N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide
Abstract
1. Introduction
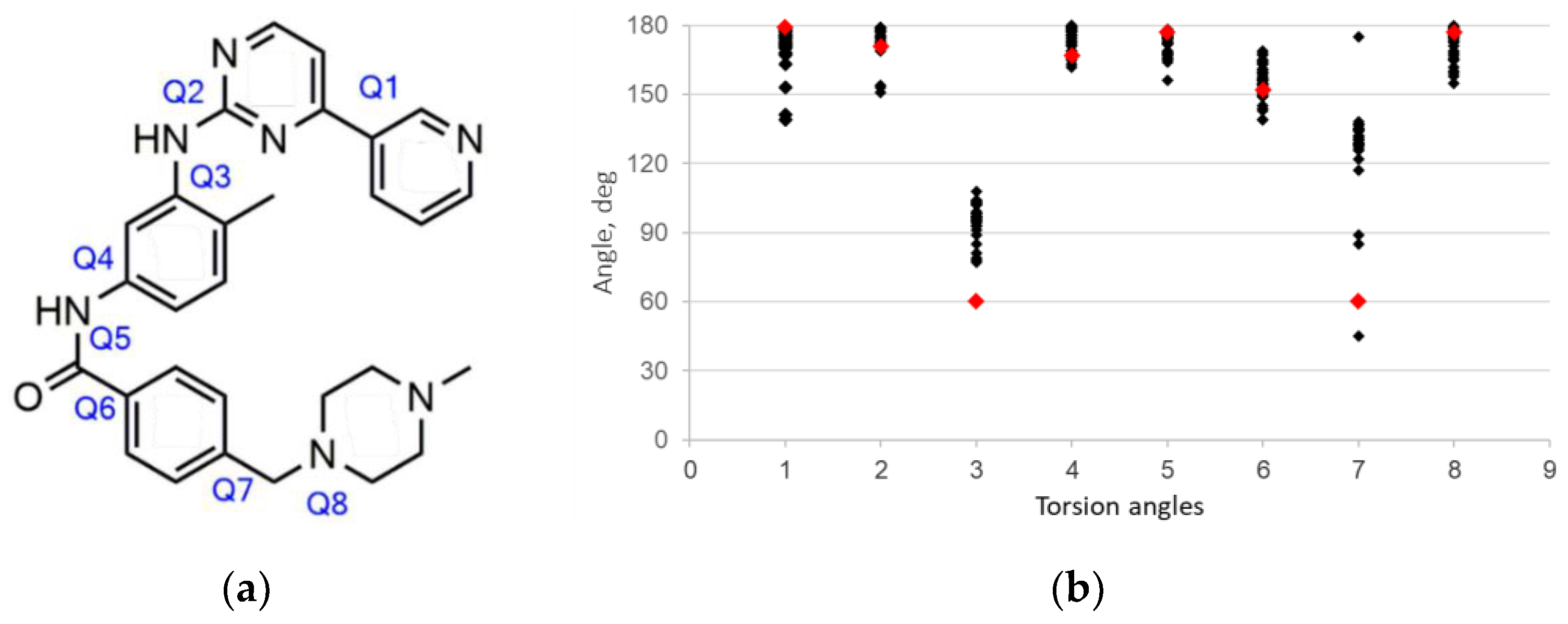
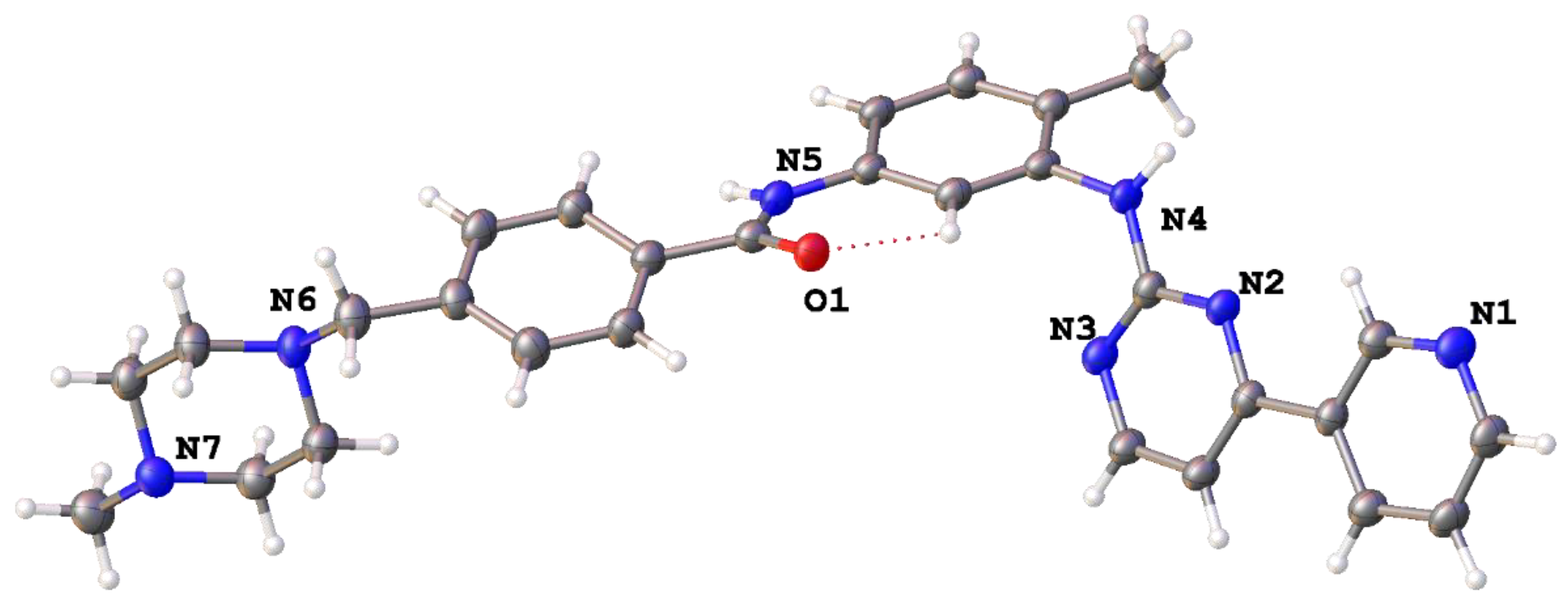
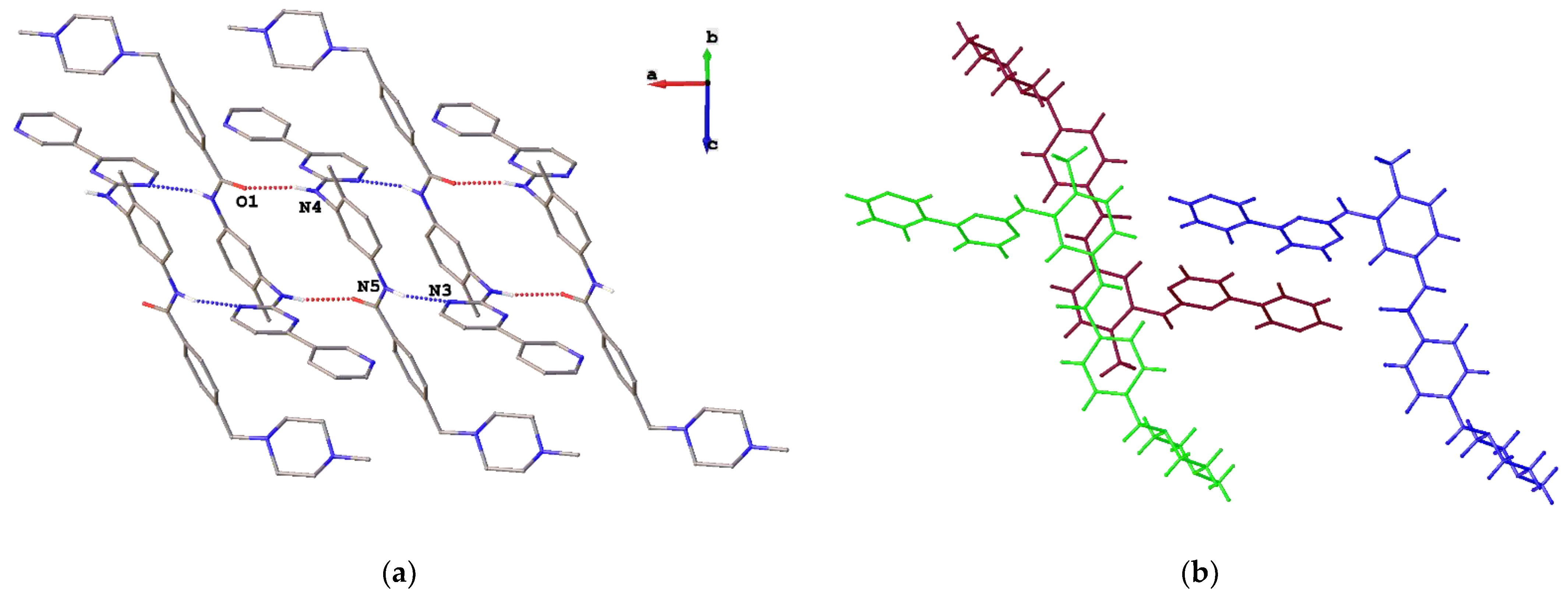
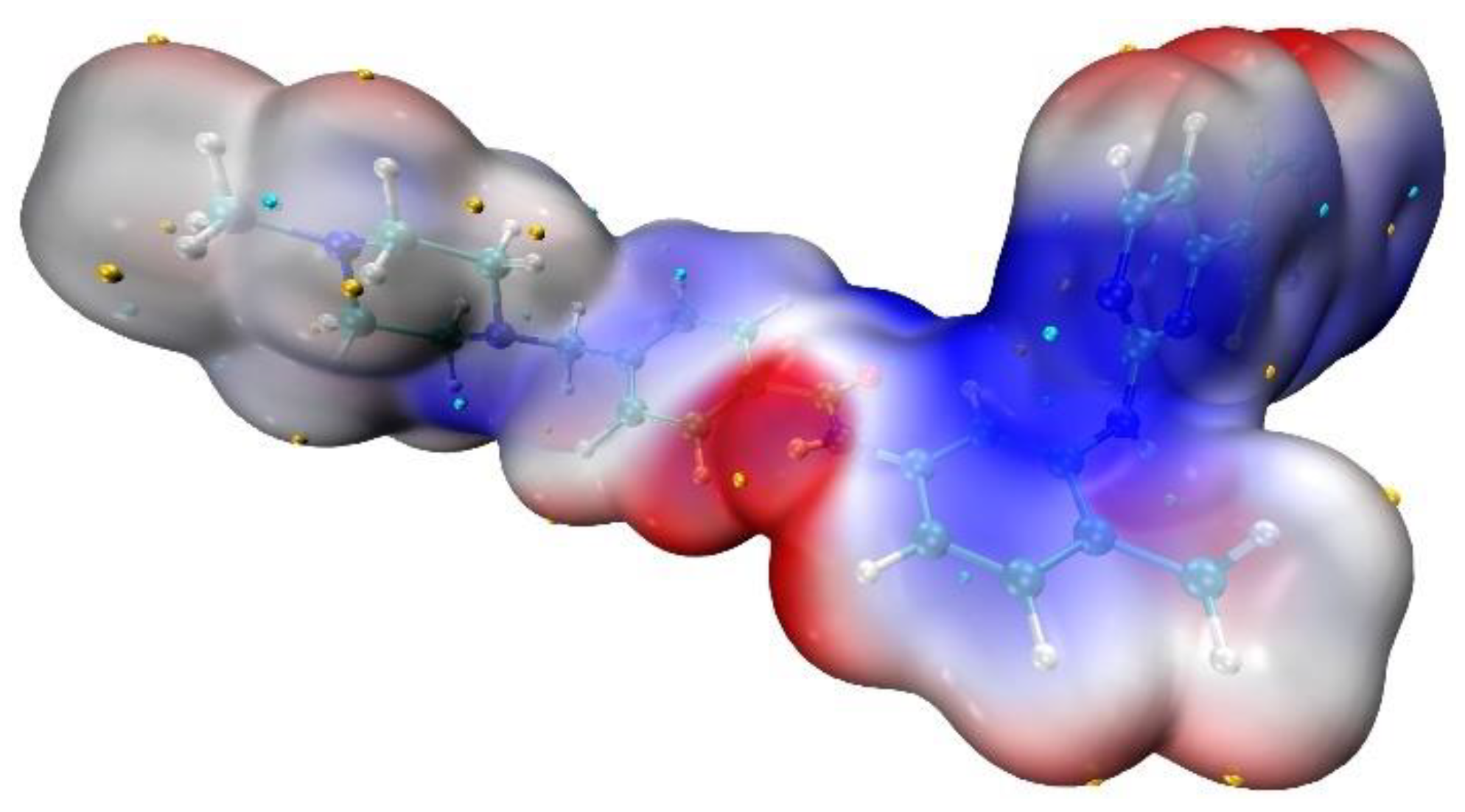
2. Results and Discussion
3. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carofiglio, F.; Lopalco, A.; Lopedota, A.; Cutrignelli, A.; Nicolotti, O.; Denora, N.; Stefanachi, A.; Leonetti, F. Bcr-Abl Tyrosine Kinase Inhibitors in the Treatment of Pediatric CML. Int. J. Mol. Sci. 2020, 21, 4469. [Google Scholar] [CrossRef]
- Deininger, M.; Buchdunger, E.; Druker, B.J. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 2005, 105, 2640–2653. [Google Scholar] [CrossRef] [PubMed]
- Nagar, B.; Hantschel, O.; Young, M.A.; Scheffzek, K.; Veach, D.; Bornmann, W.; Clarkson, B.; Superti-Furga, G.; Kuriyan, J. Structural Basis for the Autoinhibition of c-Abl Tyrosine Kinase. Cell 2003, 112, 859–871. [Google Scholar] [CrossRef]
- Schindler, T.; Bornmann, W.; Pellicena, P.; Miller, W.T.; Clarkson, B.; Kuriyan, J. Structural Mechanism for STI-571 Inhibition of Abelson Tyrosine Kinase. Science 2000, 289, 1938–1942. [Google Scholar] [CrossRef]
