Abstract
Trehalose conjugates of 3-hydroxychromone (3HC) dyes have previously been utilized as fluorescence labels to detect metabolically active mycobacteria with a view to facilitating point-of-care detection of mycobacterial pathogens, especially Mycobacterium tuberculosis. We subjected the 3HC dye 2-(6-(diethylamino)benzofuran-2-yl)-3-hydroxy-4H-chromen-4-one (3HC-2) to a combined X-ray crystallography and density functional theory (DFT) study, and conducted preliminary fluorescence labelling experiments with the model organism Mycobacterium aurum. In the crystal, 3HC-2 exhibits an s-cis conformation of the chromone and the benzofuran moieties about the central C–C bond. According to DFT calculations, the s-cis conformer is about 1.8 kcal mol−1 lower in energy than the s-trans conformer. The solid-state supramolecular structure features hydrogen-bonded dimers and π…π stacking. Fluorescence microscopy revealed fluorescence of M. aurum cells treated with the dye trehalose conjugate 3HC-2-Tre in the GFP channel. It was concluded that s-cis is the preferred conformation of 3HC-2 and that the generally considered non-pathogenic M. aurum can be labelled with the fluorescence probe 3HC-2-Tre for convenient in vitro drug screening of new antimycobacterial agents.
1. Introduction
With a total of 1.6 million deaths and an estimate of 10.6 new cases worldwide in 2021, tuberculosis (TB) remains the leading bacterial killer and a public health threat [1]. The etiologic agent of TB is primarily Mycobacterium tuberculosis. Infections caused by non-tuberculous mycobacteria (NTM), which are mostly considered opportunistic pathogens, are also on the rise globally [2,3]. Point-of-care detection of mycobacteria based on low-cost microscopy methods [4] could help prevent and combat mycobacterial infections. In this context, Kamariza et al. recently reported on 3-hydroxychromone dye trehalose conjugates for the fluorescence labelling of mycobacterial cells [5]. As illustrated in Figure 1, exogenous trehalose molecules can be mycolylated at position 6 to give trehalose monomycolates (TMM), which are incorporated into the mycomembrane. A solvatochromic 3-hydroxychromone dye appended to trehalose as a fluorophore group appears to be tolerated by the converting enzymes antigene 85 (Ag85), which enables visualization of metabolically active mycobacteria.
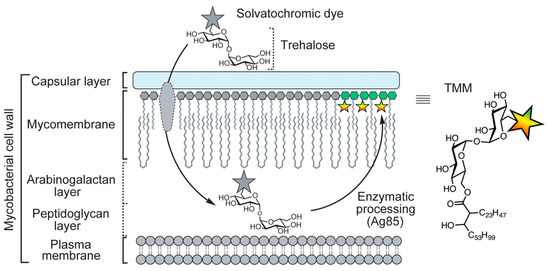
Figure 1.
Simplified representation of the mycobacterial cell wall, illustrating the conversion of dye trehalose conjugates to the corresponding trehalose monomycolates (TMM, chemical diagram on the right) and insertion into the mycomembrane. The figure was adapted from Ref. [5]. Published Open Access under the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/ (accessed on 23 April 2023)). Copyright 2021 The Authors.
Mycobacterium aurum is a fast-growing non-tuberculous mycobacterium [6], which has been used as a surrogate bacterium in anti-TB drug discovery [7,8,9], although its suitability as a model organism for M. tuberculosis has been called into question [10]. M. aurum is generally considered non-pathogenic, but a documented rare case of keratitis attributable to M. aurum has been reported in the medical literature [11]. We have also used M. aurum as a test bacterium in early-stage antimycobacterial drug discovery [12,13,14]. Therefore, we became interested in possible applications of the fluorescence probe 3HC-2-Tre (Figure 2) [5] for the labelling of M. aurum cells, as this could be a useful tool for in vitro testing of new antimycobacterial agents.
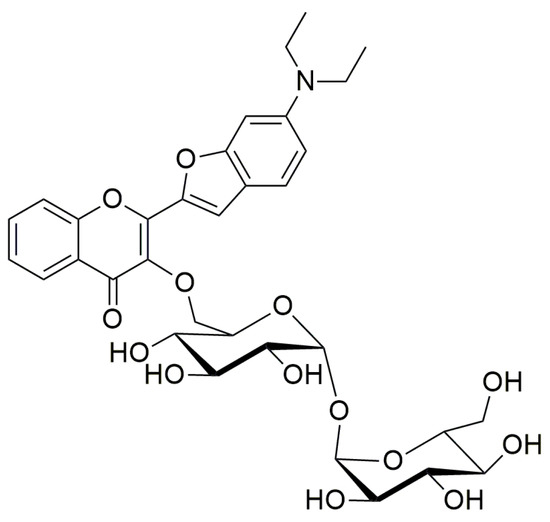
Figure 2.
