Comparative Genomic Analysis of the Biotechnological Potential of the Novel Species Pseudomonas wadenswilerensis CCOS 864T and Pseudomonas reidholzensis CCOS 865T
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Comparative Genomics
3. Results
3.1. The Genomes of P. wadenswilerensis CCOS 864T and P. reidholzensis CCOS 865T
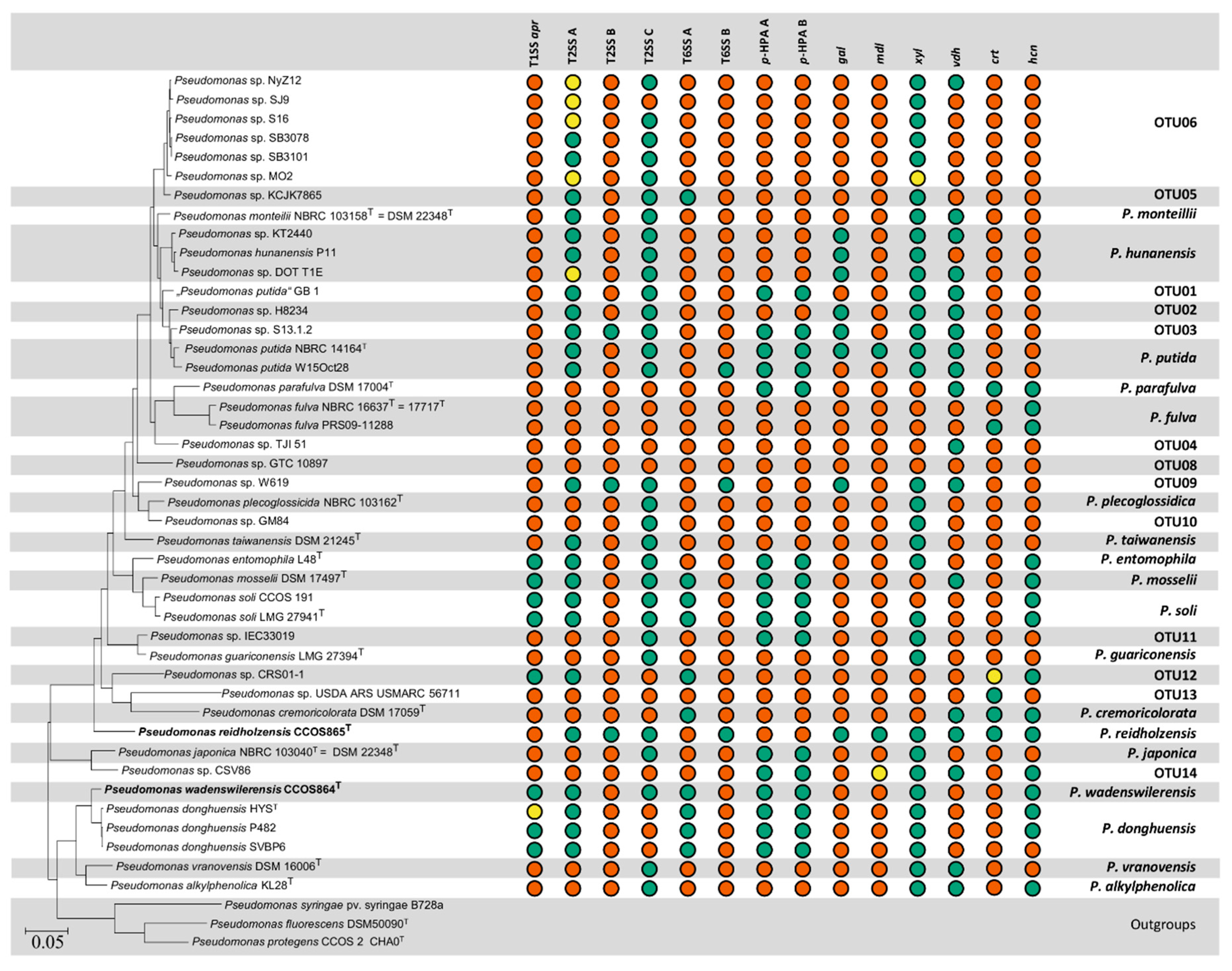
3.2. Selection of Genomes and Phylogeny
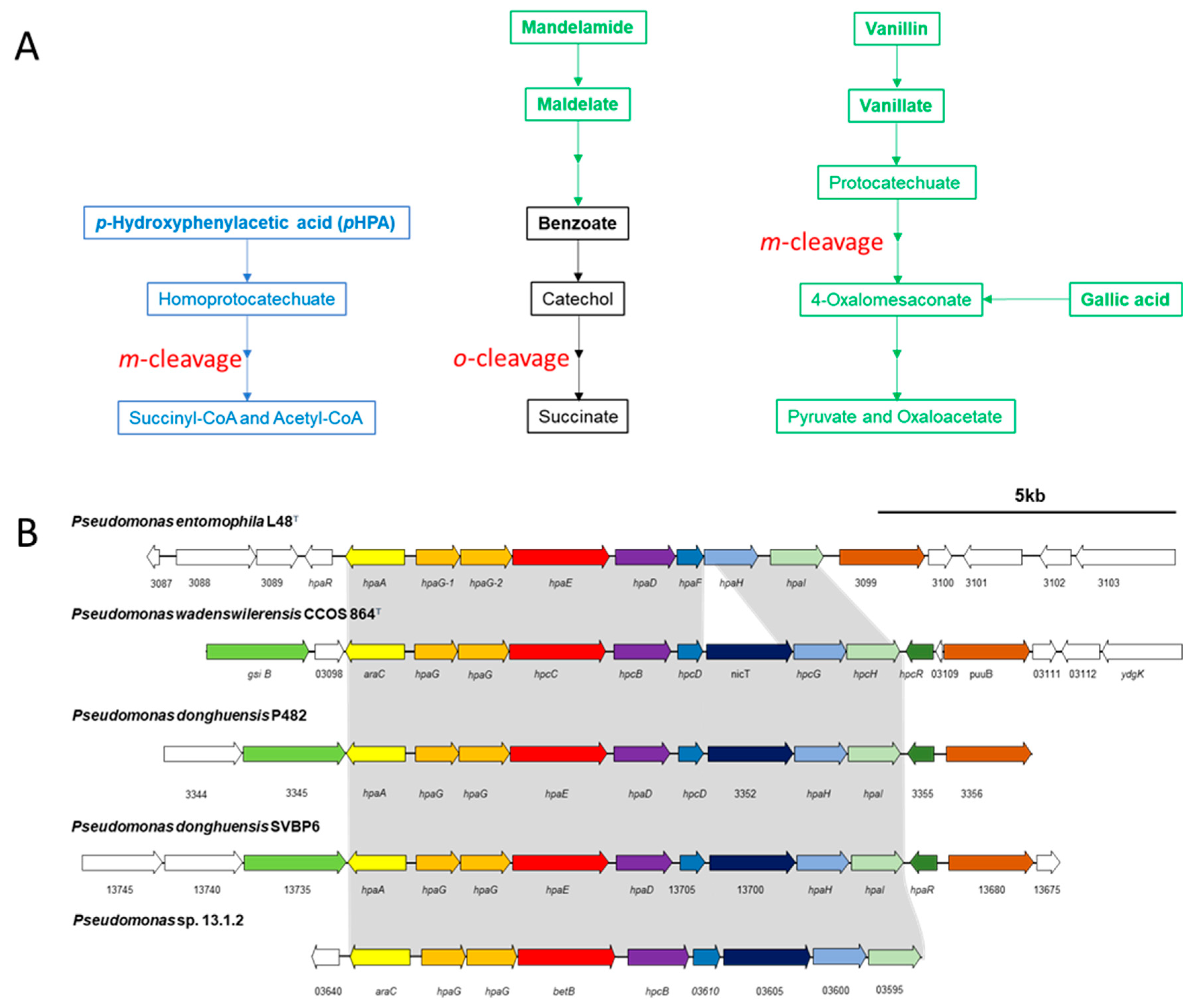
3.3. Degradation of Aromatic Compounds
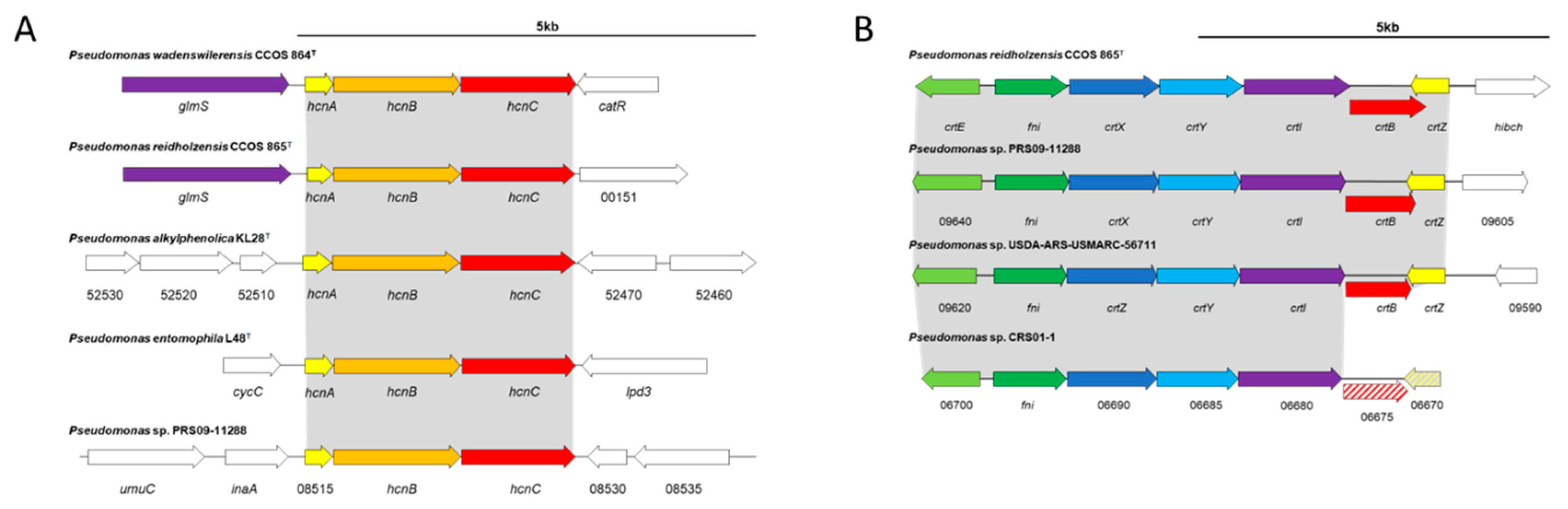
3.4. Secondary Metabolites
3.5. Secretion Systems
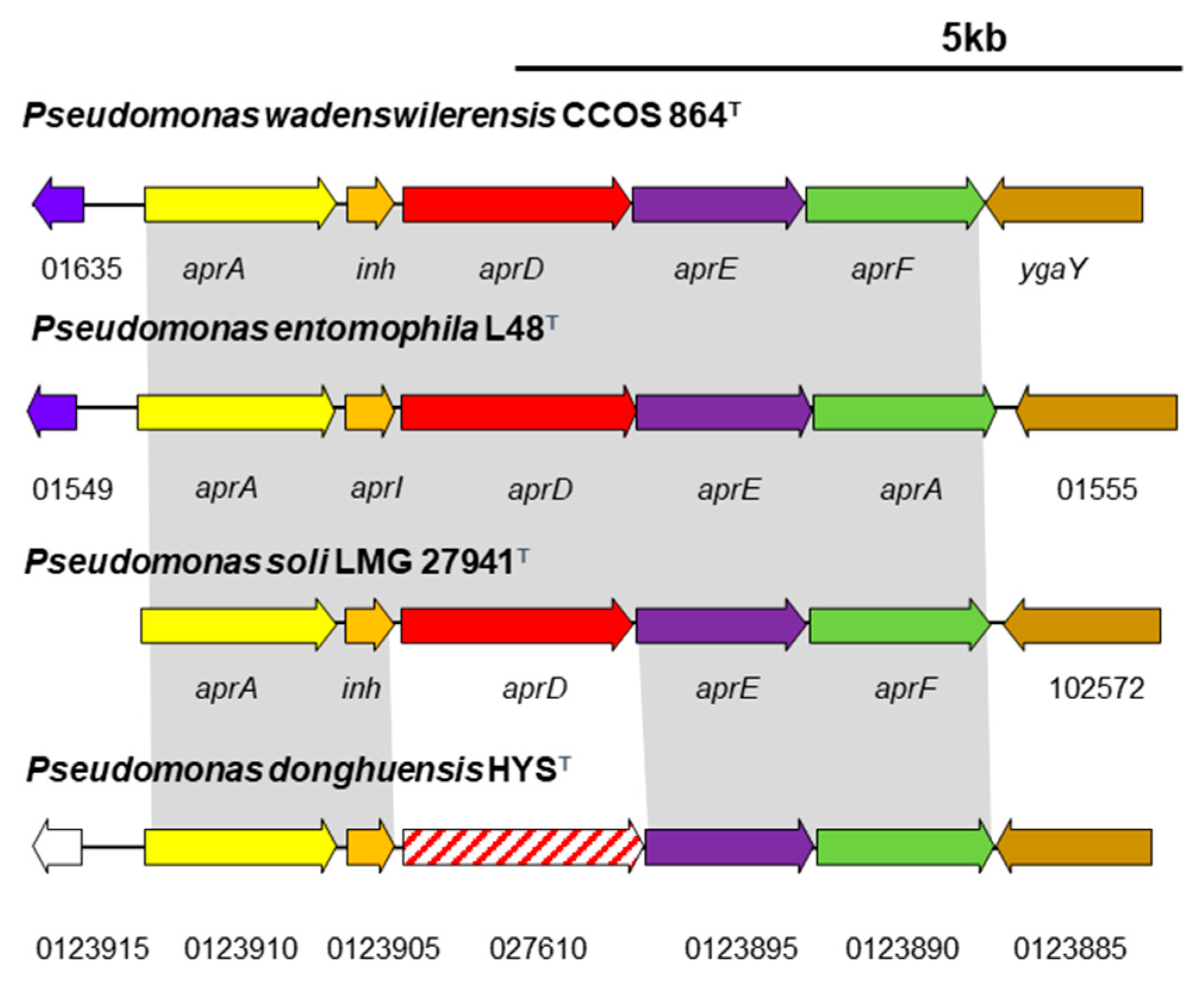
3.5.1. Type I Secretion Systems
3.5.2. Type II Secretion Systems
3.5.3. Type VI Secretion Systems
4. Discussion
4.1. Potential Ecological Role of P. wadenswilerensis CCOS 864T and P. reidholzensis CCOS 865T
4.2. Potential Role of P. wadenswilerensis CCOS 864T and P. reidholzensis CCOS 865T as a Biocatalyst
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Palleroni, N.J. Pseudomonas. In Bergey’s Manual of Systematic Bacteriology vol. 2, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; pp. 323–379. [Google Scholar]
- Timmis, K.N. Pseudomonas putida: A cosmopolitan opportunist par excellence. Environ. Microbiol. 2002, 4, 779–781. [Google Scholar] [CrossRef] [PubMed]
- Peix, A.; Ramírez-Bahena, M.-H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.B.; Singh Saini, H.; Kahlon, R.S. Pseudomonas: The Versatile and Adaptive Metabolic Network. In Pseudomonas: Molecular and Applied Biology; Kahlon, R.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 81–126. [Google Scholar]
- van Beilen, J.B.; Li, Z.; Duetz, W.A.; Smits, T.H.M.; Witholt, B. Diversity of alkane hydroxylase systems in the environment. Oil Gas. Sci. Technol. Rev. IFP 2003, 58, 427–440. [Google Scholar] [CrossRef]
- Smits, T.H.M.; Balada, S.B.; Witholt, B.; van Beilen, J.B. Functional analysis of alkane hydroxylases from Gram-negative and Gram-positive bacteria. J. Bacteriol. 2002, 184, 1733–1742. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Becker, J.; Dohnt, K.; dos Santos, V.M.; Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [Google Scholar] [CrossRef]
- Schulze, B.; Wubbolts, M.G. Biocatalysis for industrial production of fine chemicals. Curr. Opin. Biotechnol. 1999, 10, 609–615. [Google Scholar] [CrossRef]
- dos Santos, V.A.; Heim, S.; Moore, E.R.; Strätz, M.; Timmis, K.N. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004, 6, 1264–1286. [Google Scholar] [CrossRef]
- Park, J.-B.; Bühler, B.; Panke, S.; Witholt, B.; Schmid, A. Carbon metabolism and product inhibition determine the epoxidation efficiency of solvent-tolerant Pseudomonas sp. strain VLB120DC. Biotechnol. Bioeng. 2007, 98, 1219–1229. [Google Scholar] [CrossRef]