- Golzarroshan, B.; Siddegowda, M.S.; Li, H.Q.; Yathirajan, H.S.; Narayana, B.; Rathore, R.S. Imatinib (Gleevec@) conformations observed in single crystals, protein–Imatinib co-crystals and molecular dynamics: Implications for drug selectivity. J. Mol. Struct. 2012, 1018, 107–112. [Google Scholar] [CrossRef]
- Vologzhanina, A.V.; Ushakov, I.E.; Korlyukov, A.A. Intermolecular Interactions in Crystal Structures of Imatinib-Containing Compounds. Int. J. Mol. Sci. 2020, 21, 8970. [Google Scholar] [CrossRef]
- Grillo, D.; Polla, G.; Vega, D. Conformational Polymorphism on Imatinib Mesylate: Grinding Effects. J. Pharm. Sci. 2012, 101, 541–551. [Google Scholar] [CrossRef]
- Jasinski, J.P.; Butcher, R.J.; Hakim Al-Arique, Q.N.M.; Yathirajan, H.S.; Narayana, B. Imatinibium dipicrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2010, 66, o411–o412. [Google Scholar] [CrossRef]
- Fang, Z.-Y.; Zhang, B.-X.; Xing, W.-H.; Jia, H.-L.; Wang, X.; Gong, N.-B.; Lu, Y.; Du, G.-H. A series of stable, metastable and unstable salts of Imatinib with improved solubility. Chin. Chem. Lett. 2022, 33, 2159–2164. [Google Scholar] [CrossRef]
