Abstract
Microorganisms are a valuable source of pharmaceutically active chemicals, serving as scaffolds for synthesis as well as lead structures. Investigating novel biomes frequently yields intriguing chemistry; the Atacama Desert in Chile is one such example. This study reports the isolation of a new reduced anthracycline-related compound from the Atacama Desert-derived bacterium Saccharothrix S26. Structural characterisation was achieved by one-dimensional and two-dimensional NMR, HR-LCMS, and X-ray crystallography. The compound was tested against the ESKAPE pathogens, bovine mastitis-related pathogens, and the fungal strain Cryptococcus neoformans, but no antimicrobial activity was observed.
1. Introduction
Several types of chemical scaffolds useful for drug discovery can be found in natural products. However, during the past few decades, their use has decreased due to problems including restricted supply and a high incidence of rediscovery. Under-explored and harsh environments like deserts can provide novel chemical discoveries. The Atacama Desert is a hyper-arid region in the north of Chile [1], defined by extended periods of little to no precipitation, high salinity, and intense UV radiation. Often, not much macroscopic life is found. However, several structurally interesting and pharmaceutically active compounds have been discovered from the microorganisms isolated from the desert. These include chaxamycin D (1) (Figure 1) [2], which has antibacterial activity against methicillin-resistant Staphylococcus aureus, lentzeoside A (2) (Figure 1) [3], which has demonstrated anti-HIV integrase activity, and abenquine A (3) (Figure 1) [4], which has the rare combination of benzoquinones and amino acids as structural units and shows phosphodiesterase type 4b inhibitory activity.
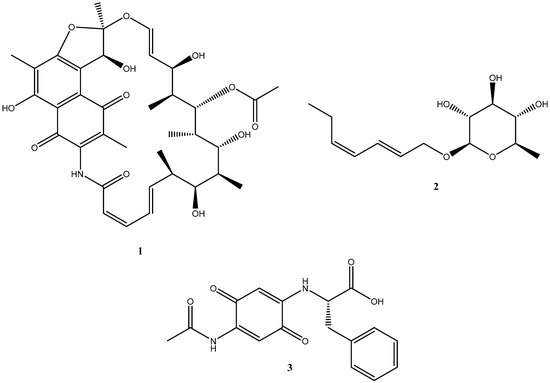
Figure 1.
Structures of chaxamycin D (1), lentzeoside A (2), and abenquine A (3) isolated from Atacama Desert microorganisms.
This paper reports the isolation and structural characterisation of a new reduced anthracycline 4 from Saccharothrix S26, an actinobacterial strain isolated from the Atacama Desert. Compound 4 is related to the reduced anthracyclines isolated from Nocardia brasiliensis [5]. Compound 4 was tested against the ESKAPE pathogens, a panel of pathogens causing severe and often fatal nosocomial infections. It was also tested against a panel of bacterial pathogens associated with bovine mastitis, a devastating disease that causes severe losses to the dairy cattle industry. In addition, it was tested against Cryptococcus neoformans.
2. Results and Discussion
2.1. Spectroscopic Structural Elucidation
Compound 4 (Figure 2) was isolated as orange needles and provided an [M + H]+ peak at m/z 333.1118 (Figure S1). The molecular formula of the compound was established as C21H16O4 (Δ = −0.9 ppm), requiring 14 degrees of unsaturation.
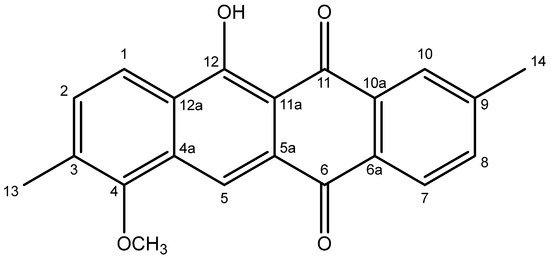
Figure 2.
Structure of compound 4.