- Anwar, A.; Saleemuddin, M. Alkaline proteases: A review. Biores. Technol. 1998, 64, 175–183. [Google Scholar] [CrossRef]
- Kahlon, R.S. Pseudomonas for industrial biotechnology. In Pseudomonas: Molecular and Applied Biology; Kahlon, R.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 281–342. [Google Scholar]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.; Davis, E.W., II; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonds. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Flury, P.; Aellen, N.; Ruffner, B.; Péchy-Tarr, M.; Fataar, S.; Metla, Z.; Dominguez-Ferreras, A.; Bloemberg, G.; Frey, J.; Goesmann, A.; et al. Insect pathogenicity in plant-beneficial pseudomonads: Phylogenetic distribution and comparative genomics. ISME J. 2016, 10, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Frasson, D.; Opoku, M.; Picozzi, T.; Torossi, T.; Balada, S.; Smits, T.H.M.; Hilber, U. Pseudomonas wadenswilerensis sp. nov. and Pseudomonas reidholzensis sp. nov., two new species within the Pseudomonas putida group isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2853–2861. [Google Scholar] [PubMed]
- Rutz, D.; Frasson, D.; Sievers, M.; Blom, J.; Rezzonico, F.; Pothier, J.F.; Smits, T.H.M. High-quality draft genome sequence of Pseudomonas wadenswilerensis CCOS 864T. Microbiol. Res. Announc. 2018, 7, e01059-18. [Google Scholar]
- Rutz, D.; Frasson, D.; Sievers, M.; Blom, J.; Rezzonico, F.; Pothier, J.F.; Smits, T.H.M. High-quality draft genome sequence of Pseudomonas reidholzensis strain CCOS 865T. Microbiol. Res. Announc. 2019, 8, e01502-18. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Kreis, J.; Spänig, S.; Juhre, T.; Bertelli, C.; Ernst, C.; Goesmann, A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic. Acids Res. 2016, 44 (W1), W22–W28. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lerat, E.; Daubin, V.; Moran, N.A. From gene trees to organismal phylogeny in prokaryotes: The case of the gamma-Proteobacteria. PLoS Biol. 2003, 1, E19. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high trhoughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 -- aproximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinforma. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Goesmann, A.; McHardy, A.C.; Bartels, D.; Bekel, T.; Clausen, J.; Kalinowski, J.; Linke, B.; Rupp, O.; Giegerich, R.; et al. GenDB - an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003, 31, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Saier Jr, M.H.; Yen, M.R.; Noto, K.; Tamang, D.G.; Elkan, C. The Transporter Classification Database: Recent advances. Nucleic Acids Res. 2009, 37, D274–D278. [Google Scholar] [CrossRef]