Chemical diagram of the dye trehalose conjugate 3HC-2-Tre used in this study.
Whereas in-depth spectroscopic investigations of the dye 3HC-2 can be found in the literature [15,16], its crystal and molecular structure appears to be hitherto unpublished, as revealed by a search of the Cambridge Structural Database (CSD) [17] via the WebCSD interface [18] in April 2023. Therefore, we subjected 3HC-2 to X-ray crystallography, density functional theory (DFT) calculations, and Hirshfeld surface analysis in order to better understand its features. In this contribution, we report the molecular structure of 3HC-2 in the crystal, a computational study of its conformational preference, its supramolecular structure in the solid state, and the preliminary results of fluorescence labelling experiments with M. aurum cells using the dye trehalose conjugate 3HC-2-Tre.
2. Results and Discussion
2.1. Structural Description of 3HC-2
Compound 3HC-2 (Figure 3a) was synthesized as described in the literature [5]. In brief, 2′-hydroxyacetophenone was reacted with 6-(diethylamino)benzofuran-2-carbaldehyde [16], followed by treatment with hydrogen peroxide to give the desired 3-hydroxychromone derivative 3HC-2. Intense yellow crystals of 3HC-2, as shown in Figure 3b, grew from a solution in heptane/ethyl acetate. X-ray crystallography revealed that the compound crystallized solvent-free in the triclinic system, centrosymmetric space group P-1, with two molecules in the unit cell (Z = 2).
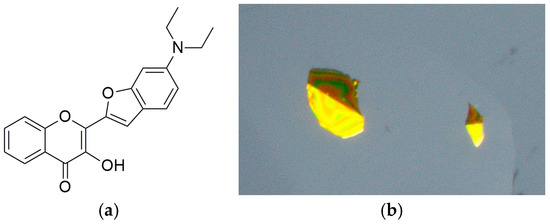
Figure 3.
Chemical diagram of 3HC-2 (a) and microscope image of crystal specimens in perfluoropolyether oil (b).
Figure 4 shows the molecular structure of 3HC-2 in the crystal. One of the ethyl groups exhibits disorder over two positions. As expected, the ten-membered chromene system is planar (r.m.s. deviation 0.0108 Å) as is the nine-membered benzofuran moiety tethered to C2 (r.m.s. deviation 0.0107 Å). The molecule adopts an s-cis conformation about the C2–C2′ bond. The O1–C2–C2′–O1′ torsion angle is −13.76(16)° in the chosen asymmetric unit, and the angle between the mean planes through the chromene and benzofuran moieties is 14.42(4)°. An s-cis conformation was encountered in the related 3-(furan-2-yl)-2-hydroxy-4H-chromen-4-one (CSD refcodes: IJUCEW and IJUCEW01) [19,20] and its nicotinic acid ester (MASGAS) [21]. Interestingly, an s-trans conformation was found in the crystal structures of the 2-thiophenyl derivatives ((5-(3-hydroxy-4-oxo-4H-chromen-2-yl)-2-thienyl)methylene)malononitrile (MUGGEC) [22] and 2-(thiophen-2-yl)-4H-chromen-4-one-3-O-2,3,4,6-O-tetraacetyl-β-d-glucopyranoside (QICLIA) [23].
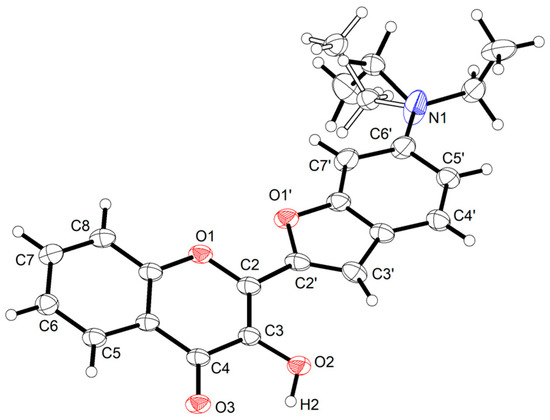
Figure 4.
Displacement ellipsoid plot of 3HC-2 drawn at the 50% probability level. Hydrogen atoms are represented by small spheres of arbitrary radius. The part of the disordered ethyl group with minor occupancy (ca. 47%) is shown with empty bonds.