It should be noted that the numbering scheme used in this discussion and in Table 1 follows that established for related anthracyclines [5], not the IUPAC numbering system used in the title. The 1H NMR spectrum of compound 4 (Figure S2) displayed a sharp singlet at δH 14.50 (12-OH), two sets of ortho-coupled doublets at δH 8.24 (H-7), 8.16 (H-1), 7.58 (H-8), and 7.48 (H-2), two singlets at δH 8.52 (H-5) and 8.15 (H-10 overlapping with H-1), one methoxy group (4-OCH3) at δH 3.88, and two methyl groups at δH 2.47 (H3-14) and 2.42 (H3-13), the assignments of which were confirmed using the HSQC spectrum (Figure S3). The COSY spectrum (Figure S4) showed two spin systems, one of which consisted of H-1 and H-2, while the other comprised H-7 and H-8 (coupling to H-10 was not observed due to overlap with H-1 and the low magnitude of the associated coupling constants). The 13C NMR spectrum (Figure S5), among others, showed two carbonyl carbon atoms at δC 188.0 (C-11) and 182.3 (C-6), a methoxy carbon atom at δC 62.1 (4-OCH3), along with two methyl carbon atoms at δC 22.0 (C-14) and 16.6 (C-13). The full NMR data are provided in Table 1.

Table 1.
NMR data of compound 4 in CDCl3 at 100/400 MHz.
In the HMBC spectrum (Figure S6, key correlations shown in Figure 3), the correlations between H3-13 and C-2 (δC 132.6), C-3 (δC 133.2), as well as C-4 (δC 156.0), H3-14, and C-8 (δC 135.4), C-9 (δC 145.1), as well as C-10 (δC 127.3) and between 4-OCH3 and C-4, established the positions of both the methyl and the methoxy groups, while the chelated 12-OH displayed correlations to C-11a (δC 109.5), C-12 (δC 163.7), as well as C-12a (δC 127.6). Individual fragments were connected by key correlations from H-5 to C-4, C-6, C-11a, and C-12a, H-7 to C-6, and H-10 to C-11. On this basis, the structure of 4 was established as 11-hydroxy-7-methoxy-2,8-dimethyltetracene-5,12-dione, representing a new natural product within the class of reduced anthracyclines.
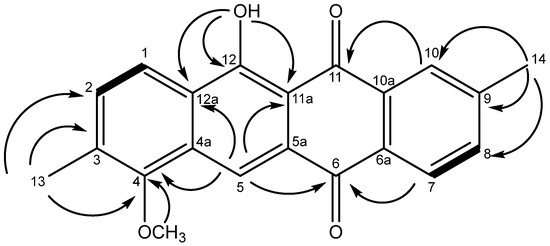
Figure 3.
Structure of 4 showing key correlations observed in the HMBC (arrows) and COSY spectra (bold bonds).
The UV spectrum of compound 4 (Figure 4) showed a broad peak at 450 nm, representing the n–π* transition of the highly conjugated system. The strong peak at 259 nm corresponds to the π–π* transition.
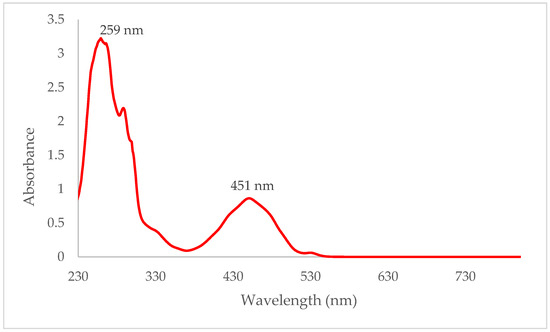
Figure 4.
UV spectrum of compound 4 in dichloromethane.
