Abstract
The 1,2,3-triazole ring system can be easily obtained by widely used copper-catalyzed click reaction of azides with alkynes. 1,2,3-triazole exhibits myriad of biological activities, including antibacterial antimalarial, and antiviral activities. We herein reported the synthesis of quinoline-based [1,2,3]-triazole hybrid derivative via Cu(I)-catalyzed click reaction of 4-azido-7-chloroquinoline with alkyne derivative of hydroxybenzotriazole (HOBt). The compound was fully characterized by proton nuclear magnetic resonance (1H-NMR), carbon-13 nuclear magnetic resonance (13C-NMR), correlated spectroscopy (1H-1H-COSY), heteronuclear single quantum coherence (HSQC) and distortionless enhancement by polarization transfer (DEPT-135 and DEPT-90) NMR, ultraviolet (UV) and Fourier-transform infrared (FTIR) spectroscopies, and high-resolution mass spectrometry (HRMS). Computational studies were enrolled to predict the interaction of the synthesized compound with acetylcholinesterase, a target of primary relevance for developing new therapeutic options to counteract neurodegeneration. Moreover, the drug-likeness of the compound was also investigated by predicting its pharmacokinetic properties.
1. Introduction
Alzheimer’s disease (AD) is the most common form of dementia worldwide and it is mainly characterized by memory loss, cognitive impairment, and other neurological symptoms. The etiology of AD is poorly understood, but what evidence proves is that neuronal damage and synaptic loss occurs in affected patients with the emergence of previously mentioned symptoms [1,2].
Acetylcholinesterase enzyme (AChE) is a protein with a pivotal role in hydrolyzing acetylcholine (ACh), an important neurotransmitter that exerts its action mainly at the level of cholinergic synapses in both central and peripheral nervous systems (CNS and PNS) [3]. During neurotransmission, ACh is released from the presynaptic neuron into the synaptic cleft. Then, ACh binds to its receptor on the post-synaptic membrane, and this event allows transmission of the signal from one nerve to the other. In physiologic conditions, AChE located on the post-synaptic membrane hydrolyses ACh and terminates the signal transmission.
Despite this important regulatory action, several research groups have demonstrated the crucial role of AChE in the evolution of pathologic conditions since it was found that it increases ACh degradation leading to impairment of cholinergic nerves, it induces accumulation of amyloid β (Aβ) plaques, neurofibrillary tangles (NFT), and it leverages oxidative stress [4]. Given the very high prevalence of AD in the world, different molecules and targets were identified and analyzed with the aim of resolving the condition. Nowadays, few are the drugs that were approved by Food and Drug Administration (FDA), and they generally only alleviate symptoms. Among them is donepezil (2-[(1-benzylpiperidin-4-yl)methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one), which inhibits AChE and leads to a neuroprotective effect by regulating ACh levels at brain synapses [5,6]. The FDA also approved three other AChE inhibitors, such as galantamine, rivastigmine, and tacrine, as well as the N-methyl-d-aspartate (NMDA) receptor antagonist memantine, but the quest for novel remedies is still wide open [7,8,9,10].
The structure of AChE was determined by high resolution X-ray diffraction experiments both in unliganded form and in complex with donepezil [11]. Its binding site is composed of the peripheral anionic site (PAS) and of the catalytic anionic site (CAS) which is made up of catalytic triad: Ser203, His447, and Glu334. Donepezil binds both PAS and CAS through hydrogen bonds, π–π interactions and hydrophobic interactions [5,6,12]. In view of the beneficial effects given by donepezil, many attempts are still being made to identify novel molecules which could be structural analogues or chemically different derivatives with better performances in targeting AChE. In this connection, the nitrogen containing heterocycles are found in abundance in most of the medicinal compounds. In particular, triazoles are five-membered rings, which contain two carbon and three nitrogen atoms and many covalently linked triazole hybrids exhibits diverse biological activities, such as anti-inflammatory [13], anticancer [14], antibacterial [15], and antitubercular [16] (Figure 1a–l).
Previously, we have reported the synthesis of a novel semi-synthetic isoflavones as BACE-1 inhibitors against AD [17]. The current work is based on the preparation of compound 5 (Figure 1) which was designed, synthesized, and fully characterized by NMR, MS, IR, and UV. Then, the interaction of the molecule with AChE and its drug-likeness were assayed by means of computational tools, considering donepezil as reference in the study.
