Abstract
We previously established that polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis using partial plastid rbcL and mitochondrial trnC–trnP gene sequences can be used to distinguish the six representative Pyropia species produced via mariculture in Korea. In this study, we develop progressive InDel markers by comparing seven complete Pyropia chloroplast genomes obtained from The National Center of Biotechnology Informnation (NCBI) GenBank. Comparative analyses of nucleotide diversity among the genomes revealed seven hypervariable sites (cemA, rps13, trnM-argB, petD-petB, trnR-trnQ, ccs1-orf24, and ycf12-ftrB) among 637 sliding windows with nucleotide diversity > 0.025 (Pi). These sites included two genes and five gene-intergenic regions, three of which (cemA, trnM-argB, trnR-trnQ) showed complete amplification for all six test species. Finally, trnM-argB, an InDel-variable locus with high discriminatory power, was selected as a DNA barcode candidate. These results suggest that the obtained trnM-argB region can be used for the effective exploration of the variation present in six Korean Pyropia and for further evolutionary, phylogenetic, barcoding and genetic engineering studies of Pyropia species.
1. Introduction
Pyropia, a genus of red algae (Bangiales, Rhodophyta), is an economically important mariculture crop and food source with a long history of consumption in China, Japan, and South Korea []. This genus is widely used as a laver or dried sheet product, “zicai” in China, “nori” in Japan, and “gim” in South Korea [].
Among twelve species of Pyropia distributed in South Korea, Pyropia yezoensis, Pyropia seriata, Pyropia dentata, and Pyropia suborbiculata are extensively cultured in the Korean southwest sea, whereas Pyropia pseudolinearis is distributed across the Korean East Sea []. Pyropia haitanensis is one of the Chinese indigenous laver species and has been utilized as a potential genetic source for breeding to develop improved cultivars that are optimized for the warm temperature zones in Korean mariculture industries.
In Korean aquaculture industry, Pyropia species are identified using morphological and anatomical features. Morphological characteristics remain an important tool for phylogenetic studies, even in the current age of molecular systematics; however, the use of classical morphological features to distinguish Pyropia species has some limitations. In addition, for morphologically similar Pyropia species such as P. dentata and P. haitanensis species-specific identification and discrimination in the field is difficult and require a using a common DNA barcoding method.
Thus, in our previous study, we established a PCR-restriction fragment length polymorphism (RFLP) method for the rapid and accurate identification of six representative Korean Pyropia species []. On the basis of that study, a high level of expertise is required to correctly identify species with the accuracy required in the aquaculture field and marine industry.
In the present study, we identified hypervariable loci by comparing the chloroplast genomes of seven Pyropia species, which enabled us to develop valuable chloroplast-based InDel markers for authenticating six Korean Pyropia species.
2. Materials and Methods
2.1. Chloroplast Genome Comparison and Identification of Hypervariable InDels
We downloaded from GenBank all of the chloroplast genome sequences in the Pyropia genus with complete genome sequence information (Table 1). The sequences were first aligned using the Clustal W algorithm of MEGA7.0 []. The chloroplast genome gene distribution and similarities of Pyropia species were compared and visualized using mVISTA software in Shuffle-LAGAN mode with the annotation of P. haitanensis NC021189 as a reference []. The variability of the aligned genomes was evaluated using the sliding window method in DNAsp ver. 5.0 []. The window length was set to 600 bp, the typical length of DNA markers. The step size was set to 250 for relatively accurate positioning of hyper-variable InDels. We only considered regions with nucleotide diversity (Pi) of a value > 0.2. Hypervariable sites and genetic distances among the chloroplast genomes were calculated using MEGA 7.0. Based on the aligned sequence matrix, the InDel events were checked manually.

Table 1.
Complete and partial chloroplast genomes of the seven Pyropia species used in this study.
2.2. Sample Collection and DNA Isolation
All samples were identified based on our previous study [] (Table 2). Prior to DNA extraction, conchocelis-stage wet samples of all strains (~200 mg wet weight) were washed twice with distilled water and centrifuged for 20 min at 3000× g twice. The remaining wet phase was dried between sheets of filter papers at room temperature, and 50 mg dry weight of each sample was added to steal bead tubes (2.38 mm diameter) from the PowerPlant Pro DNA isolation Kit (Qiagen, Valencia, Spain), and then the mixtures were subjected to bead beating using a Precellys Evolution homogenizer (Bertin Technologies, Paris, France). Genomic DNA was extracted from ground conchocelis-stage samples using the PowerPlant Pro DNA isolation Kit (Qiagen, Valencia, Spain) according to the manufacturer’s instructions.

Table 2.
Pyropia samples used in this study.