2.2. Crystal Structure
The asymmetric unit of compound 4, which crystallises in space group Cc (No. 9), comprises two C21H16O4 molecules, viz., A (containing atom C1) and B (containing C22). As might be expected, the essentially rigid 18-atom fused tetra-cycle ring systems have similar geometrical parameters, and the main difference between the molecules arises from the orientations of the methoxy groups: when viewed in the same orientation (Figure 5), the methyl group in molecule A points ‘backwards’, as indicated by the C1–C2–O1–C21 torsion angle of 98.4 (11)°, whereas, for molecule B, the methyl group points forwards, with a corresponding C22–C23–O5–C42 torsion angle of −97.5 (11)°. The C1–C18 fused ring system (r.m.s. deviation = 0.110 Å) of molecule A is distinctly bowed, as indicated by the dihedral angle between the C1–C3/C16–C18 and C7–C12 rings of 9.2 (5)°. Molecule B is much closer to being planar, as indicated by the dihedral angle between the terminal rings of 1.4 (5)° and a value of 0.020 Å for the r.m.s. deviation for the 18 carbon atoms of the tetra-cycle. Otherwise, all the bond lengths and angles in 4 may be regarded as normal and are similar to those observed in unsubstituted tetracene-5,12-dione, C18H10O2 [6].
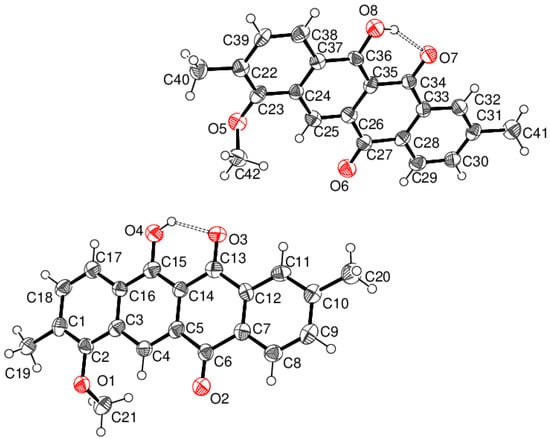
Figure 5.
Molecular structure of 4 showing 50% displacement ellipsoids. The hydrogen bonds are indicated by double-dashed lines.
Both molecules of 4 feature an intramolecular O–H⋅⋅⋅O hydrogen bond from the hydroxyl group to the adjacent ketone O atom, which generates an S (6) ring in each case. The key geometrical data are molecule A: H4o⋅⋅⋅O3 = 1.83 Å, O4–H4o⋅⋅⋅O3 = 135°; molecule B: H4o⋅⋅⋅O3 = 1.72 Å and O4–H4o⋅⋅⋅O3 = 149°. These differences are probably a reflection of the rather low precision of the refinement.
The extended structure of 4 is largely devoid of directional interactions, with just two weak C—H⋅⋅⋅O links arising from the methyl H-atoms identified in a PLATON [7] analysis of the packing. A view of the unit-cell packing (Figure 6) shows that the C1 and C22 molecules stack in separate [010] columns, with numerous slipped weak aromatic π–π stacking interactions between the adjacent molecules related by translation: the centroid–centroid separations vary between 3.660 (6) and 4.013 (7) Å with slippages of between 1.3 and 2.1 Å. The mean plane of the C1–C18 ring system of molecule A subtends an angle of 31.3° with respect to the (010) plane with an equivalent value of 30.7° for the C22–C39 plane of molecule B; the dihedral angle between the C1–C18 and C22–C39 planes for the molecules in the asymmetric unit is 60.9 (1)°. It may be noted that C18H10O2 [6] has a short α unit-cell axis of ~4 Å, and the molecules are canted at about 30° to the (100) plane.
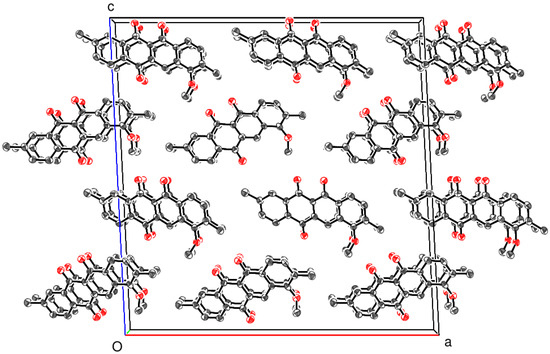
Figure 6.