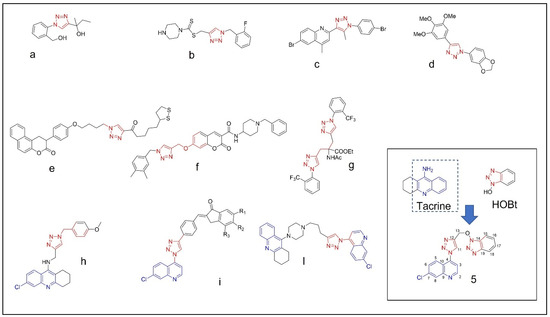
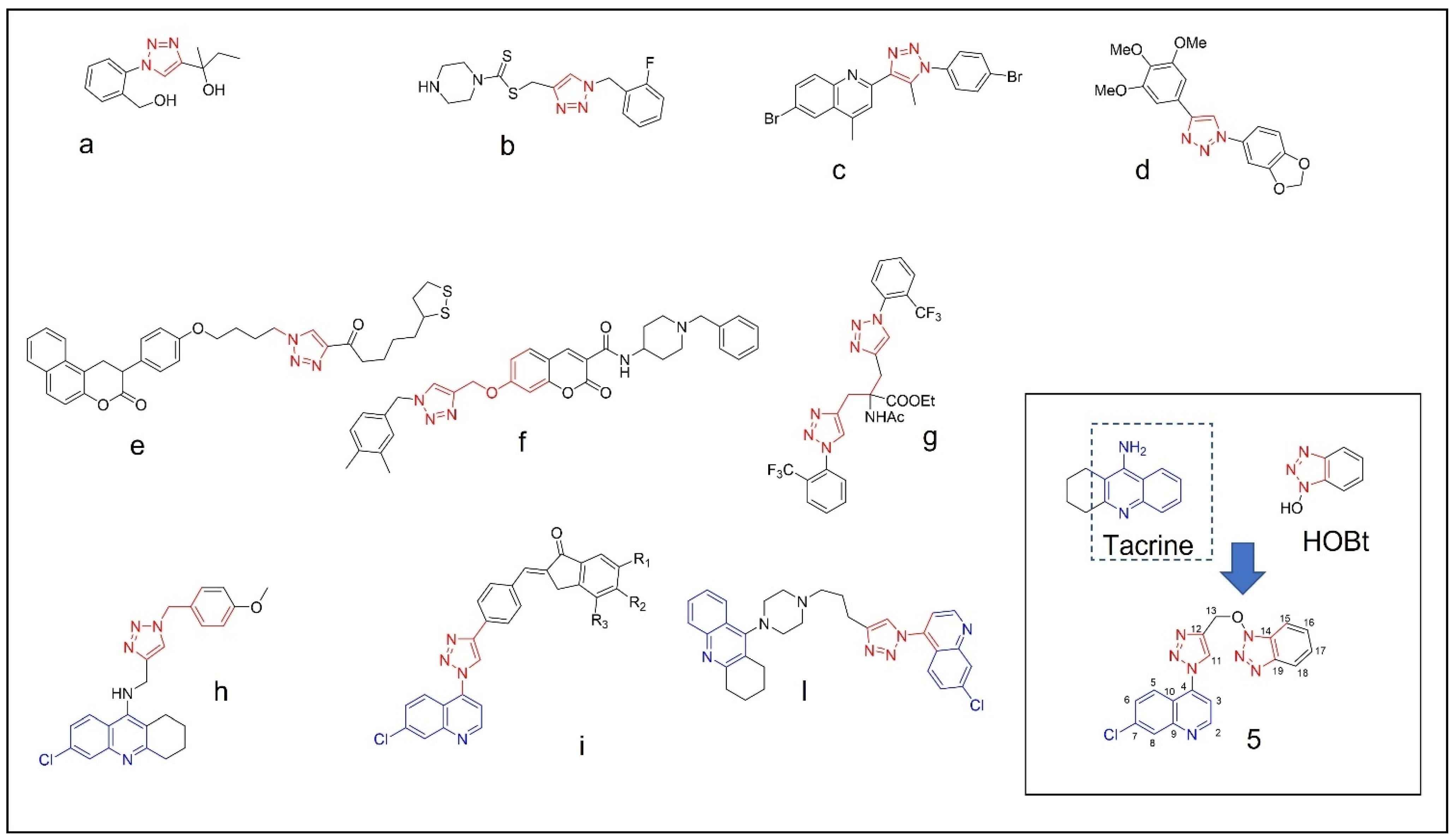

Figure 1.
Example of 1,2,3-triazole (red) containing bioactive molecules reported in literature (a–d) [13,14,15,16], 1,2,3-triazole (e–g) [18,19,20] and 1,2,3 triazole containing quinoline ring (blue) active against cholinesterases (h–l) [21,22,23]; the structure of compound (5), structurally related to tacrine and HOBt, is reported in the inset of the figure.
Figure 1.
Example of 1,2,3-triazole (red) containing bioactive molecules reported in literature (a–d) [13,14,15,16], 1,2,3-triazole (e–g) [18,19,20] and 1,2,3 triazole containing quinoline ring (blue) active against cholinesterases (h–l) [21,22,23]; the structure of compound (5), structurally related to tacrine and HOBt, is reported in the inset of the figure.

2. Results and Discussion
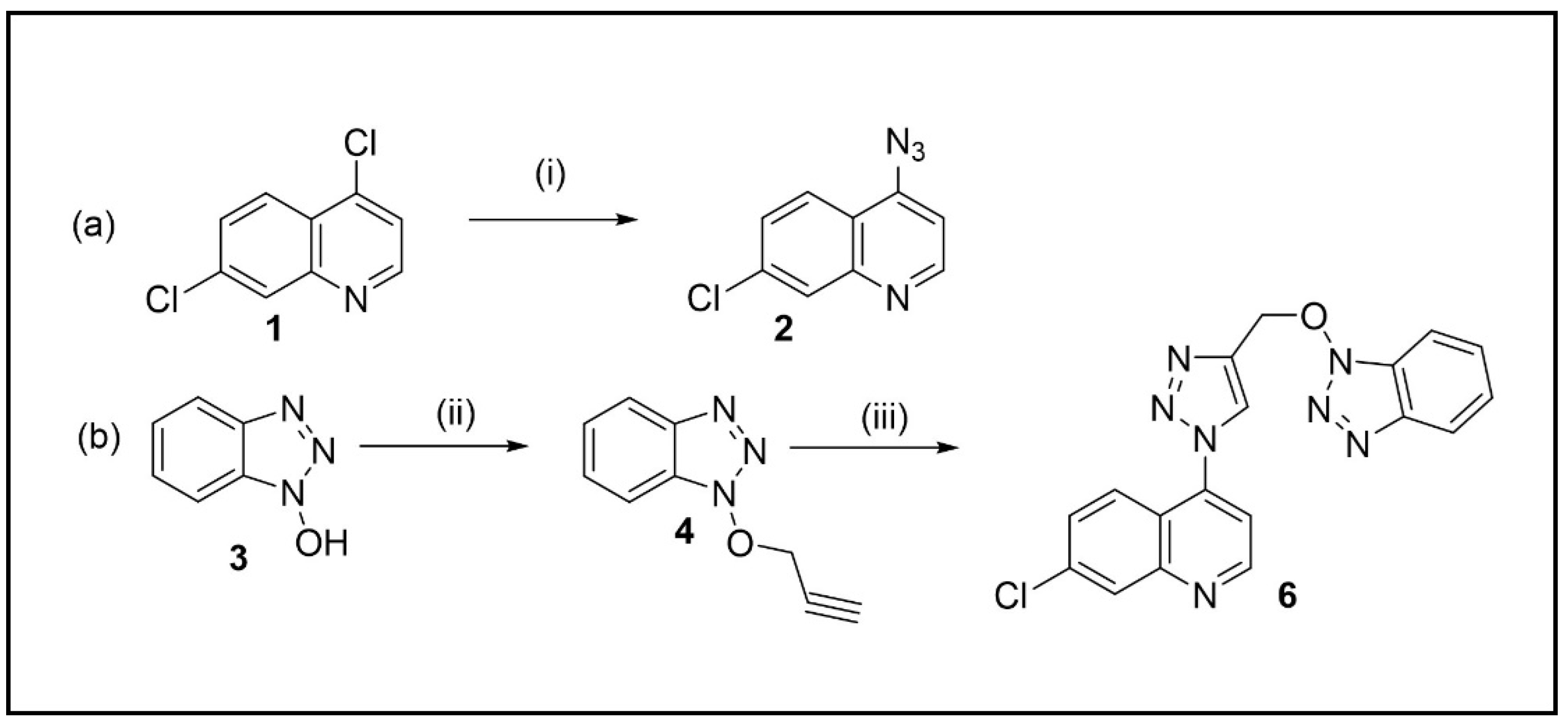
The synthetic route of the triazole hybrid compound, 4-(4-(((1H-benzo[d][1,2,3]triazol-1-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)-7-chloroquinoline 5, started from the preparation of precursors, 4-azido-7-chloroquinoline 2 and O-acetylenic derivative 4 (Scheme 1).
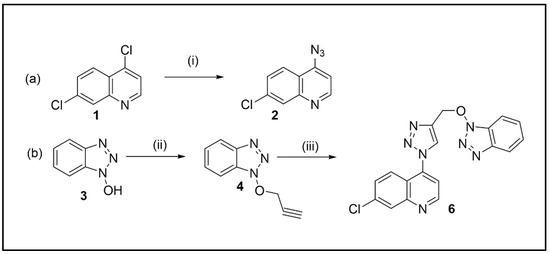
Scheme 1.
(a) Synthesis of 4-azido-7-chloro-quinoline (i) NaN3, DMF; (b) Synthesis of 4-(4-(((1H-benzo[d][1,2,3]triazol-1-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)-7-chloroquinoline (ii) propargyl bromide, DMF, K2CO3; (iii) 2, CuSO4, sodium ascorbate, tBuOH/water (1:1).
Similar to the protocol reported in our paper [24], quinoline 2 was prepared by the reaction of 4,7-dichloroquinoline 1 with NaN3 (2 equiv.) in anhydrous DMF at 65 C for 6 h (Scheme 1a). The crude product was purified by recrystallization from CH2Cl2/hexane to afford 2 in 78% yield.
The other cycloaddition partner, acetylenic intermediate 4, was generated by O-alkylation of 1H-benzo[d][1,2,3-triazol-1-ol 3 (HOBt) with propargyl bromide (1.5 equiv.) and anhydrous K2CO3 in anhydrous DMF with a good yield (82%) after recrystallization from CH2Cl2/hexane (Scheme 1b).