- Tanabe, M.; Kanehisa, M. Using the KEGG database resource. Curr. Prot. Bioinform. 2012, 38, 1.12.1–1.12.43. [Google Scholar] [CrossRef]
- Gao, J.; Ellis, L.B.M.; Wackett, L.P. The University of Minnesota Biocatalysis/Biodegradation Database: Improving public access. Nucleic Acids Res. 2010, 38, D488–D491. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef]
- Yonezuka, K.; Shimodaira, J.; Tabata, M.; Ohji, S.; Hosoyama, A.; Kasai, D.; Yamazoe, A.; Fujita, N.; Ezaki, T.; Fukuda, M. Phylogenetic analysis reveals the taxonomically diverse distribution of the Pseudomonas putida group. J. Gen. Appl. Microbiol. 2017, 63, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds — from one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Thotsaporn, K.; Tinikul, R.; Maenpuen, S.; Phonbuppha, J.; Watthaisong, P.; Chenprakhon, P.; Chaiyen, P. Enzymes in the p-hydroxyphenylacetate degradation pathway of Acinetobacter baumannii. J. Mol. Catal. B Enzym. 2016, 134, 353–363. [Google Scholar] [CrossRef]
- Prieto, M.A.; Díaz, E.; García, J.L. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: Engineering a mobile aromatic degradative cluster. J. Bacteriol. 1996, 178, 111–120. [Google Scholar] [CrossRef]
- Paliwal, V.; Raju, S.C.; Modak, A.; Phale, P.S.; Purohit, H.J. Pseudomonas putida CSV86: A candidate genome for genetic bioaugmentation. PLoS ONE 2014, 9, e84000. [Google Scholar] [CrossRef]
- Fewson, C.A. Microbial metabolism of mandelate: A microcosm of diversity. FEMS Microbiol. Rev. 1988, 54, 85–110. [Google Scholar] [CrossRef]
- Tsou, A.Y.; Ransom, S.C.; Gerlt, J.A.; Buechter, D.D.; Babbitt, P.C.; Kenyon, G.L. Mandelate pathway of Pseudomonas putida: Sequence relationships involving mandelate racemase, (S)-mandelate dehydrogenase, and benzoylformate decarboxylase and expression of benzoylformate decarboxylase in Escherichia coli. Biochemistry 1990, 29, 9856–9862. [Google Scholar] [CrossRef]
- Gopalakrishna, K.N.; Stewart, B.H.; Kneen, M.M.; Andricopulo, A.D.; Kenyon, G.L.; McLeish, M.J. Mandelamide hydrolase from Pseudomonas putida: Characterization of a new member of the amidase signature family. Biochemistry 2004, 43, 7725–7735. [Google Scholar] [CrossRef]
- Ladino-Orjuela, G.; Gomes, E.; da Silva, R.; Salt, C.; Parsons, J.R. Metabolic pathways for degradation of aromatic hydrocarbons by bacteria. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer International Publishing: Cham, Switzerland, 2016; Volume 237, pp. 105–121. [Google Scholar]
- Zhang, B.; Cai, J.; Duan, C.-Q.; Reeves, M.J.; He, F. A review of polyphenolics in oak wood. Int. J. Mol. Sci. 2015, 16, 6978–7014. [Google Scholar] [CrossRef]