A CSD search (April 2023) for crystal structures containing a 2,3-diether-butadiene moiety with a central acyclic C-C single bond revealed 107 entries. Of these, 80 structures exhibited an O–C–C–O torsion angle between 163 and −164°, and for 21 structures the O–C–C–O torsion angle was between −26 and 20°, as observed for 3HC-2. Intermediate torsion angle values in six structures can likely be attributed to packing effects or the steric bulk of substituents (NENLAT −60.2° [24], AHAYEO −47.3° [25], CIVXUC 37.9° [26], SILXAP 49.5° [27], HEJRUL 76.3° [28], DADHOJ 127.2° [29]). The relatively small number of structures with an O–C–C–O torsion angle around 0° prompted us to calculate the optimized structure of the free molecule of 3HC-2 using DFT methods. The minimum energy structure of the free molecule exhibited an O1–C2–C2′–O1′ torsion angle of 0°. Figure 5 shows a superposition of the 3-hydroxychromone moieties of the molecular structure in the crystal and the DFT-optimized molecular structure of the free molecule of 3HC-2, illustrating the conformational difference between the two structures, which can be attributed to packing effects in the crystal (vide infra). A relaxed surface scan was subsequently calculated in order to explore the conformational flexibility of 3HC-2 about the central C2–C2′ bond. This revealed a rotational barrier of ca. 7.5 kcal mol−1 and the energy of the s-trans conformer was about 1.8 kcal mol−1 higher than that of the local ground state structure adopting the s-cis conformation (Figure 6).
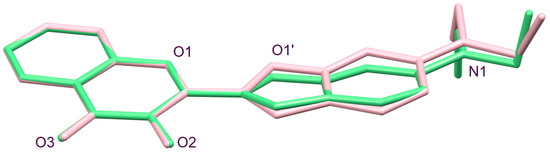
Figure 5.
Structure overlay of the 3-hydroxychromone moieties of the molecular structure of the major disorder part structure (green) and DFT-optimized structure (pink) of the free molecule of 3HC-2.
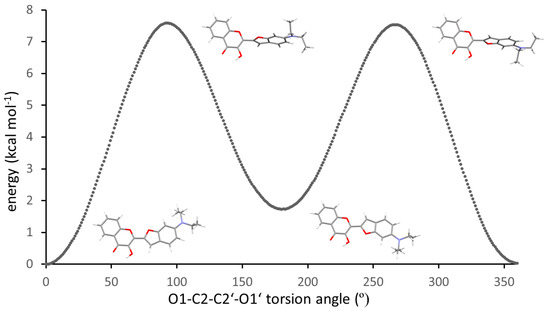
Figure 6.
Calculated relaxed surface scan about the central C2–C2′ single bond between the chromenone and benzofuran moieties in 3HC-2. Color scheme C, grey; H, white; N, blue; O, red.
The supramolecular structure of 3HC-2 in the solid state features centrosymmetric O–H⋯O hydrogen-bonded dimers (Figure 7). The 3-hydroxy groups each act as a hydrogen bond donor towards the chromone carbonyl oxygen atom of the other molecule in a dimer, resulting in a motif [30]. The corresponding hydrogen bond parameters (Table 1) are characteristic of strong hydrogen bonds [31]. The same intermolecular hydrogen bond motif was observed in the crystal structures of the unsubstituted 3-hydroxychromone (HOHHIW) [32] and in the aforementioned IJUCEW and MUGGEC. Whereas the chromone oxygen atom O1 does not exhibit contacts shorter than the sum of van der Waals radii in the crystal, the benzofuran oxygen atom O1′ is approached by a methyl hydrogen atom of the disordered ethyl group of an adjacent molecule. Figure 8 shows the Hirshfeld surface of 3HC-2 and the corresponding 2D fingerprint plot, revealing the dominance of short out of the plane O⋯H contacts, resulting from the intermolecular O–H⋯O hydrogen bonding described above and the π⋯π stacking of the molecules.
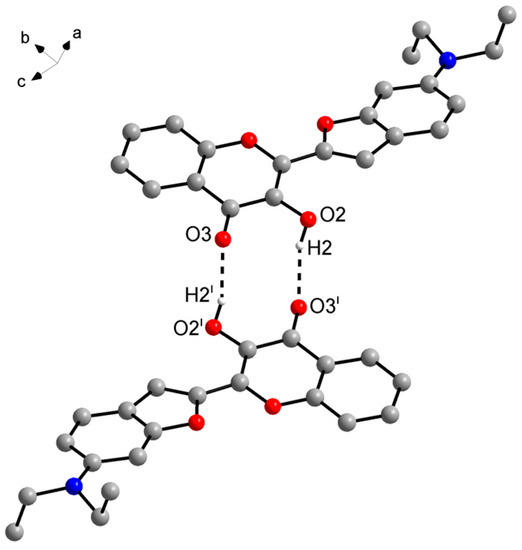
Figure 7.