Finally, the hybrid compound 5 was then furnished by using a modified protocol of click reaction reported by Feldman et al. [25]. Equimolars of quinoline 2 and acetylene 4 in tBuOH/water (1:1) were subjected to sequential additions of sodium ascorbate (0.4 equiv.) and CuSO4 (20 mol%) (Scheme 1b). After stirring at 65 °C for 24 h, compound 5 was isolated by column chromatography in a high yield (86%).
This approach was adopted due to fast procedure, high yields, and easy separation of products. Moreover, starting materials (1 and HOBt) are cheap and this is an advantage also for future modifications starting from same products.
The structure of 5 was determined by 1H and 13C NMR spectra (Supplementary Materials, Figures S3 and S4). The 1H-NMR spectrum showed a singlet at 5.38 ppm corresponding to C13-methylene group.
For the 13C-NMR signals, the disappearance of characteristic peaks of acetylenic group at 81.6 and 76 ppm, appearance of C-13 methylene signal at 72.1 ppm and C-11 vinylic signal at 127 ppm were the proposed relevant characteristics to recognize the synthesis of a triazole moiety.
Heteronuclear single quantum coherence spectroscopy (HSQC), distortionless enhancement by polarization transfer (DEPT-135 and DEPT-90), and Correlated spectroscopy (1H-1H-COSY) were also used to assign 1H and 13C signals of compound 5 as shown in Table S1 (see Supplementary Materials for 2D spectra, Figures S5–S8). The 13C-NMR spectrum of 5 exhibited 18 carbon signals, which were classified by DEPT-135 and 90 experiments, including one methylene, ten olefinic methines, and seven quaternary carbons including olefinic carbons.
HRMS of 5 was also obtained for further characterization, validating the proposed structure determined by NMR spectra (Supplementary Materials, Figure S9).
The IR spectrum of compound 5 showed characteristic C-C stretching and C=C stretching signals respectively at 3093 and 2924 cm−1 (Supplementary Materials, Figure S10). Moreover, the absence of azide stretching peak of 2 at 2123 cm−1 in IR spectrum of 5 confirmed the conversion of the intermediate.
The UV spectrum of compound 5 showed an absorption peak at 237 nm and a broad absorption peak from 260 nm to 310nm (π → π transition) (Supplementary Materials, Figure S11).