- Ushakov, I.E.; Lenenko, N.D.; Goloveshkin, A.S.; Korlyukov, A.A.; Golub, A.S. Influence of noncovalent intramolecular and host–guest interactions on imatinib binding to MoS2 sheets: A PXRD/DFT study. CrystEngComm 2022, 24, 639–646. [Google Scholar] [CrossRef]
- Kabova, E.A.; Blundell, C.D.; Muryn, C.A.; Whitehead, G.F.S.; Vitorica-Yrezabal, I.J.; Ross, M.J.; Shankland, K. SDPD-SX: Combining a single crystal X-ray diffraction setup with advanced powder data structure determination for use in early stage drug discovery. CrystEngComm 2022, 24, 4337–4340. [Google Scholar] [CrossRef]
- Goloveshkin, A.S.; Korlyukov, A.A.; Vologzhanina, A.V. Novel Polymorph of Favipiravir—An Antiviral Medication. Pharmaceutics 2021, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Galek, P.T.A.; Allen, F.H.; Fábián, L.; Feeder, N. Knowledge-based H-bond prediction to aid experimental polymorph screening. CrystEngComm 2009, 11, 2634–2639. [Google Scholar] [CrossRef]
- Vologzhanina, A.V. Intermolecular Interactions in Functional Crystalline Materials: From Data to Knowledge. Crystals 2019, 9, 478. [Google Scholar] [CrossRef]
- Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef]