Centrosymmetric hydrogen-bonded dimer of 3HC-2 in the crystal. Dashed lines represent hydrogen bonds. Carbon-bound hydrogen atoms and the minor disorder part are omitted for clarity. Color scheme: C, grey; H, white; N, blue; O, red. Symmetry code: (i) −x, −y, −z + 2.

Table 1.
Hydrogen bond geometry for 3HC-2 (Å, °).
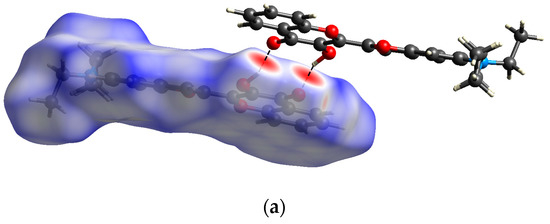
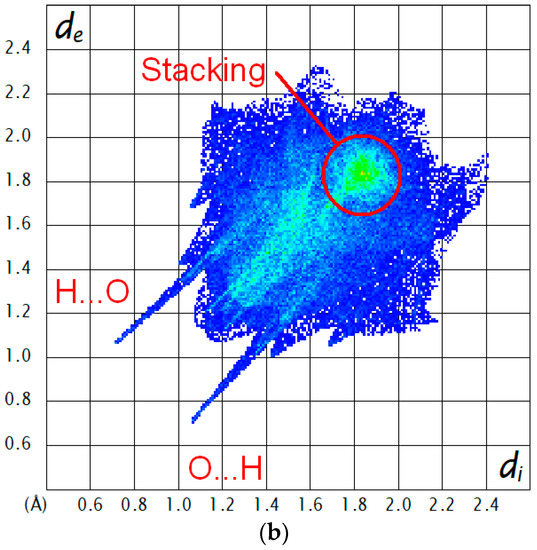
Figure 8.
(a) Hirshfeld surface mapped with dnorm (red areas indicate short contacts) and (b) the corresponding 2D fingerprint plot for 3HC-2; di and de are the respective interior and exterior distances of the nearest atom to the Hirshfeld surface over the range 0.4–2.6 Å (blue, few points; green, moderate fraction; red, many points). Color scheme for the atoms: C, grey; H, white; N, blue; O, red.
2.2. Labelling of Mycobacterium aurum Cells
The dye trehalose conjugate 3HC-2-Tre (Figure 2) for the labelling of M. aurum cells was prepared from anhydrous trehalose and 3HC-2 as described by Kamariza et al. [5]. In brief, trehalose was brominated in the 6-position by a variation of the Appel reaction using N-bromosuccinimide and triphenylphosphine, followed by acetylation. Subsequently, a nucleophilic substitution reaction with 3HC-2, after deprotonation of the 3-hydroxy group, followed by deacetylation afforded the anticipated 3HC-2-Tre. We then incubated M. aurum cells with 100 µM 3HC-2-Tre in liquid growth medium for 3 h at 36 °C and subjected the sample to fluorescence microscopy. As shown in Figure 9, our preliminary results demonstrate fluorescence of 3HC-2-Tre-labelled M. aurum cells with λex = 485 nm and λem = 510–531 nm filter sets. It is interesting to note that Kamariza et al. came to the conclusion that 3HC-2-Tre-labelling of Mycobacterium smegmatis, another generally considered non-pathogenic, fast-growing mycobacterium and model organism for the pathogen M. tuberculosis [33], was not specific to the trehalose pathway (vide supra) [5]. Exploring the mechanism of the labelling of M. aurum cells with 3HC-2-Tre, however, is beyond the scope of this preliminary investigation.

Figure 9.
Microscope imaging (20× magnification) of M. aurum cells treated with 100 µM 3HC-2-Tre (GFP channel: λex = 485 nm, λem = 510–531 nm).
3. Materials and Methods
3.1. General
Compounds 3HC-2 and 3HC-2-Tre were prepared following the procedures reported by Kamariza et al. [5]. The synthesis of the aldehyde precursor to 3HC-2, i.e., 6-(diethylamino)benzofuran-2-carbaldehyde, from 3-dimethylaminophenol is described in Ref. [16]. Experimental details of the preparation of 3HC-2-Tre can be found in the Supplementary Materials. The starting materials 3-dimethylaminophenol (97.69%, BLD Pharmatech GmbH, Kaiserslautern, Germany), 2′-hydroxyacetophenone (99.94%, BLD Pharmatech GmbH), and anhydrous trehalose (>98.0%, TCI, Chuo-ku, Tokyo) were purchased and used as received.