In order to investigate whether compound 5 could be a good candidate as AChE inhibitor, we firstly performed molecular modeling studies. To achieve this goal, the X-ray crystallographic structure of AChE in complex with its inhibitor donepezil (PDB ID: 4EY7) [26] was used. The effectiveness of site-specific molecular docking protocol was checked by re-docking donepezil to AChE into its binding site and by comparing the docked pose (−12.2 kcal/mol) with the co-crystallized ligand, and a root-mean-square deviation (RMSD) value of 0.328 Å was obtained, thus supporting the accuracy of the method.
Then, site-specific molecular docking of compound 5 was performed into the previously identified binding site (see Section 3) and we observed that, as donepezil did, compound 5 fitted in an appropriate way into the identified binding pocket without displaying unfavorable interactions or steric clashes. In support of this, the calculated binding energy value for compound 5 with AChE was −12.1 kcal/mol, suggesting that compound 5 could represent a promising starting point for the development of AChE binders.
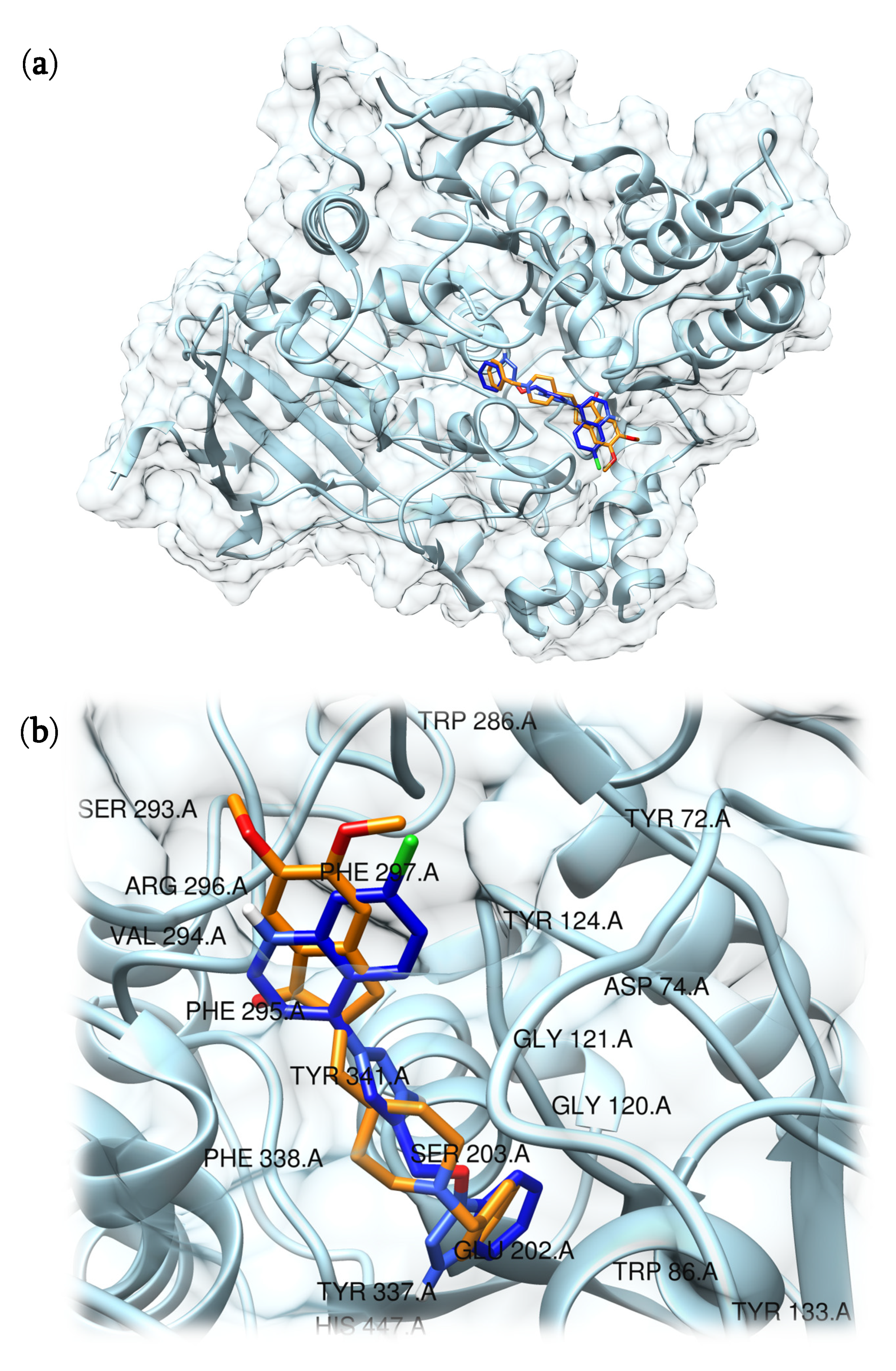
Then, to evaluate the binding mode of compound 5 into the active site we used the Protein-Compound Interaction Profiler tool (PLIP) [27]. Although PAS and CAS are fundamental to achieve interactions able to inhibit AChE, it has been reported that the presence of two tryptophan residues located near to the peripheral site (Trp286) and to the catalytic cleft (Trp86) is equally important in establishing the simultaneous bond in both sides of the active site and in contributing to the inhibitory effects [22,28]. Interestingly the benzotriazole moiety of compound 5 interacted with Trp86 residue through four π–π bonds (4.26 Å; 4.15 Å, 4.62 Å, and 3.87 Å). Furthermore, hydrogen bonds were settled through the -OH group of the benzotriazole moiety with and His447 residue (2.24 Å). Next, the triazole group, which acts as a linker between benzotriazole an chloroquinoline components, showed a π–π interaction with Tyr341 residue (4.85 Å) and two hydrogen bonds with –OH located on Tyr124 (1.96 Å and 2.51 Å). Furthermore, even the chloroquinoline moiety gave rise to a π–π interaction with Trp286 (3.94 Å) and three hydrophobic interactions with Tyr72 (3.65 Å), Phe338 (3.59 Å), and Tyr341 (3.44 Å). Together, these interactions support that compound 5 could be a good ligand candidate for AChE since both tryptophan residues (Trp86 and Trp286), relevant for binding of both CAS and PAS sites, are involved (Figure 2). The benzotriazole moiety gave rise to a hydrogen bond with His447 present in the catalytic triad, and the presence of the triazole group promotes the formation of two additional hydrogen bonds with Tyr124 that strengthens the binding between compound 5 and AChE. An overview of the detected interactions for the compounds is reported in Tables S2 and S3 and depicted in Figures S12–S14 in the Supplementary Materials.