- Ow, Y.-Y.; Stupans, I. Gallic acid and gallic acid derivatives: Effects on drug metabolizing enzymes. Curr. Drug Metabol. 2003, 4, 241–248. [Google Scholar] [CrossRef]
- Nogales, J.; Canales, Á.; Jiménez-Barbero, J.; Serra, B.; Pingarrón, J.M.; García, J.L.; Díaz, E. Unravelling the gallic acid degradation pathway in bacteria: The gal cluster from Pseudomonas putida: Aerobic gallic acid degradation. Mol. Microbiol. 2011, 79, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra Rao, S.; Ravishankar, G.A. Vanilla flavour: Production by conventional and biotechnological routes. J. Sci. Food Agric. 2000, 80, 289–304. [Google Scholar] [CrossRef]
- Priefert, H.; Rabenhorst, J.; Steinbüchel, A. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 1997, 179, 2595–2607. [Google Scholar] [CrossRef] [PubMed]
- Blumer, C.; Haas, D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 2000, 173, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.H.M.; Pothier, J.F.; Ruinelli, M.; Blom, J.; Frasson, D.; Koechli, C.; Fabbri, C.; Brandl, H.; Duffy, B.; Sievers, M. Complete genome of the cyanogenic phosphate-solubilizing Pseudomonas sp. strain CCOS 191, a close relative of Pseudomonas mosselii. Genome Announc. 2015, 3, e00616-15. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A.; Frapolli, M.; Défago, G.; Moënne-Loccoz, Y. Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant.-Microbe Interact. 2003, 16, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Sagar, A.; Dhusiya, K.; Shukla, P.K.; Singh, A.; Lawrence, R.; Ramteke, P.W. Comparative analysis of production of hydrogen cyanide with production of siderophore and phosphate solubilization activity in plant growth promoting bacteria. Vegetos 2018, 31, 130–135. [Google Scholar] [CrossRef]
- Ramette, A.; Moënne-Loccoz, Y.; Défago, G. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 2003, 44, 35–43. [Google Scholar] [CrossRef]
- Zdor, R.E. Bacterial cyanogenesis: Impact on biotic interactions. J. Appl. Microbiol. 2014, 118, 267–274. [Google Scholar] [CrossRef]
- Fukaya, Y.; Takemura, M.; Koyanagi, T.; Maoka, T.; Shindo, K.; Misawa, N. Structural and functional analysis of the carotenoid biosynthesis genes of a Pseudomonas strain isolated from the excrement of Autumn Darter. Biosci. Biotechnol. Biochem. 2017, 82, 1043–1052. [Google Scholar] [CrossRef]
- Rezzonico, F.; Smits, T.H.M.; Born, Y.; Blom, J.; Frey, J.E.; Goesmann, A.; Cleenwerck, I.; de Vos, P.; Bonaterra, A.; Duffy, B.; et al. Erwinia gerundensis sp. nov., a cosmopolitan epiphyte originally isolated from pome fruit trees. Int. J. Syst. Evol. Microbiol. 2016, 66, 1583–1592. [Google Scholar] [CrossRef]
- Johler, S.; Stephan, R.; Hartmann, I.; Kuehner, K.A.; Lehner, A. Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl. Environ. Microbiol. 2010, 76, 1053–1061. [Google Scholar] [CrossRef]