3.2. X-ray Crystallography
A few crystals of 3HC-2 suitable for single-crystal X-ray diffraction were obtained by chance when the remainder of a heptane/ethyl acetate solution resulting from flash chromatography evaporated to dryness in a tube. The crystals were coated with perfluoropolyether PFO-XR75 and mounted using a MiTeGen cryo-loop. The X-ray diffraction data were measured on a Bruker AXS D8 Venture diffractometer, equipped with an Incoatec IµS Diamond microfocus X-ray source, Incoatec multilayer optics, and a CMOS Photon III detector. The APEX4 software was used to control the diffractometer [34]. The raw data were processed with the SAINT software [35] and corrected for absorption effects with SADABS-2016/2 [36] using the Gaussian method based on indexed crystal faces.
The crystal structure was solved with SHELXT [37] and refined with SHELXL-2019/3 [38]. Anisotropic atomic displacement parameters (ADPs) were introduced for all non-hydrogen atoms. The diethylamino group was affected by positional disorder, which was taken into account by a split model for one ethyl group. Refinement of the ratio of occupancies by means of a free variable resulted in 0.531(4):0.469(4). Similar distance (SADI) and enhanced rigid-bond restraints (RIGU) [39] were applied to the disordered ethyl group. Except for the hydroxy hydrogen atom H2 and H3′ on the benzofuran moiety, hydrogen atoms were placed in geometrically calculated positions with Caromat–H = 0.95 Å, Cmethylene–H = 0.99 Å, and Cmethyl–H = 0.98 Å and refined using a riding model with Uiso(H) = 1.2 Ueq(C) (1.5 for methyl groups). The initial torsion angle of the methyl group of C10 was determined in a circular Fourier calculation and subsequently refined while maintaining a tetrahedral structure. H2 and H3′ were located in difference Fourier maps and refined with the O2–H2 and C3′–H3′ distance restrained to target values of 0.84(2) and 0.95(2) Å, respectively, and with Uiso(H) = 1.2 Ueq(C, O). Crystal data and refinement details are listed in Table 2. Structure pictures were drawn with Diamond [40]. Hirshfeld surface analysis was carried out using CrystalExplorer [41].

Table 2.
Crystal data and refinement details for 3HC-2.
3.3. Computational Methods
DFT calculations were performed with ORCA (version 5.0) [42] with a B3LYP/G VWN1 hybrid functional (20% HF exchange) [43,44,45], using a def2-TZVPP basis set [45]. Optimization of the structure was completed using the BFGS method from an initial Hessian according to Almoef’s model with a very tight self-consistent field convergence threshold [46]. Calculations were made on the free molecule of 3HC-2. A relaxed surface scan was carried out varying the crystallographic O1–C2–C2′–O1′ torsion angle from 0 to 359° in 1° steps using a def2-TZVPP basis set and optimizing as above with a tight self-consistent field convergence criterion. Structures near the transition states were subsequently optimized using a def2-TZVPP basis set and a very tight self-consistent field convergence criterion, as for the optimized structure. The optimized local minimum-energy structures exhibited only positive modes and the transition states each exhibited one imaginary mode. Cartesian coordinates of the DFT-optimized structure of 3HC-2 can be found in the Supplementary Materials. Structure pictures were generated with Mercury [47].
3.4. Microbiology
M. aurum DSM 43999 was cultivated via the inoculation of a single colony from a fresh streak plate (Middlebrook 7H10 agar) into 10 mL of Middlebrook 7H9 liquid medium supplemented with 10% ADS [5% (m/v) bovine serum albumin fraction V, 0.81% (m/v) sodium chloride, 2% (m/v) dextrose in purified water] and 0.05% polysorbate 80. The culture was grown to an OD600 between 0.2 and 0.8 for the labelling experiment by incubation at 36 °C with shaking (50 rpm). After dilution to an initial concentration of 5 × 107 CFU/mL (OD600 0.1 = 1 × 108 CFU/mL) with growth medium, the culture was transferred to a clear flat-bottom 96-well plate (Sarstedt, 83.3924.500) with 100 µL per well. Subsequently, 1 µL of 3HC-2-Tre (10 mM in DMSO) was added to each well to achieve a final dye concentration of 100 µM. After homogenization by pipetting, the plate was incubated for 3 h at 36 °C with shaking (50 rpm).