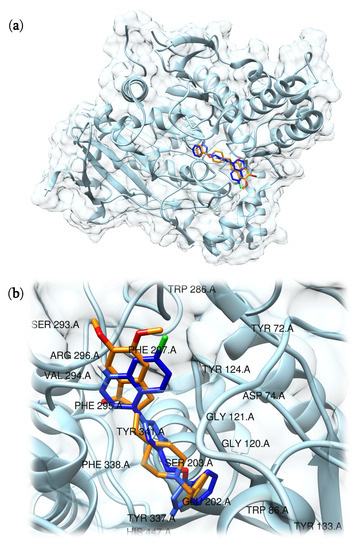
Figure 2.
Predicted interaction motif for compound 5 (blue) with AChE and co-crystallized pose of donepezil within AChE (orange, PDB ID: 4EY7) (a); magnification of compound 5 (blue) in the binding site of AChE and comparison with donepezil (orange), the residues present in a zone of 5 Å radius from the center of the ligand have been labeled (b).
Further modifications are in progress increasing carbons length of linker among triazole and quinoline and/or benzotriazole and could furnish more information relatively to binding mode with AChE.
Subsequently, absorption, distribution, metabolism, and excretion (ADME) properties of compound 5 were computed and evaluated (see Section 3). Its drug-likeness was determined according to Lipinski’s rule of five which is a set of criteria needed to evaluate drug-likeness that would make it a likely orally active drug in humans. Usually, a compound with one or no violations is considered a good candidate [29]. The criteria on which this rule is based are: (1) molecular weight ≤ 500 g/mol, (2) an octanol-water partition coefficient logP ≤ 5, (3) hydrogen bond acceptors ≤ 10, and (4) hydrogen bond donors ≤ 5. According to the prediction, performed with SwissADME [30], compound 5 does not violate any criteria because the following descriptors were calculated: (1) molecular weight of 377.79 g/mol, (2) an octanol-water partition coefficient logP = 2.97, (3) hydrogen bond acceptors are 6, and (4) hydrogen bond donors are 0. Then Veber’s et al. rule was also evaluated [31], since it introduces as criteria the topological polar surface area (TPSA), that has to be ≤140 Å2, and the number of rotatable bonds, that have to be ≤10. These criteria are important for drug adsorption, and in the case of compound 5 no violations were found, since a TPSA of 83.54 Å2 and four rotatable bonds were calculated (reported in Table S4).
Regarding the prediction of pharmacokinetic properties, according to such physico-chemical descriptors, we observed that as for donepezil the absorption could highly occur at the level of the gastrointestinal tract (GI tract) while, in contrast to donepezil, the passage of compound 5 through blood–brain barrier (BBB) is predicted by a BOILED-Egg model to be less feasible (Figure S15) [32]. This is one of the aspects that must be taken into account and improved in future compound developments since the target of 5 is located within the CNS.
3. Materials and Methods
3.1. Chemistry
Silica gel (FCP 230-400 mesh) was used for column chromatography. Thin-layer chromatography was carried out on E. Merck precoated silica gel 60 F254 plates and visualized with phosphomolybdic acid, iodine, or a UV-visible lamp.
All chemicals were purchased from Bide Pharmatech., Ltd. (Shanghai, China) and J & K scientific (Hong Kong, China). 1H-NMR and 13C-NMR spectra were collected in CDCl3 at 25 °C on a Bruker Ascend®-600 NMR spectrometer (600 MHz for 1H and 150 MHz for 13C). All chemical shifts were reported in the standard δ notation of parts per million using the peak of residual proton signals of CDCl3 or DMSO-d6 as an internal reference (CDCl3, δC 77.2 ppm, δH 7.26 ppm; DMSO-d6, δC 39.5 ppm, δH 2.50 ppm). High resolution mass spectra (HRMS) were measured using electrospray ionization (ESI). The measurements were conducted in a positive ion mode (interface capillary voltage 4500 V); the mass ratio was from m/z 50 to 3000 Da; external/internal calibration was done with Electrospray Calibration Solution.
HRMS analyses were performed by an Agilent 6230 electrospray ionization (ESI) time-of-flight (TOF) mass spectrometer with Agilent C18 column (4.6 mm × 150 mm, 3.5 μm). The mobile phase was isocratic (water + 0.01% TFA; CH3CN) at a flow rate of 0.35 mL/min. The peaks were determined at 254 nm under UV.
UV analysis was performed by a Shimadzu UV—2600 with 1 cm quartz cell and a slit width of 2.0 nm. The analysis was carried out using wavelength in the range of 200–400 nm.
IR analysis (KBr) was performed by a Shimadzu IRAffinity-1S with frequency range of 4000–500 cm−1.
3.1.1. Synthesis of 4-Azido-7-chloro-quinoline (2)
4,7-Dichloroquinoline (2.0 g, 10 mmol) was dissolved in 5 mL anhydrous DMF. NaN3 (1.3 g, 20 mmol) was then added in one portion, and the resulting mixture was stirred at 65 °C for 6 h, whereupon TLC indicated the completion of reaction. The reaction mixture was then allowed to cool to room temperature, after which it was diluted with 100 mL CH2Cl2, washed with water (3 × 30 mL), dried over anhydrous Na2SO4, and evaporated to dryness. The resulting residue was recrystallized from a 1:1 mixture of CH2Cl2/hexane to yield the final pure product 2 as a colorless, needle-like crystal in 78% yield.