- Tseng, T.-T.; Tyler, B.M.; Setubal, J.C. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009, 9 (Suppl. 1), S2. [Google Scholar] [CrossRef]
- Guzzo, J.; Duong, F.; Wandersman, C.; Murgier, M.; Lazdunski, A. The secretion genes of Pseudomonas aeruginosa alkaline protease are functionally related to those of Erwinia chrysanthemi proteases and Escherichia coli a-haemolysin. Mol. Microbiol. 1991, 5, 447–453. [Google Scholar] [CrossRef]
- Duong, F.; Bonnet, E.; Géli, V.; Lazdunski, A.; Murgier, M.; Filloux, A. The AprX protein of Pseudomonas aeruginosa: A new substrate for the Apr type I secretion system. Gene 2001, 262, 147–153. [Google Scholar] [CrossRef]
- Delepelaire, P. Type I secretion in Gram-negative bacteria. Biochim. Biophys. Acta 2004, 1694, 149–161. [Google Scholar] [CrossRef]
- Suter, S. The role of bacterial proteases in the pathogenesis of cystic fibrosis. Am. J. Respir. Crit. Care Med. 1994, 150, S118–S122. [Google Scholar] [CrossRef]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Duong, F.; Lazdunski, A.; Cami, B.; Murgier, M. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: Relationships to other secretory pathways. Gene 1992, 121, 47–54. [Google Scholar] [CrossRef]
- Sandkvist, M. Type II secretion and pathogenesis. Infect. Immun. 2001, 69, 3523–3535. [Google Scholar] [CrossRef]
- Jani, A.J.; Cotter, P.A. Type VI secretion: Not just for pathogenesis anymore. Cell Host Microbe 2010, 8, 2–6. [Google Scholar] [CrossRef] [PubMed]
- De Maayer, P.; Venter, S.N.; Kamber, T.; Duffy, B.; Coutinho, T.A.; Smits, T.H.M. Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genomics 2011, 12, 576. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Hood, R.D.; Mougous, J.D. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010, 18, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Allsopp, L.P.; Filloux, A.; Llamas, M.A. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017, 11, 972–987. [Google Scholar] [CrossRef] [PubMed]
- Kamber, T.; Pothier, J.F.; Pelludat, C.; Rezzonico, F.; Duffy, B.; Smits, T.H.M. Role of the type VI secretion systems during disease interactions of Erwinia amylovora with its plant host. BMC Genomics 2017, 18, 628. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 2018, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef]
- Zaks, A. Industrial biocatalysis. Curr. Opin. Chem. Biol. 2001, 5, 130–136. [Google Scholar] [CrossRef]
- Smits, T.H.M. The importance of genome sequence quality to microbial comparative genomics research. BMC Genomics 2019, 20, 662. [Google Scholar] [CrossRef]
- Luo, X.-J.; Yu, H.-L.; Xu, J.-H. Genomic data mining: An efficient way to find new and better enzymes. Enzyme Eng. 2012, 1, 104. [Google Scholar] [CrossRef]