2.3. Development and Validation of the InDel Molecular Marker
In order to validate interspecies polymorphisms within the chloroplast genomes and develop DNA genetic markers for identifying the six Pyropia species studied here, primers were designed using Primer 3 and NCBI Primer-BLAST based on the mutational hotspot regions (hypervariable regions) found in the Pyropia chloroplast genomes. PCR amplifications were performed in a reaction volume of 50 μL containing 25 μL premix (Ex Taq Version 2.0, TaKaRa, Japan), 10 ng genomic DNA template, and 1 μL (10 pM) forward and reverse primers. The mixtures were denatured at 95 °C for 5 min and amplified with 40 cycles of 95 °C for 30 s, 55 °C for 20 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. For the detection of PCR amplicons, PCR products were separated by capillary electrophoresis (QIAxcel, Qiagen, Valencia, Spain) using the high-resolution kit and the 0M500 method (Qiagen, Valencia, Spain). The target DNA was extracted and purified using a MinElute PCR Purification Kit (Qiagen, Valencia, Spain). Purified PCR products were then sent to CosmoGenetech for sequencing (Seoul, Korea) with both forward and reverse primers. Sequencing results were analyzed by a BLAST search of the GenBank database. Alignment and graphical representation was carried out in CLC Sequence Viewer 8.0.
3. Results
The divergent regions of seven Pyropia chloroplast genomes were examined with the aim of distinguishing six common Pyropia species. First, interspecific comparisons of sequence identity among the seven chloroplast genomes were conducted with mVISTA using the annotated P. haitanensis sequence as a reference (Figure 1).
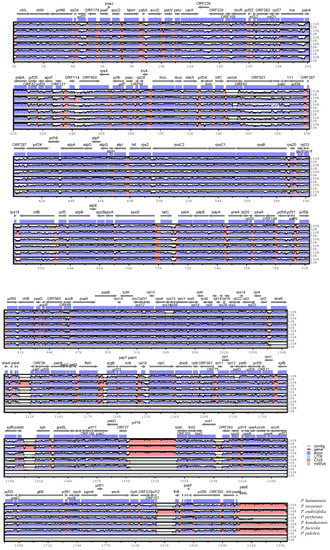
Figure 1.
Comparison of seven Pyropia chloroplast genomes using mVISTA. The complete chloroplast genomes of seven Pyropia species obtained from GenBank were compared using P. haitanensis as a reference. Blue block: conserved genes; red block: conserved non-coding sequences (CNS). White areas represent regions with sequence variation among the six Pyropia species.
The mVISTA results showed that the seven chloroplast genomes were highly conserved; however, the coding gene regions appeared to be more variable than the non-coding sequence (CNS) regions. The overall sequence divergence estimated based on the p-distance among the seven genomes was only 0.1335. The pairwise p-distances between the species ranged from 0.1205 to 0.1416. The highest p-distance value was due to insufficient regions analyzed between the partial genome sequences of two species, P. fucicola KJ776837 and P. kanakaensis KJ776836. Furthermore, sliding window analysis using DnaSP detected highly variable regions among the Pyropia chloroplast genomes (Figure 2).
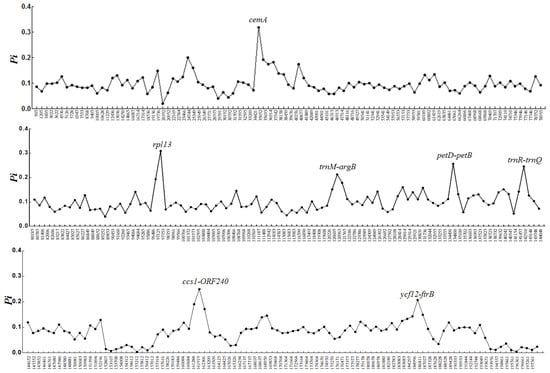
Figure 2.
Sliding window analysis of the whole chloroplast genomes of seven Pyropia species. Window length: 600 bp; step size: 250 bp. X-axis: position of the midpoint of a window; Y-axis: nucleotide diversity of each window (Pi).
The average value of nucleotide diversity (Pi) was 0.09601. Among the 637 windows, there were seven mutational hotspots that showed remarkably higher Pi values (>0.2), including two gene regions (cemA, 0.31105; rps13, 0.31989) and five intergenic regions (trnM-argB, 0.21322; petD-petB, 0.24061; trnR-trnQ, 0.24611; ccs1-ORF240, 0.24944; ycf12-chlI, 0.20689) (Figure 2).
Among these seven hypervariable regions, we identified InDel regions by PCR amplification and sequencing to discriminate between six of the Pyropia species (Table 3).

Table 3.
Primers for amplifying and sequencing seven highly variable loci.