δH (600 MHz, CDCl3) 8.82 (1H, d, J = 4.9 Hz, H-2), 8.09 (1H, d, J = 2.4 Hz, H-8), 8.01 (1H, d, J 9.3, H-5), 7.49 (1H, dd, J 2.4 and 9.3, H-6) 7.12 (1H, d, J 4.9, H-3) ppm; δC (150 MHz, CDCl3) 150.9, 149.1, 146.8, 136.9, 127.9, 123.8, 119.9, 108.7 ppm. The spectral characteristics are consistent with those of 2 in the literature [15].
3.1.2. Synthesis of 1-(Prop-2-yn-1-yloxy)-1H-benzo[d][1,2,3]triazole (4)
1H-benzo[d][1,2,3]triazol-1-ol (13 mmol) was dissolved in 10 mL of anhydrous DMF. Anhydrous K2CO3 (2.7 g, 19.5 mmol) was then added to the solution, and the mixture was stirred at 30 °C for 30 min. Propargyl bromide (3-bromopropyne, 2.2 mL, 19.5 mmol) was then added slowly to the reaction mixture, and subsequently stirred at 30 °C for 6 h upon which TLC indicated completion of the reaction. The reaction mixture was then diluted with 50 mL water and extracted with ethyl acetate (3 × 50 mL). These extracts were then combined, washed with water (2 × 50 mL), dried over anhydrous Na2SO4 and evaporated under vacuum to yield the product residue that was then recrystallized from CH2Cl2/hexane 1:1 mixture to yield the compound 4 in 86% yield. δH (600 MHz, DMSO-d6) 3.82 (H, s), 5.38 (2H, s, CH2), 7.49 (H, t, J = 8.04 Hz, HAr), 7.64 (1H, t, J = 7.3 Hz, HAr), 7.85 (1H, d, J = 8.3 Hz, HAr), 8.09 (1H, d, J = 8.4 Hz, HAr) ppm; δC (150MHz, CDCl3) 67.3, 76.0, 81.6,109, 110.9, 124.4, 127.0, 127.9, 141.9 ppm.
3.1.3. Synthesis of 4-(4-(((1H-Benzo[d][1,2,3]triazol-1-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)-7-chloroquinolin (5)
The O-acetylenic derivative 4 (1 mmol) and azide 2 were dissolved in 5 mL tBuOH/water (1:1) and, while stirring at 65 °C, 1 M sodium ascorbate (0.4 mL, 0.4 mmol) and 1 M CuSO4 (0.2 mL, 20 mol%) were added sequentially, in that order. The reaction mixture was then stirred at 65 °C for 24 h. The crude product was then precipitated out by slowly adding cold water to the reaction mixture, after which it was filtered, washed with water, air dried, and purified by silica column chromatography (eluents: from CH2Cl2 to 5% MeOH). Yield 73%, δH (600 MHz, CDCl3) 5.87 (2H, s, CH2-O), 7.29 (1H, d, J = 4.6 Hz, H-3), 7.35 (1H, d, J = 9 Hz, H-18), 7.38–7.41 (1H, m, H-15), 7.43–7.48 (2H, m, H-16, H-17), 7.51–7.53 (1H, dd, J = 8.7 and 2 Hz, H-6), 7.81 (1H, s, H-11), 8.02 (1H, d, J = 8.4 Hz, H-5), 8.24 (1H, d, J = 2 Hz, H-8), 9.03 (1H, d, J = 4.6 Hz, H-2) ppm; δC (150MHz, CDCl3) 72.1 (C-13), 108.7, 116.3, 120.2, 120.4, 123.7, 124.8, 126.8, 128.0, 128.4, 128.9, 129.7, 137.2, 140.4, 141.0, 143.2 (C-19), 150 (C-9), 151.2 (C-2), ppm; HRMS-ESI m/z 378.0871 [M + H]+ (calcd. for C18H12ClN7O1, m/z 378.0865); UV (CH2Cl2) peaks 237, 288, and 326 nm.
3.2. Computational Studies
The 3D X-ray crystal structure of recombinant human acetylcholinesterase (AChE) complexed with donepezil, was retrieved from the RCSB Protein Data Bank (www.rcsb.org; accessed on 24 May 2022; PDB ID 4EY7, resolution 2.35 Å). The PDB file was selected in agreement with a previous work [33].
Prior to site-specific docking studies, AchE was prepared, and chain was isolated and considered. Further, the obtained structure was processed through the DockPrep tool of Chimera to fix potentially missing residues in the structure [34].
In order to identify the binding site region, PLIP tool was used to highlight residues involved in the binding between donepezil and AchE. PLIP tool was also used to evaluate interaction patterns data [27].
To perform a multiple site-specific molecular docking study, PyRX software based on AutoDock Vina was used [35,36]. From this point, a grid-enclosing box was centered on the identified site and a receptor search volume was created with the following coordinates and dimensions: x = −13.2324, y = −43.1299, z = 30.3805; size: 28.9740 × 24.7203 × 27.4155 Å. Number of generated docking poses was set to 8 and docking energy conformation value was expressed in −kcal/mol. The best scoring pose was selected for further analyses. Residue numbering used in the PDB file was adopted. Compound 5 was docked into the previously defined binding site of AchE. The best pose obtained was selected for further studies. To verify the site-specific molecular docking protocol, co-crystallized donepezil compound was re-docked and root-mean square deviation (RMSD) value was calculated using the Chimera MatchMarker tool [34].
UCSF Chimera molecular viewer [34] and PyMol software (The PyMOL Molecular Graphics System, Version 2.3.5, Schrödinger, LLC.) were used to produce the artworks.
ADME properties for compound 5 were retrieved using the SwissADME tool [30].