- Kuhn, D.; Blank, L.M.; Schmid, A.; Bühler, B. Systems biotechnology–rational whole-cell biocatalyst and bioprocess design. Eng. Life Sci. 2010, 10, 384–397. [Google Scholar] [CrossRef]
- Schmid, A.; Blank, L.M. Hypothesis-driven omics integration. Nat. Chem. Biol. 2010, 6, 485–487. [Google Scholar] [CrossRef]
- Felfer, U.; Goriup, M.; Koegl, M.F.; Wagner, U.; Larissegger-Schnell, B.; Faber, K.; Kroutil, W. The substrate spectrum of mandelate racemase: Minimum structural requirements for substrates and substrate model. Adv. Synth. Catal. 2005, 347, 951–961. [Google Scholar] [CrossRef]
- Ahmed, M.; Kelly, T.; Ghanem, A. Applications of enzymatic and non-enzymatic methods to access enantiomerically pure compounds using kinetic resolution and racemisation. Tetrahedron 2012, 68, 6781–6802. [Google Scholar] [CrossRef]
- Bi, M.-C.; Rosen, R.; Zha, R.-Y.; McCormick, S.A.; Song, E.; Hu, D.-N. Zeaxanthin induces apoptosis in human uveal melanoma cells through Bcl-2 family proteins and intrinsic apoptosis pathway. Evid. Based Complement. Alternat. Med. 2013, 2013, 205082. [Google Scholar] [CrossRef]
- Álvarez, R.; Vaz, B.; Gronemeyer, H.; de Lera, Á.R. Functions, therapeutic aplications, and synthesis of retinoids and carotenoids. Chem. Rev. 2014, 114, 1–125. [Google Scholar] [CrossRef]


| Strain Name (NCBI) | Taxonomic Group | Name Used in this Study | Assembly Level | Contigs/Scaffolds | Genome Size [bp] | Genes | G+C [%] | GenBank Accession No. |
|---|---|---|---|---|---|---|---|---|
| P. alkylphenolica KL28T | P. alkylphenolica | P. alkylphenolica KL28T | Complete | 1 | 5′764′622 | 5′454 | 60.63 | CP009048 |
| P. cremoricolorata DSM 17059T | P. cremoricolorata | P. cremoricolorata DSM 17059T | Draft | 26 | 4′655′082 | 4′072 | 63.35 | AUEA00000000 |
| P. donghuensis P482 | P. donghuensis | P. donghuensis P482 | Draft | 69 | 5′623′997 | 5′259 | 62.40 | JHTS00000000 |
| P. donghuensis HYST | P. donghuensis | P. donghuensis HYST | Draft | 64 | 5′646′028 | 5′239 | 62.40 | AJJP01000000 |
| P. donghuensis SVBP6 | P. donghuensis | P. donghuensis SVBP6 | Draft | 71 | 5′701′342 | 5′355 | 62.40 | NWCB01000000 |
| P. entomophila L48T | P. entomophila | P. entomophila L48T | Complete | 1 | 5′888′780 | 5′223 | 64.20 | NC_008027 |
| P. fulva NBRC 16637T = DSM 17717T | P. fulva | P. fulva NBRC 16637 T = DSM 17717T | Draft | 46 | 4′768′229 | 4′331 | 61.80 | BBIQ00000000 |
| P. guariconensis LMG 27394T | P. guariconensis | P. guariconensis LMG 27394T | Draft | 29 | 5′079′034 | 4′703 | 62.20 | FMYX00000000 |
| P. hunanensis P11 | P. hunanensis | P. hunanensis P11 | Draft | 171 | 6′644′424 | 6′469 | 61.20 | PISL00000000 |
