrometry- and MALDI biotyper-based identification of cultured biphenyl-metabolizing bacteria from contaminated horseradish rhizosphere soil. Appl. Environ. Microbiol. 2011, 77, 6858–6866. [Google Scholar] [CrossRef]
- Angolini, C.F.F.; Pilau, E.J.; Lopes-Oliveira, P.F.; Garcia, I.N.S.; Gozzo, F.C.; De Oliveira, V.M.; Marsaioli, A.J. Classification and identification of petroleum microorganisms by MALDI-TOF mass spectrometry. J. Braz. Chem. Soc. 2015, 26, 513–520. [Google Scholar] [CrossRef]
- Ziegler, D.; Mariotti, A.; Pflüger, V.; Saad, M.; Vogel, G.; Tonolla, M.; Perret, X. In situ identification of plant-invasive bacteria with MALDI-TOF mass spectrometry. PLoS ONE 2012, 7, e37189. [Google Scholar] [CrossRef]
| Culture Media | Endo-Rhizosphere (Log CFUs g−1) | Endo-Phyllosphere (Log CFUs g−1) | ||||
|---|---|---|---|---|---|---|
| Incubation at 28 °C (Days) | ||||||
| 2 Days | 7 Days | 14 Days | 2 Days | 7 Days | 14 Days | |
| ½ modified R2A | 7.86 abc | 7.96 a | 7.94 ab | 6.43 b | 6.59 a | 6.46 ab |
| ½ CCM | 7.83 abcd | 7.90 abc | 7.76 bcde | 6.19 cd | 6.22 cd | 6.22 cd |
| WB b | 7.16 f | 7.72 cde | 7.62 e | 6.19 cd | 6.24 cd | 6.27 c |
| WPT b | 7.64 de | 7.92 ab | 7.88 abc | 6.11 d | 6.22 cd | 6.17 cd |
| HSD (p value ≤ 0.05) = | 0.20 | 0.15 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsawey, H.; Patz, S.; Nemr, R.A.; Sarhan, M.S.; Hamza, M.A.; Youssef, H.H.; Abdelfadeel, M.R.; Daanaa, H.-S.A.; El-Tahan, M.; Abbas, M.; et al. Plant Broth- (Not Bovine-) Based Culture Media Provide the Most Compatible Vegan Nutrition for In Vitro Culturing and In Situ Probing of Plant Microbiota. Diversity 2020, 12, 418. https://doi.org/10.3390/d12110418
Elsawey H, Patz S, Nemr RA, Sarhan MS, Hamza MA, Youssef HH, Abdelfadeel MR, Daanaa H-SA, El-Tahan M, Abbas M, et al. Plant Broth- (Not Bovine-) Based Culture Media Provide the Most Compatible Vegan Nutrition for In Vitro Culturing and In Situ Probing of Plant Microbiota. Diversity. 2020; 12(11):418. https://doi.org/10.3390/d12110418
Chicago/Turabian StyleElsawey, Hend, Sascha Patz, Rahma A. Nemr, Mohamed S. Sarhan, Mervat A. Hamza, Hanan H. Youssef, Mohamed R. Abdelfadeel, Hassan-Sibroe A. Daanaa, Mahmoud El-Tahan, Mohamed Abbas, and et al. 2020. "Plant Broth- (Not Bovine-) Based Culture Media Provide the Most Compatible Vegan Nutrition for In Vitro Culturing and In Situ Probing of Plant Microbiota" Diversity 12, no. 11: 418. https://doi.org/10.3390/d12110418
APA StyleElsawey, H., Patz, S., Nemr, R. A., Sarhan, M. S., Hamza, M. A., Youssef, H. H., Abdelfadeel, M. R., Daanaa, H.-S. A., El-Tahan, M., Abbas, M., Fayez, M., Witzel, K., Ruppel, S., & Hegazi, N. A. (2020). Plant Broth- (Not Bovine-) Based Culture Media Provide the Most Compatible Vegan Nutrition for In Vitro Culturing and In Situ Probing of Plant Microbiota. Diversity, 12(11), 418. https://doi.org/10.3390/d12110418