3.5. Fluorescence Microscopy
After incubation (see Section 3.4), the contents of the wells were homogenized by pipetting and 1 µL of each well was transferred to a black clear flat-bottom 96-well plate (Greiner bio one, 6550909) filled with 200 µL phosphate-buffered saline per well. Fluorescence microcopy was performed with a Thermo Scientific (Waltham, MA, USA) CellInsight CX5 instrument. Samples were excited at 485 nm and imaged with the GFP channel (510–531 nm).
4. Conclusions
We have structurally characterized the 3-hydroxychromone dye 3HC-2 in the solid state by X-ray crystallography. In the crystal, the molecule exhibits an s-cis conformation with an O–C–C–O torsion angle of ±13.76(16)°. DFT calculations on the free molecule gave an O–C–C–O torsion angle of ca. 0°, and revealed that the s-cis conformer was approximately 1.8 kcal mol−1 more stable than the s-trans conformer, with a rotational barrier of ca. 7.5 kcal mol−1. Preliminary labelling experiments demonstrated that M. aurum cells incubated in the presence of the dye trehalose conjugate 3HC-2-Tre could be detected by fluorescence microscopy in the GFP channel. We expect that this will facilitate in vitro testing of new antimycobacterial agents using this test bacterium, which is generally regarded as non-pathogenic.
Supplementary Materials
The following materials are available online: Description of the synthesis of 3HC-2-Tre and cartesian coordinates of the DFT-optimized structures of the s-cis and s-trans conformers of 3HC-2.
Author Contributions
Conceptualization, A.R. and R.W.S.; methodology, A.R., R.G., F.S., L.M. and R.W.S.; validation, A.R., R.G., F.S., L.M. and R.W.S.; formal analysis, A.R., R.G., F.S., L.M. and R.W.S.; investigation, A.R., R.G., F.S. and L.M.; resources, A.R. and R.G.; data curation, R.W.S.; writing—original draft preparation, F.S. and R.W.S.; writing—review and editing, A.R., R.G. and L.M.; visualization, A.R., R.G., F.S., L.M. and R.W.S.; supervision, A.R.; project administration, A.R. and R.W.S.; funding acquisition, A.R. and F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—432291016 (to A.R.), Mukoviszidose Institut gGmbH (Bonn, Germany) project number 2202 (to A.R.), the research and development arm of the German Cystic Fibrosis Association Mukoviszidose e. V. and by a scholarship from the Vereinigung der Freunde und Förderer des Institutes für Pharmazie der Martin-Luther-Universität Halle-Wittenberg (VFFIP) to F.S.
Data Availability Statement
CCDC 2258616 contains the supplementary crystallographic data for this paper. The data can be obtained free of charge from the Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/structures. Cartesian coordinates of the DFT-calculated s-cis and s-trans conformers of 3HC-2 can be found in the Supplementary Materials.
Acknowledgments
We would like to thank Christian W. Lehmann for providing access to the X-ray diffraction facility at the Max-Planck-Institut für Kohlenforschung and Heike Schucht for technical assistance with the X-ray intensity data collection.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Sample Availability
Not Applicable.
References
- World Health Organization. Global Tuberculosis Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General Overview of Nontuberculous Mycobacteria Opportunistic Pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef] [PubMed]
- Dahl, V.N.; Molhave, M.; Floe, A.; van Ingen, J.; Schon, T.; Lillebaek, T.; Andersen, A.B.; Wejse, C. Global trends of pulmonary infections with nontuberculous mycobacteria: A systematic review. Int. J. Infect. Dis. 2022, 125, 120–131. [Google Scholar] [CrossRef]