| P. japonica NBRC 103040T = DSM 22348 T | P. japonica | P. japonica NBRC 103040T = DSM 22348 T | Draft | 162 | 6′663′130 | 5′845 | 64.20 | BBIR00000000 |
| P. monteilii MO2 | OTU06 | Pseudomonas sp. MO2 | Draft | 1′589 | 6′240′608 | 5′947 | 62.00 | JFBC00000000 |
| P. monteilii SB3078 | OTU06 | Pseudomonas sp. SB3078 | Complete | 1 | 6′000′087 | 5′620 | 62.50 | CP006978 |
| P. monteilii SB3101 | OTU06 | Pseudomonas sp. SB3101 | Complete | 1 | 5′945′120 | 5′546 | 62.50 | CP006979 |
| P. monteilii GTC 10897 | OTU08 | Pseudomonas sp. GTC 10897 | Draft | 149 | 5′547′282 | 5′313 | 60.40 | BCAO00000000 |
| P. monteilii NBRC 103158 = DSM 14164T | P. monteilii | P. monteilii NBRC 103158 = DSM 14164T | Draft | 132 | 6′299′985 | 6′005 | 61.50 | BBIS00000000 |
| P. monteilii USDA-ARS-USMARC-56711 | OTU13 | Pseudomonas sp. USDA-ARS-USMARC-56711 | Complete | 1 | 4′714′359 | 4′100 | 64.40 | CP013997 |
| P. mosselii DSM 17497T | P. mosselii | P. mosselii DSM 17497T | Draft | 55 | 6′260′844 | 5′916 | 64.00 | JHYW00000000 |
| P. parafulva DSM 17004T | P. parafulva | P. parafulva DSM 17004T | Draft | 32 | 4′956′622 | 4′536 | 62.50 | AUEB00000000 |
| P. parafulva PRS09-11288 | P. fulva | P. fulva PRS09-11288 | Complete | 1 | 4′690′783 | 4′246 | 61.70 | CP019952 |
| P. parafulva CRS01-1 | OTU12 | Pseudomonas sp. CRS01-1 | Complete | 1 | 5′087′619 | 4′457 | 63.50 | CP009747 |
| P. plecoglossicida KCJK7865 | OTU05 | Pseudomonas sp. KCJK7865 | Draft | 205 | 5′806′309 | 5′606 | 63.00 | QANO00000000 |
| P. plecoglossicida NyZ12 | OTU06 | Pseudomonas sp. NyZ12 | Complete | 1 | 6′233′254 | 5′843 | 62.40 | CP010359 |
| P. plecoglossicida NBRC 103162T | P. plecoglossicida | P. plecoglossicida NBRC 103162T | Draft | 97 | 5′341′796 | 4′946 | 63.00 | BBIV00000000 |
| P. putida DOT-T1E | P. hunanensis | Pseudomonas sp. DOT-T1E | Complete | 1 | 6′260′702 | 5′803 | 61.40 | CP003734 |
| P. putida KT2440 | P. hunanensis | Pseudomonas sp. KT2440 | Complete | 1 | 6′181′873 | 5′420 | 62.30 | AE015451 |
| P. putida GB-1 | OTU01 | Pseudomonas sp. GB-1 | Complete | 1 | 6′078′430 | 5′586 | 61.90 | AAXR01000000 |
| P. putida H8234 | OTU02 | Pseudomonas sp. H8234 | Complete | 1 | 5′956′110 | 6′512 | 61.60 | NC_021491 |
| P. putida S13.1.2 | OTU03 | Pseudomonas sp. S13.1.2 | Complete | 1 | 6′621′848 | 5′993 | 62.30 | CP010979 |
| P. putida NBRC 14164T | P. putida | P. putida NBRC 14164T | Complete | 1 | 6′156′701 | 5′610 | 62.30 | NC_021505 |
| P. putida W15Oct28 | P. putida | P. putida W15Oct28 | Draft | 119 | 6′320′510 | 5′703 | 62.80 | JENB00000000 |
| P. putida S16 | OTU06 | Pseudomonas sp. S16 | Complete | 1 | 5′984′790 | 5′585 | 62.30 | NC_015733 |
| P. putida IEC33019 | OTU11 | Pseudomonas sp. IEC33019 | Complete | 1 | 5′847′120 | 5′445 | 62.27 | CP016634 |