- Kleemiss, F.; Dolomanov, O.V.; Bodensteiner, M.; Peyerimhoff, N.; Midgley, L.; Bourhis, L.J.; Genoni, A.; Malaspina, L.A.; Jayatilaka, D.; Spencer, J.L.; et al. Accurate crystal structures and chemical properties from NoSpherA2. Chem. Sci. 2021, 12, 1675–1692. [Google Scholar] [CrossRef] [PubMed]
- Allen, F.H.; Bruno, I.J. Bond lengths in organic and metal-organic compounds revisited: X–H bond lengths from neutron diffraction data. Acta Crystallogr. B 2010, 66, 380–386. [Google Scholar] [CrossRef]
- Zimmermann, J.D. Pyrimidin Derivatives and Process for Their Preparation 1993. Patent EP0564409A1, 25 March 1993. [Google Scholar]
- Lazarenko, V.A.; Dorovatovskii, P.V.; Zubavichus, Y.V.; Burlov, A.S.; Koshchienko, Y.V.; Vlasenko, V.G.; Khrustalev, V.N. High-Throughput Small-Molecule Crystallography at the ‘Belok’ Beamline of the Kurchatov Synchrotron Radiation Source: Transition Metal Complexes with Azomethine Ligands as a Case Study. Crystals 2017, 7, 325. [Google Scholar] [CrossRef]
- Svetogorov, R.D.; Dorovatovskii, P.V.; Lazarenko, V.A. Belok/XSA Diffraction Beamline for Studying Crystalline Samples at Kurchatov Synchrotron Radiation Source. Cryst. Res. Technol. 2020, 55, 1900184. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Blatov, V.A.; Shevchenko, A.P.; Proserpio, D.M. Applied Topological Analysis of Crystal Structures with the Program Package ToposPro. Cryst. Growth Des. 2014, 14, 3576–3586. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, C.; Cao, Y.; Yuan, Z.; Xu, L.; Zhou, Y. Continuous flow processing: In situ preparation of imatinib freebase. Lat. Am. J. Pharm. 2018, 37, 1251–1256. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korlyukov, A.A.; Dorovatovskii, P.V.; Vologzhanina, A.V. N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide. Molbank 2022, 2022, M1461. https://doi.org/10.3390/M1461
Korlyukov AA, Dorovatovskii PV, Vologzhanina AV. N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide. Molbank. 2022; 2022(4):M1461. https://doi.org/10.3390/M1461
Chicago/Turabian StyleKorlyukov, Alexander A., Pavel V. Dorovatovskii, and Anna V. Vologzhanina. 2022. "N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide" Molbank 2022, no. 4: M1461. https://doi.org/10.3390/M1461
APA StyleKorlyukov, A. A., Dorovatovskii, P. V., & Vologzhanina, A. V. (2022). N-(4-Methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)-4-((4-methylpiperazin-1-yl)methyl)benzamide. Molbank, 2022(4), M1461. https://doi.org/10.3390/M1461