The specific primer pairs for three loci (cemA, trnM-argB, trnR-trnQ) successfully amplified their target in all six species. For the other loci, we failed to amplify sequences as follows: rpl13 of P. yezoensis; petD-petB of P. yezoensis, P. haitanensis, and P. pseudolinearis; ccs1-ORF240 of P. yezoensis and P. pseudolinearis; ycf12-ftrB of P. yezoensis, P. dentata, P. haitanensis, and P. seriata (Figure 3).
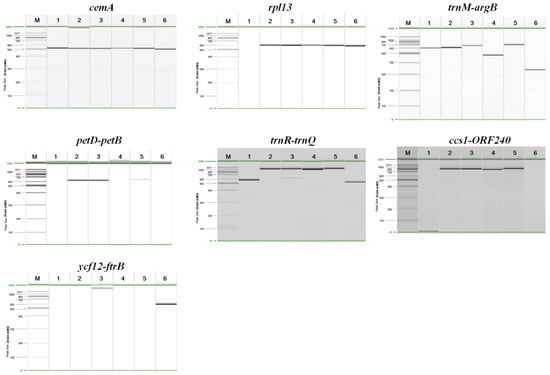
Figure 3.
Gel profiles of fragments amplified from six species using seven pairs of primers derived from hypervariable regions of seven Pyropia chloroplast genomes. M indicates the lane containing the molecular weight marker. The numbers correspond to the species listed in Table 2.
The cemA primers resulted in PCR products of 529, 522, 520, 527, 523, and 506 bp from P. yezoensis, P. dentata, P. suborbiculata, P. haitanensis, P. seriata, and P. pseudolinearis, respectively, and the trnM-argB primers resulted in products of 597, 629, 655, 484, 696, and 341 bp, respectively. The product sizes for the trnR-trnQ primers were as follows: 579 bp from P. yezoensis; 1166 bp from P. dentata; 1198 bp from P. suborbiculata; 1061 bp from P. haitanensis; 1260 bp from P. seriata; and 534 bp from P. Pseudolinearis (Table 3 and Figure 3).
We considered 600 bp to be the acceptable length of the sequence read of a given PCR product. To evaluate the sequence divergence between six Pyropia species at the three hypervariable InDel regions, the average pairwise distance values were calculated for each marker using MEGA7.0. The trnR-trnQ locus was the most divergent with a maximum pairwise distance of 0.572, followed by trnM-argB (0.295). The lowest genetic distance was observed at cemA (0.108), indicating that there are no differences between the six species at that locus. Therefore, the species-specific primers derived from the trnR-trnQ and trnM-argB regions are considered useful markers for elucidating the phylogenetic relationships of the six Pyropia species analyzed. However, when selecting attractive InDel markers, the length of the amplified regions must also be considered. The length of only one region, trnM-argB, is considered relatively short and sufficient to reproduce the nucleotide variation in Pyropia taxa (Figure 4).
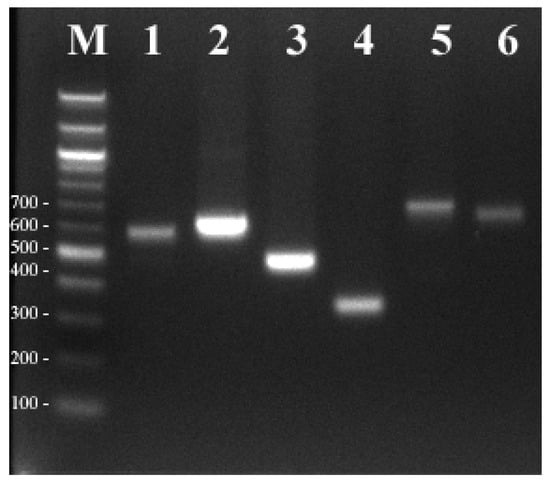
Figure 4.
PCR verification of trnM-argB InDel markers for six Pyropia species. M, 1 kb DNA ladder; 1, P. yezoensis; 2, P. dentata; 3, P. suborbiculata; 4, P. haitanensis; 5, P. seriata; 6, P. pseudolinearis.
4. Discussion
In our previous work, we found that six restriction enzymes were predicted to show species-specific RFLP patterns and could be used to identify the six Pyropia species using rbcL in P. yezoensis and rps11-sdh3 in P. seriata, P. pseudolinearis, P. dentata, P. suborbiculata, and P. haitanensis []. This work demonstrated that the PCR-RFLP method is an efficient tool for discriminating between these six Korean Pyropia species, and that it avoids confusion caused by surface contamination from symbiotic and invasive species. However, the RFLP method has limited target sensitivity and is time consuming, due to the researcher-dependent nature of the method. Nowadays, the PCR-RFLP method is being replaced by more practical and faster methods utilizing chloroplast genome-based markers.