| P. putida CSV86 | OTU14 | Pseudomonas sp.CSV86 | Draft | 209 | 6′469′780 | 5′906 | 63.10 | AMWJ01000000 |
| P. putida W619 | OTU09 | Pseudomonas sp. W619 | Complete | 1 | 5′774′330 | 5′378 | 61.40 | NC_010501 |
| P. reidholzensis CCOS 865T | P. reidholzensis | P. reidholzensis CCOS 865T | Draft | 45 | 6′163′129 | 5′441 | 64.09 | UIDD01000000 |
| P. soli CCOS 191 | P. soli | P. soli CCOS 191 | Complete | 1 | 6′012′947 | 5′301 | 64.19 | LN847264 |
| P. soli LMG 27941T | P. soli | P. soli LMG 27941T | Draft | 34 | 5′644′910 | 5′188 | 64.00 | FOEQ00000000 |
| Pseudomonas sp. TJI-51 | OTU04 | Pseudomonas sp. TJI-51 | Draft | 208 | 5′805′096 | 5′508 | 62.10 | AEWE00000000 |
| Pseudomonas sp. GM84 | OTU10 | Pseudomonas sp. GM84 | Draft | 384 | 5′818′772 | 5′299 | 63.20 | AKJC00000000 |
| P. taiwanensis DSM 21245T | P. taiwanensis | P. taiwanensis DSM 21245T | Draft | 67 | 5′415′134 | 5′056 | 61.90 | AUEC00000000 |
| P. taiwanensis SJ9 | OTU06 | Pseudomonas sp. SJ9 | Draft | 736 | 6′253′055 | 6′075 | 61.80 | AXUP00000000 |
| P. vranovensis DSM 16006T | P. vranovensis | P. vranovensis DSM 16006T | Draft | 36 | 5′697′807 | 5′295 | 61.50 | AUED00000000 |
| P. wadenswilerensis CCOS 864T | P. wadenswilerensis | P. wadenswilerensis CCOS 864T | Draft | 18 | 5′966′942 | 5′437 | 62.39 | UNOZ01000000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutz, D.; Frasson, D.; Sievers, M.; Blom, J.; Rezzonico, F.; Pothier, J.F.; Smits, T.H.M. Comparative Genomic Analysis of the Biotechnological Potential of the Novel Species Pseudomonas wadenswilerensis CCOS 864T and Pseudomonas reidholzensis CCOS 865T. Diversity 2019, 11, 204. https://doi.org/10.3390/d11110204
Rutz D, Frasson D, Sievers M, Blom J, Rezzonico F, Pothier JF, Smits THM. Comparative Genomic Analysis of the Biotechnological Potential of the Novel Species Pseudomonas wadenswilerensis CCOS 864T and Pseudomonas reidholzensis CCOS 865T. Diversity. 2019; 11(11):204. https://doi.org/10.3390/d11110204
Chicago/Turabian StyleRutz, Dominik, David Frasson, Martin Sievers, Jochen Blom, Fabio Rezzonico, Joël F. Pothier, and Theo H. M. Smits. 2019. "Comparative Genomic Analysis of the Biotechnological Potential of the Novel Species Pseudomonas wadenswilerensis CCOS 864T and Pseudomonas reidholzensis CCOS 865T" Diversity 11, no. 11: 204. https://doi.org/10.3390/d11110204
APA StyleRutz, D., Frasson, D., Sievers, M., Blom, J., Rezzonico, F., Pothier, J. F., & Smits, T. H. M. (2019). Comparative Genomic Analysis of the Biotechnological Potential of the Novel Species Pseudomonas wadenswilerensis CCOS 864T and Pseudomonas reidholzensis CCOS 865T. Diversity, 11(11), 204. https://doi.org/10.3390/d11110204