- Singhal, R.; Myneedu, V.P. Microscopy as a diagnostic tool in pulmonary tuberculosis. Int. J. Mycobacteriology 2015, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kamariza, M.; Keyser, S.G.L.; Utz, A.; Knapp, B.D.; Ealand, C.; Ahn, G.; Cambier, C.J.; Chen, T.; Kana, B.; Huang, K.C.; et al. Toward Point-of-Care Detection of Mycobacterium tuberculosis: A Brighter Solvatochromic Probe Detects Mycobacteria within Minutes. JACS Au. 2021, 1, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E.; Brown-Elliott, B.A.; Chalmers, J.D.; Cirillo, D.M.; Daley, C.L.; Emler, S.; Floto, R.A.; Garcia, M.J.; Hoefsloot, W.; Koh, W.-J.; et al. Same meat, different gravy: Ignore the new names of mycobacteria. Eur. Respir. J. 2019, 54, 1900795, Mycobacterium aurum has been reclassified into the new genus Mycolicibacterium, but the new names of mycobacteria have not widely been accepted. [Google Scholar] [CrossRef] [PubMed]
- Phelan, J.; Maitra, A.; McNerney, R.; Nair, M.; Gupta, A.; Coll, F.; Pain, A.; Bhakta, S.; Clark, T.G. The draft genome of Mycobacterium aurum, a potential model organism for investigating drugs against Mycobacterium tuberculosis and Mycobacterium leprae. Int. J. Mycobacteriology 2015, 4, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Namouchi, A.; Cimino, M.; Favre-Rochex, S.; Charles, P.; Gicquel, B. Phenotypic and genomic comparison of Mycobacterium aurum and surrogate model species to Mycobacterium tuberculosis: Implications for drug discovery. BMC Genom. 2017, 18, 530. [Google Scholar] [CrossRef]
- Gupta, A.; Bhakta, S.; Kundu, S.; Gupta, M.; Srivastava, B.S.; Srivastava, R. Fast-growing, non-infectious and intracellularly surviving drug-resistant Mycobacterium aurum: A model for high-throughput antituberculosis drug screening. J. Antimicrob. Chemother. 2009, 64, 774–781. [Google Scholar] [CrossRef]
- Sood, S.; Yadav, A.; Shrivastava, R. Mycobacterium aurum is Unable to Survive Mycobacterium tuberculosis Latency Associated Stress Conditions: Implications as Non-suitable Model Organism. Indian J. Microbiol. 2016, 56, 198–204. [Google Scholar] [CrossRef]
- Honarvar, B.; Movahedan, H.; Mahmoodi, M.; Sheikholeslami, F.M.; Farnia, P. Mycobacterium aurum keratitis: An unusual etiology of a sight-threatening infection. Braz. J. Infect. Dis. 2012, 16, 204–208. [Google Scholar] [CrossRef]
- Madikizela, B.; Eckhardt, T.; Goddard, R.; Richter, A.; Lins, A.; Lehmann, C.; Imming, P.; Seidel, R.W. Synthesis, structural characterization and antimycobacterial evaluation of several halogenated non-nitro benzothiazinones. Med. Chem. Res. 2021, 30, 1523–1533. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, T.; Goddard, R.; Lehmann, C.; Richter, A.; Sahile, H.A.; Liu, R.; Tiwari, R.; Oliver, A.G.; Miller, M.J.; Seidel, R.W.; et al. Crystallographic evidence for unintended benzisothiazolinone 1-oxide formation from benzothiazinones through oxidation. Acta Crystallogr. Sect. C 2020, 76, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Seidel, R.W.; Goddard, R.; Eckhardt, T.; Lehmann, C.; Dörner, J.; Siersleben, F.; Sondermann, T.; Mann, L.; Patzer, M.; et al. BTZ-Derived Benzisothiazolinones with In Vitro Activity against Mycobacterium tuberculosis. ACS Med. Chem. Lett. 2022, 13, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Klymchenko, A.S.; Ozturk, T.; Pivovarenko, V.G.; Demchenko, A.P. A 3-hydroxychromone with dramatically improved fluorescence properties. Tetrahedron Lett. 2001, 42, 7967–7970. [Google Scholar] [CrossRef]
- Klymchenko, A.S.; Pivovarenko, V.G.; Ozturk, T.; Demchenko, A.P. Modulation of the solvent-dependent dual emission in 3-hydroxychromones by substituents. New J. Chem. 2003, 27, 1336–1343. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef]
- Thomas, I.R.; Bruno, I.J.; Cole, J.C.; Macrae, C.F.; Pidcock, E.; Wood, P.A. WebCSD: The online portal to the Cambridge Structural Database. J. Appl. Crystallogr. 2010, 43, 362–366. [Google Scholar] [CrossRef]
- Wera, M.; Pivovarenko, V.G.; Sikorski, A.; Lis, T.; Blazejowski, J. 2-(Furan-2-yl)-3-hydroxy-4H-chromen-4-one. Acta Crystallogr. Sect. E 2011, 67, o266. [Google Scholar] [CrossRef]
- Camargo, M.L.M.; dos Santos, F.A.; Pizzuti, L.; Abram, U.; Schwade, V.D. Complexes with Furyl-Substituted 3-Hydroxychromone: Synthesis, Characterization and Fluorescence Studies. J. Braz. Chem. Soc. 2021, 32, 1519–1530. [Google Scholar] [CrossRef]