In recent years, mitochondrial CO1 and plastid rbcL have become the most commonly used genes for red algal barcoding [,]. However, the resolution of these markers may not be optimal in some lineages, particularly at the population level, with rbcL known to provide lower resolution than CO1 in some cases [,]. A comprehensive comparison and evaluation of new species-specific markers and their potential ability to discriminate species, subspecies, and populations is significant, but few have been assessed []. Many researchers began by comprehensively investigating new or combined species-specific variable markers in the chloroplast region of Pyropia. Xu et al. 2018 utilized a set of chloroplast whole genomes to explore the phylogenetic relationships of 10 Pyropia species []. Research has increased the availability of genetic resources for Pyropia, such as useful molecular markers or DNA barcodes for species identification using the chloroplast genomes. However, systematic molecular investigations and genomic information on Pyropia remain incomplete compared to those of many land plants. In public databases such as NCBI GenBank, only seven complete or partial chloroplast (plastid) genomes within the Pyropia genus have been reported for Pyropia species, including P. yezoensis, P. haitanensis, P. fucicola, P. pulchra, P. kanakaensis, P. perforata, and P. endiviifolia. Information on the chloroplast genomes of Korean red algae, including representative P. dentata and P. seriata, is very limited.
Using seven previously identified Pyropia chloroplast genomes to assess the genetic nucleotide diversity between six Pyropia species, we identified and validated three hypervariable regions, from which we developed species-specific InDels in the trnM-argB region. Although the present study analyzed a limited number of Pyropia species and chloroplast genomes, further studies should include various other species and chloroplast genomes to further elucidate the phylogenetic relationships of the Pyropia genus. The markers developed here can now be employed to explore the variations of Pyropia populations for further evolutionary, phylogenetic, and barcoding studies in marine fields.
Author Contributions
Conceptualization, S.-J.C., Y.K., and C.C.; methodology, Y.K.; investigation, S.-J.C. and Y.K.; resources, S.-J.C.; writing-original draft preparation, Y.K. and S.-J.C.; writing-review and editing, Y.K.
Funding
This research received no external funding.
Acknowledgments
This paper is dedicated to the memory of Heungsu Kim for inspiring and mentoring our work and teaching us as a great inventor and scientist with a passion for plant biology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vergés, A.; Sánchez, N.; Peteiro, C.; Polo, L.; Brodie, J. Pyropiasuborbiculata (Bangiales, Rhodophyta): First records from the northeastern Atlantic and Mediterranean of this North Pacific species. Phycologia 2013, 52, 121–129. [Google Scholar] [CrossRef]
- Hallmann, A. Algae biotechnology–green cell-factories on the rise. Curr. Biotechnol. 2015, 4, 389–415. [Google Scholar] [CrossRef]
- Hwang, M.-S.; Lee, I.G. Character analysis and numerical taxonomy of Porphyra (Bangiales, Rhodophyta) from Korea. Algae 2002, 17, 217–233. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, S.J.; Choi, C. An Efficient PCR-RFLP Method for the Rapid Identification of Korean Pyropia Species. Molecules 2017, 22, 2182. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Saunders, G.W. Applying DNA barcoding to red macroalgae: A preliminary appraisal holds promise for future application. Philos. Trans. R. Soc. Ser. B 2005, 9, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Hughey, J.; Silva, P.; Hommersand, M. Solving taxonomic and nomenclatural problems in Pacific Gigartinaceae (Rhodophyta) using DNA from type material. J. Phycol. 2001, 1109, 1091–1109. [Google Scholar] [CrossRef]
- Freshwater, D.W.; Tudor, K.; O’shaughnessy, K.; Wysor, B. DNA barcoding in the red algal order Gelidiales: Comparison of COI with rbcL and verification of the barcoding gap. Cryptogam. Algol. 2010, 31, 435–449. [Google Scholar]
- Yang, E.C.; Kim, M.S.; Geraldino, P.J.L.; Sahoo, D.; Shin, J.A.; Boo, S.M. Mitochondrial cox1 and plastid rbcL genes of Gracilariavermiculophylla (Gracilariaceae, Rhodophyta). J. Appl. Phycol. 2008, 20, 161–168. [Google Scholar] [CrossRef]
- Yang, E.C.; Boo, S.M. A red alga-specific phycoerythrin gene for biodiversity surveys of callithamnioid red algae. Mol. Ecol. Notes 2006, 6, 533–535. [Google Scholar] [CrossRef]
- Xu, K.; Tang, X.; Bi, G.; Cao, M.; Wang, L.; Mao, Y. The first complete organellar genomes of an Antarctic red alga, Pyropiaendiviifolia: Insights into its genome architecture and phylogenetic position within genus Pyropia (Bangiales, Rhodophyta). J. Oceanol. Limnol. 2018, 36, 1315–1328. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).