Unit-cell packing for 4 viewed down [010] with H atoms omitted for clarity.
2.3. Antimicrobial Tests
Compound 4 was tested against the ESKAPE pathogens (i.e., Enterococcus faecium (DSM 17050), Staphylococcus aureus (DSM 2569), Klebsiella pneumoniae (DSM 681), Acinetobacter baumannii (DSM 30008), Pseudomonas aeruginosa (DSM 1117), and Enterobacter cloacae subsp. Cloacae (DSM 30054)) and bovine mastitis bacterial pathogens (i.e., Enterococcus pseudoavium (NCIMB 13084), Escherichia coli (NCIMB 701266), Klebsiela oxytoca (NCIMB 701361), Staphylococcus aureus subsp. Aureus (NCIMB 701494), and Streptococcus bovis (NCIMB 702087)) at 100 μg mL−1 using disc-diffusion assays but was found to be inactive against them.
It was also tested against the fungal strain Cryptococcus neoformans (DSM 11959) at the same concentration as mentioned above but was also found to be inactive.
3. Materials and Methods
3.1. Cultivation of Bacterial Strain and Isolation of Compound
First, 200 g of rice was added to 200 mL of tryptic soy broth (TSB) in a 2 L Erlenmeyer flask and autoclaved at 121 °C for 20 min. Then, 10 mL of a week-old culture of Saccharothrix S26, a strain whose Isolation, characterisation, and phylogenetic tree have previously been described [8], in TSB (30 g L−1) was added to the rice/TSB medium under sterile conditions and left to grow for one month. The culture was extracted three times with 300 mL of methanol, followed by three times with 300 mL dichloromethane. The extract (4 g) was dried under low-pressure conditions and dissolved in 200 mL of water, after which it was subjected to liquid–liquid partitioning using hexane. The hexane fraction (800 mg) was dissolved in 75% chloroform/methanol and fractionated using Sephadex LH20, which revealed five fractions. The 3rd fraction (100 mg) obtained was dissolved in methanol and filtered thoroughly using a Whatman filter paper, revealing 8 mg of compound 4.
The compound was crystallised in the form of orange needles by dissolving it in dichloromethane and allowing evaporation at room temperature.
3.2. Structural Elucidation
LC–MS was carried out using an Agilent 1290 infinity UHPLC (Edinburgh, UK) with a Phenomenex Kinetex XB-C18 (2.6 µM, 100 × 2.1 mm2) column. The mobile phases were 5% acetonitrile + 0.1% formic acid and 94.9% water to 100% acetonitrile + 0.1% formic acid over 15 min. The MS was carried out using a Bruker Maxis Q-tof II (Coventry, UK), with the following settings: 4.5 kV capillary voltage, 4 bar for the nebuliser gas, 9 L min−1 for the dry gas, and 220 °C for the dry temperature. The Auto MS/MS scan mode was used for the MS/MS studies, and the step collision energy range was 80–200%. The NMR spectra were acquired at 25 °C using a Bruker Avance III HD 400 MHz system (Coventry, UK) at a concentration of 10 mg mL−1 in deuterated chloroform.
3.3. Antimicrobial Assay
The antimicrobial activity of compound 4 was evaluated against ESKAPE pathogens (i.e., Enterococcus faecium (DSM 17050), Staphylococcus aureus (DSM 2569), Klebsiella pneumoniae (DSM 681), Acinetobacter baumannii (DSM 30008), Pseudomonas aeruginosa (DSM 1117), and Enterobacter cloacae subsp. Cloacae (DSM 30054) was procured from the DSMZ-German Collection of Microorganisms and Cell Cultures GmbH using the disc diffusion method previously described [9], with slight modifications. Filter paper disks containing gentamicin (10 μg) were used as positive controls, and the same solvent system used to dissolve the test compounds (90% methanol/dichloromethane) was used as the negative control.