- Camargo, M.L.M.; Schwalm, C.S.; Bortolotto, T.; de Freitas Daudt, N.; Rossi, G.G.; Anraku de Campos, M.M.; D’Oliveira, K.A.; Cuin, A.; Schwade, V.D. MII (M = Mn, Fe, Co, Ni and Cu) complexes with a chromone-derived neutral ligand: Synthesis, structural characterization, photocatalytic and mycobacterial activity studies. New J. Chem. 2022, 46, 2534–2545. [Google Scholar] [CrossRef]
- Tseng, H.-W.; Shen, J.-Y.; Kuo, T.-Y.; Tu, T.-S.; Chen, Y.-A.; Demchenko, A.P.; Chou, P.-T. Excited-state intramolecular proton-transfer reaction demonstrating anti-Kasha behavior. Chem. Sci. 2016, 7, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Mughal, E.U.; Javid, A.; Sadiq, A.; Murtaza, S.; Zafar, M.N.; Khan, B.A.; Sumrra, S.H.; Tahir, M.N.; Kanwal; Khan, K.M. Synthesis, structure-activity relationship and molecular docking studies of 3-O-flavonol glycosides as cholinesterase inhibitors. Bioorganic Med. Chem. 2018, 26, 3696–3706. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.-M.; Peters, K.; Meints, C.; Tochtermann, W. Crystal structure of (pM*,pM*)-(±)-bi-(dimethyl-3,6-decanooxepine-4,5-dicarboxylate), [C6HO(CH2)IO(COOCH3)2]2. Z. Für Krist.-New Cryst. Struct. 2001, 216, 315–316. [Google Scholar] [CrossRef]
- Alcaide, B.; Almendros, P.; Carrascosa, R.; Torres, M.R. Synthesis of a New Class of C2-Symmetrical Biheteroaryls by Ammonium Cerium(IV) Nitrate Mediated Dimerization of 2-(Furan-3-yl)pyrroles. Eur. J. Org. Chem. 2010, 2010, 823–826. [Google Scholar] [CrossRef]
- Fan, Y.-S.; Das, U.; Hsiao, M.-Y.; Liu, M.-H.; Lin, W. Chemoselective Intramolecular Wittig Reactions for the Synthesis of Oxazoles and Benzofurans. J. Org. Chem. 2014, 79, 11567–11582. [Google Scholar] [CrossRef]
- Mulay, S.V.; Bogoslavky, B.; Galanti, I.; Galun, E.; Gidron, O. Bifuran-imide: A stable furan building unit for organic electronics. J. Mater. Chem. C 2018, 6, 11951–11955. [Google Scholar] [CrossRef]
- Ookubo, Y.; Wakamiya, A.; Yorimitsu, H.; Osuka, A. Synthesis of a Library of Fluorescent 2-Aryl-3-trifluoromethylnaphthofurans from Naphthols by Using a Sequential Pummerer-Annulation/Cross-Coupling Strategy and their Photophysical Properties. Chem. A Eur. J. 2012, 18, 12690–12697. [Google Scholar] [CrossRef]
- Wang, C.-H.; Gao, Z.-C.; Sun, W.; Guo, X.; Zhang, F.-B. P⋯O noncovalent conformational locks for constructing highly planar Bis(diphenylphosphanyl) Bi(benzofurano). Dye. Pigment. 2021, 184, 108820. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.L. Patterns in Hydrogen Bonding: Functionality and Graph Set Analysis in Crystals. Angew. Chem. Int. Ed. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- Thakuria, R.; Sarma, B.; Nangia, A. 7.03-Hydrogen Bonding in Molecular Crystals. In Comprehensive Supramolecular Chemistry II; Atwood, J.L., Ed.; Elsevier: Oxford, UK, 2017; pp. 25–48. [Google Scholar] [CrossRef]
- Binbuga, N.; Schultz, T.P.; Henry, W.P. Intra- and intermolecular hydrogen bonding in 3-hydroxy- and 5-hydroxychromone. Tetrahedron Lett. 2008, 49, 5762–5765. [Google Scholar] [CrossRef]
- T, J.A.S.; J, R.; Rajan, A.; Shankar, V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Infect. Public Health 2020, 13, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- APEX4; Bruker AXS Inc.: Madison, WI, USA, 2017.
- SAINT; Bruker AXS Inc.: Madison, WI, USA, 2012.
- SADABS; Bruker AXS Inc.: Madison, WI, USA, 2012.
- Sheldrick, G.M. SHELXT-integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Thorn, A.; Dittrich, B.; Sheldrick, G.M. Enhanced rigid-bond restraints. Acta Crystallogr. Sect. A 2012, 68, 448–451. [Google Scholar] [CrossRef]
- Brandenburg, K. Diamond; Crystal Impact GbR: Bonn, Germany, 2018. [Google Scholar]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Hertwig, R.H.; Koch, W. On the parameterization of the local correlation functional. What is Becke-3-LYP? Chem. Phys. Lett. 1997, 268, 345–351. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Fletcher, R. Practical Methods of Optimization, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).