4. Conclusions
The synthesis and complete characterization of a triazole-based quinoline was presented in this work. To date, the search for compounds capable of treating AD is one of the main modern challenges in the field of drug discovery. From here, among the different approaches used, we attempted the one that aims to inhibit the enzyme through interactions occurring in multiple regions (PAS and CAS) of the same binding site, which is the strategy pursued by donepezil. Along the lines of these properties, we have synthesized compound 5, which has been shown to be a good candidate for the inhibitory action of AchE according to computational site-specific molecular docking experiments. Although the predicted physico-chemical properties suggest that the compound is drug-like, in line with donepezil, the crossing of compound 5 through the blood-brain barrier (BBB) appears to be less feasible according to abovementioned calculations. Thus, compound 5 represents a promising starting point for further optimization.
Supplementary Materials
The following are available online, Figure S1: 1-H NMR compound 4, Figure S2: 13-C NMR compound 4, Figure S3: 1-H NMR compound 5, Figure S4: 13-C NMR compound 5, Figure S5: HSQC compound 5, Figures S6 and S7: DEPT compound 5, Figure S8a,b: 1H-1H COSY compound 5, Figure S9: HRMS of 5, Figure S10: IR spectrum, Figure S11: UV spectrum, Figures S12–S14: detailed views of the predicted interaction of ligands with AchE, Figure S15: BOILED-Egg graph, Table S1 1H and 13C-nuclear magnetic spectroscopy (NMR) chemical shifts, Tables S2 and S3: overview of the residues interaction with ligands according to molecular modeling studies, Table S4: physicochemical properties of compound 5 calculated by SwissADME.
Author Contributions
Conceptualization, P.C.; methodology, G.R. and P.C.; investigation, L.K.F., M.A., J.A. and Y.X.; data curation, J.A.; writing—original draft preparation, L.K.F., M.A. and J.A.; writing—review and editing, A.G., G.R. and P.C.; supervision, G.R. and P.C.; project administration, G.R. and P.C.; funding acquisition, G.R. and P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FDCT grants from Macao Science and Technology Development Fund to PC (Project Code: 0096/2020/A) and by University of Brescia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Palmer, A.M. Neuroprotective Therapeutics for Alzheimer’s Disease: Progress and Prospects. Trends Pharmacol. Sci. 2011, 32, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; LeVine, H. Alzheimer’s Disease and the Amyloid-β Peptide. JAD 2010, 19, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colovic, M.B.; Krstic, D.Z.; Lazarevic-Pasti, T.D.; Bondzic, A.M.; Vasic, V.M. Acetylcholinesterase Inhibitors: Pharmacology and Toxicology. CN 2013, 11, 315–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, C.; Yadav, D.K.; Subedi, L.; Venkatesan, R.; Venkanna, A.; Afzal, S.; Lee, E.; Yoo, J.; Ji, E.; Kim, S.Y.; et al. Identification of Novel Acetylcholinesterase Inhibitors Designed by Pharmacophore-Based Virtual Screening, Molecular Docking and Bioassay. Sci. Rep. 2018, 8, 14921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarian, M.; Gonzalez, M.; Salvador, S.M.; Lorzadeh, S.; Hudson, P.K.; Pecic, S. Synthesis, Kinetic Evaluation and Molecular Docking Studies of Donepezil-Based Acetylcholinesterase Inhibitors. J. Mol. Struct. 2022, 1247, 131425. [Google Scholar] [CrossRef]
- Silva, M.A.; Kiametis, A.S.; Treptow, W. Donepezil Inhibits Acetylcholinesterase via Multiple Binding Modes at Room Temperature. J. Chem. Inf. Model. 2020, 60, 3463–3471. [Google Scholar] [CrossRef]
- Shega, J.W.; Ellner, L.; Lau, D.T.; Maxwell, T.L. Cholinesterase Inhibitor and N-Methyl-D-Aspartic Acid Receptor Antagonist Use in Older Adults with End-Stage Dementia: A Survey of Hospice Medical Directors. J. Palliat. Med. 2009, 12, 779–783. [Google Scholar] [CrossRef]
- Zanforlin, E.; Zagotto, G.; Ribaudo, G. An Overview of New Possible Treatments of Alzheimer’s Disease, Based on Natural Products and Semi-Synthetic Compounds. CMC 2017, 24, 3749–3773. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Therapeutic Potential of Phosphodiesterase Inhibitors against Neurodegeneration: The Perspective of the Medicinal Chemist. ACS Chem. Neurosci. 2020, 11, 1726–1739. [Google Scholar] [CrossRef]
- Ribaudo, G.; Memo, M.; Gianoncelli, A. A Perspective on Natural and Nature-Inspired Small Molecules Targeting Phosphodiesterase 9 (PDE9): Chances and Challenges against Neurodegeneration. Pharmaceuticals 2021, 14, 58. [Google Scholar] [CrossRef]
- Cheung, J.; Rudolph, M.J.; Burshteyn, F.; Cassidy, M.S.; Gary, E.N.; Love, J.; Franklin, M.C.; Height, J.J. Structures of Human Acetylcholinesterase in Complex with Pharmacologically Important Ligands. J. Med. Chem. 2012, 55, 10282–10286. [Google Scholar] [CrossRef] [PubMed]