The antimicrobial activity of the compound was evaluated against bovine mastitis bacterial pathogens [specifically, Enterococcus pseudoavium (NCIMB 13084), Escherichia coli (NCIMB 701266), Klebsiella oxytoca (NCIMB 701361), Staphylococcus aureus subsp. Aureus (NCIMB 701494), and Streptococcus bovis (NCIMB 702087), all procured from NCIMB Ltd., Aberdeen, UK] using the disc diffusion method previously described [9]. Positive and negative controls were the same as mentioned above.
The antifungal activity of the compound was determined against Cryptococcus neoformans (DSM 11959) with sterile discs loaded with 25 μg of fluconazole.
3.4. X-ray Crystallography
The crystal structure of compound 4 (orange needle, ~0.10 × 0.005 × 0.005 mm, recrystallized from DCM) (Figure 6) was established using intensity data collected on a Rigaku XtalLAB P200K area-detector diffractometer (Cu Kα radiation, λ = 1.54184 Å) at 100 K. An empirical (multi-scan) absorption correction was applied, and the structure was routinely solved by dual-space methods using SHELXT [10] and the structural model completed and optimised by refinement against |F|2 with SHELXL-2019 [11]. The O-bound H atoms were located in different Fourier maps and refined as riding in their as-found relative positions. The C-bound H atoms were placed geometrically (C–H = 0.95–0.98 Å) and refined as riding atoms: the methyl groups were allowed to rotate, but not to tip, to best fit the electron density. The constraint Uiso(H) = 1.2Ueq(carrier) or 1.5Ueq (methyl carrier) was applied in all cases. Full details of the structure and refinement are available in the deposited cif.
Crystal data for compound 0 (C21H16O4): Mr = 332.34, monoclinic, space group Cc (9), a = 27.5410 (14) Å, b = 4.0128 (2) Å, c = 27.9199 (16) Å, β = 92.829 (5)°, V = 3081.9 (3) Å3, Z = 8, T = 100 K, μ = 0.808 mm−1, ρcalc = 1.433 g cm−3, 22964 reflections measured (6.3 ≤ 2θ ≤ 129.9°), 4586 unique (Rint = 0.131), R(F) = 0.080 [2719 reflections with I > 2σ(I)], wR(F2) = 0.221 (all data), Δρmin, max (e Å−3) = −0.29, +0.34, and CCDC deposition number 2343162.
4. Conclusions
11-Hydroxy-7-methoxy-2,8-dimethyltetracene-5,12-dione (4), a new reduced anthracycline, has been isolated from an actinobacterial strain isolated from the Atacama Desert, whose structure was elucidated using one-dimensional and two-dimensional NMR data and X-ray crystallography. The crystal structure shows that it crystallises in space group Cc (No. 9) and shows intramolecular hydrogen bonding between the hydroxyl group and the adjacent ketone O atom. The compound was tested against two panels of pathogenic bacteria: the ESKAPE pathogens and five strains associated with bovine mastitis and a fungal strain, but it showed no activity against any of them.
Supplementary Materials
The following supporting information is available online. Figure S1: LCMS spectrum of compound 4; Figure S2: 1H spectrum of compound 4 in CDCl3 at 400 MHz; Figure S3: HSQC spectrum of compound 4 in CDCl3 at 400 MHz; Figure S4: COSY spectrum of compound 4 in CDCl3 at 400 MHz; Figure S5: 13C spectrum of compound 4 in CDCl3 at 100 MHz; Figure S6: HMBC spectrum of compound 4 in CDCl3 at 400 MHz.
Author Contributions
Conceptualisation, R.V.A., G.P. and A.H.H.; methodology, R.V.A., A.H.H. and R.D.; software, R.V.A.; validation, R.V.A., G.P. and A.H.H.; formal analysis, R.V.A. and W.T.A.H.; investigation, R.V.A. and W.T.A.H.; resources, R.V.A. and A.H.H.; data curation, R.V.A.; writing—original draft preparation, R.V.A. and A.H.H.; writing—review and editing, R.V.A., G.P., A.H.H., R.D., J.A.A., B.A., W.T.A.H., R.E. and M.J.; visualisation, R.V.A., G.P. and A.H.H.; supervision, J.A.A., B.A., W.T.A.H., R.E. and M.J.; project administration, M.J.; funding acquisition, A.H.H. and M.J. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the Deanship of Research at Jordan University of Science and Technology (JUST) for their generous financial support. We are grateful for support from the Basal Programme of ANID (Chile) to the Centre for Biotechnology and Bioengineering (CeBiB, Project FB0001).
Data Availability Statement
Not Applicable.
Acknowledgments
We thank David Cordes and Aidan McKay (University of St Andrews) for collecting the single-crystal X-ray data. We thank Emmanuel T. Oluwabusola and Russell Gray for their continued support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gomez-Silva, B.; Rainey, F.; Warren-Rhodes, K.; McKay, C.; Navarro-Gonzalez, R. Atacama Desert soil microbiology. Microbiol. Extrem. Soils 2008, 13, 117–132. [Google Scholar] [CrossRef]
- Rateb, M.E.; Houssen, W.E.; Arnold, M.; Abdelrahman, M.H.; Deng, H.; Harrison, W.T.A.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Ferguson, G.; et al. Chaxamycins A-D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J. Nat. Prod. 2011, 74, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Wichner, D.; Idris, H.; E Houssen, W.; McEwan, A.R.; Bull, A.T.; A Asenjo, J.; Goodfellow, M.; Jaspars, M.; Ebel, R.; E Rateb, M. Isolation and anti-HIV-1 integrase activity of lentzeosides A–F from extremotolerant Lentzea sp. H45, a strain isolated from a high-altitude Atacama Desert soil. J. Antibiot. 2017, 70, 448–453. [Google Scholar] [CrossRef][Green Version]
- Schulz, D.; Beese, P.; Ohlendorf, B.; Erhard, A.; Zinecker, H.; Dorador, C.; Imhoff, J.F. Abenquines A–D: Aminoquinone derivatives produced by Streptomyces sp. strain DB634. J. Antibiot. 2011, 64, 763–768. [Google Scholar] [CrossRef]
- Maeda, A.; Nagai, H.; Yazawa, K.; Tanaka, Y.; Imai, T.; Mikami, Y.; Kuramochi, T.; Yamazaki, C. Three new reduced anthracycline related compounds from pathogenic Nocardia brasiliensis. J. Antibiot. 1994, 47, 976–981. [Google Scholar] [CrossRef]
- Zheng, X.; Jiang, J.; Liu, Y. Crystal structure of tetracene-5,12-dione, C18H10O2. Z. Für Krist.-New Cryst.Struct. 2023, 238, 717–718. [Google Scholar] [CrossRef]
- Spek, L. checkCIF validation ALERTS: What they mean and how to respond. Acta Crystallogr. E Crystallogr. Commun. 2020, 76, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Astakala, R.V.; Preet, G.; Milne, B.F.; Tibyangye, J.; Razmilic, V.; Castro, J.F.; Asenjo, J.A.; Andrews, B.; Ebel, R.; Jaspars, M. Mutactimycin AP, a new mutactimycin isolated from an actinobacteria from the Atacama Desert. Molecules 2022, 27, 7185. [Google Scholar] [CrossRef] [PubMed]
- Siddharth, S.; Vittal, R.R. Isolation, characterization, and structural elucidation of 4-methoxyacetanilide from marine actinobacteria Streptomyces sp. SCA29 and evaluation of its enzyme inhibitory, antibacterial, and cytotoxic potential. Arch. Microbiol. 2019, 201, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).