- Dvir, H.; Silman, I.; Harel, M.; Rosenberry, T.L.; Sussman, J.L. Acetylcholinesterase: From 3D Structure to Function. Chem.-Biol. Interact. 2010, 187, 10–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angajala, K.K.; Vianala, S.; Macha, R.; Raghavender, M.; Thupurani, M.K.; Pathi, P.J. Synthesis, Anti-Inflammatory, Bactericidal Activities and Docking Studies of Novel 1,2,3-Triazoles Derived from Ibuprofen Using Click Chemistry. SpringerPlus 2016, 5, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Zhao, S.-J.; Liu, Y. 1,2,3-Triazole-Containing Hybrids as Potential Anticancer Agents: Current Developments, Action Mechanisms and Structure-Activity Relationships. Eur. J. Med. Chem. 2019, 183, 111700. [Google Scholar] [CrossRef]
- El Malah, T.; Nour, H.F.; Satti, A.A.E.; Hemdan, B.A.; El-Sayed, W.A. Design, Synthesis, and Antimicrobial Activities of 1,2,3-Triazole Glycoside Clickamers. Molecules 2020, 25, 790. [Google Scholar] [CrossRef] [Green Version]
- Ghiano, D.G.; de la Iglesia, A.; Liu, N.; Tonge, P.J.; Morbidoni, H.R.; Labadie, G.R. Antitubercular Activity of 1,2,3-Triazolyl Fatty Acid Derivatives. Eur. J. Med. Chem. 2017, 125, 842–852. [Google Scholar] [CrossRef] [Green Version]
- Ribaudo, G.; Coghi, P.; Zanforlin, E.; Law, B.Y.K.; Wu, Y.Y.J.; Han, Y.; Qiu, A.C.; Qu, Y.Q.; Zagotto, G.; Wong, V.K.W. Semi-Synthetic Isoflavones as BACE-1 Inhibitors against Alzheimer’s Disease. Bioorg. Chem. 2019, 87, 474–483. [Google Scholar] [CrossRef]
- Jalili-Baleh, L.; Nadri, H.; Forootanfar, H.; Samzadeh-Kermani, A.; Küçükkılınç, T.T.; Ayazgok, B.; Rahimifard, M.; Baeeri, M.; Doostmohammadi, M.; Firoozpour, L.; et al. Novel 3-Phenylcoumarin–Lipoic Acid Conjugates as Multi-Functional Agents for Potential Treatment of Alzheimer’s Disease. Bioorg. Chem. 2018, 79, 223–234. [Google Scholar] [CrossRef]
- Rastegari, A.; Nadri, H.; Mahdavi, M.; Moradi, A.; Mirfazli, S.S.; Edraki, N.; Moghadam, F.H.; Larijani, B.; Akbarzadeh, T.; Saeedi, M. Design, Synthesis and Anti-Alzheimer’s Activity of Novel 1,2,3-Triazole-Chromenone Carboxamide Derivatives. Bioorg. Chem. 2019, 83, 391–401. [Google Scholar] [CrossRef]
- Kaur, A.; Mann, S.; Kaur, A.; Priyadarshi, N.; Goyal, B.; Singhal, N.K.; Goyal, D. Multi-Target-Directed Triazole Derivatives as Promising Agents for the Treatment of Alzheimer’s Disease. Bioorg. Chem. 2019, 87, 572–584. [Google Scholar] [CrossRef]
- Najafi, Z.; Mahdavi, M.; Saeedi, M.; Karimpour-Razkenari, E.; Asatouri, R.; Vafadarnejad, F.; Moghadam, F.H.; Khanavi, M.; Sharifzadeh, M.; Akbarzadeh, T. Novel Tacrine-1,2,3-Triazole Hybrids: In Vitro, in Vivo Biological Evaluation and Docking Study of Cholinesterase Inhibitors. Eur. J. Med. Chem. 2017, 125, 1200–1212. [Google Scholar] [CrossRef] [PubMed]
- Mantoani, S.; Chierrito, T.; Vilela, A.; Cardoso, C.; Martínez, A.; Carvalho, I. Novel Triazole-Quinoline Derivatives as Selective Dual Binding Site Acetylcholinesterase Inhibitors. Molecules 2016, 21, 193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Gao, Y.; Kang, D.; Huang, B.; Huo, Z.; Liu, H.; Poongavanam, V.; Zhan, P.; Liu, X. Design, Synthesis and Biological Evaluation of Tacrine-1,2,3-Triazole Derivatives as Potent Cholinesterase Inhibitors. Med. Chem. Commun. 2018, 9, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coghi, P.; Ng, J.P.L.; Nasim, A.A.; Wong, V.K.W. N-[7-Chloro-4-[4-(Phenoxymethyl)-1H-1,2,3-Triazol-1-Yl]Quinoline]-Acetamide. Molbank 2021, 2021, M1213. [Google Scholar] [CrossRef]
- Feldman, A.K.; Colasson, B.; Fokin, V.V. One-Pot Synthesis of 1,4-Disubstituted 1,2,3-Triazoles from In Situ Generated Azides. Org. Lett. 2004, 6, 3897–3899. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, S.; Arora, S.; Attri, S.; Kaur, P.; Kaur Gulati, H.; Bhagat, K.; Kumar, N.; Singh, H.; Vir Singh, J.; et al. New Coumarin-Benzotriazole Based Hybrid Molecules as Inhibitors of Acetylcholinesterase and Amyloid Aggregation. Bioorg. Med. Chem. Lett. 2020, 30, 127477. [Google Scholar] [CrossRef]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Res. 2021, 49, W530–W534. [Google Scholar] [CrossRef]
- Pourshojaei, Y.; Abiri, A.; Eskandari, K.; Haghighijoo, Z.; Edraki, N.; Asadipour, A. Phenoxyethyl Piperidine/Morpholine Derivatives as PAS and CAS Inhibitors of Cholinesterases: Insights for Future Drug Design. Sci. Rep. 2019, 9, 19855. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelli, R.; Giacomini, A.; Anselmi, M.; Bozza, N.; Vacondio, F.; Rivara, S.; Matarazzo, S.; Presta, M.; Mor, M.; Ronca, R. Synthesis, Structural Elucidation, and Biological Evaluation of NSC12, an Orally Available Fibroblast Growth Factor (FGF) Ligand Trap for the Treatment of FGF-Dependent Lung Tumors. J. Med. Chem. 2016, 59, 4651–4663. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology; Methods in Molecular Biology; Hempel, J.E., Williams, C.H., Hong, C.C., Eds.; Springer: New York, NY, USA, 2015; Volume 1263, pp. 243–250. ISBN 978-1-4939-2268-0. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).