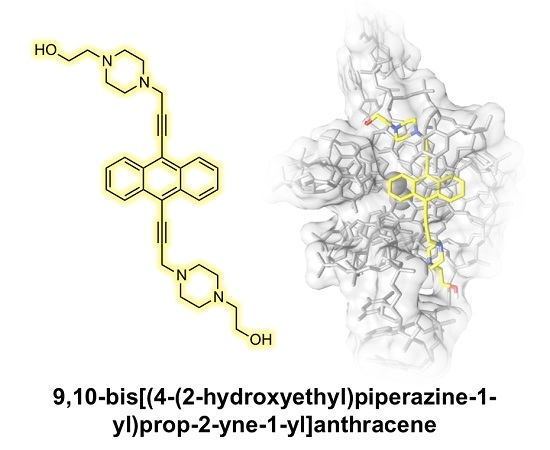
9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene: Synthesis and G-quadruplex Selectivity
Abstract
1. Introduction
2. Results and Discussion
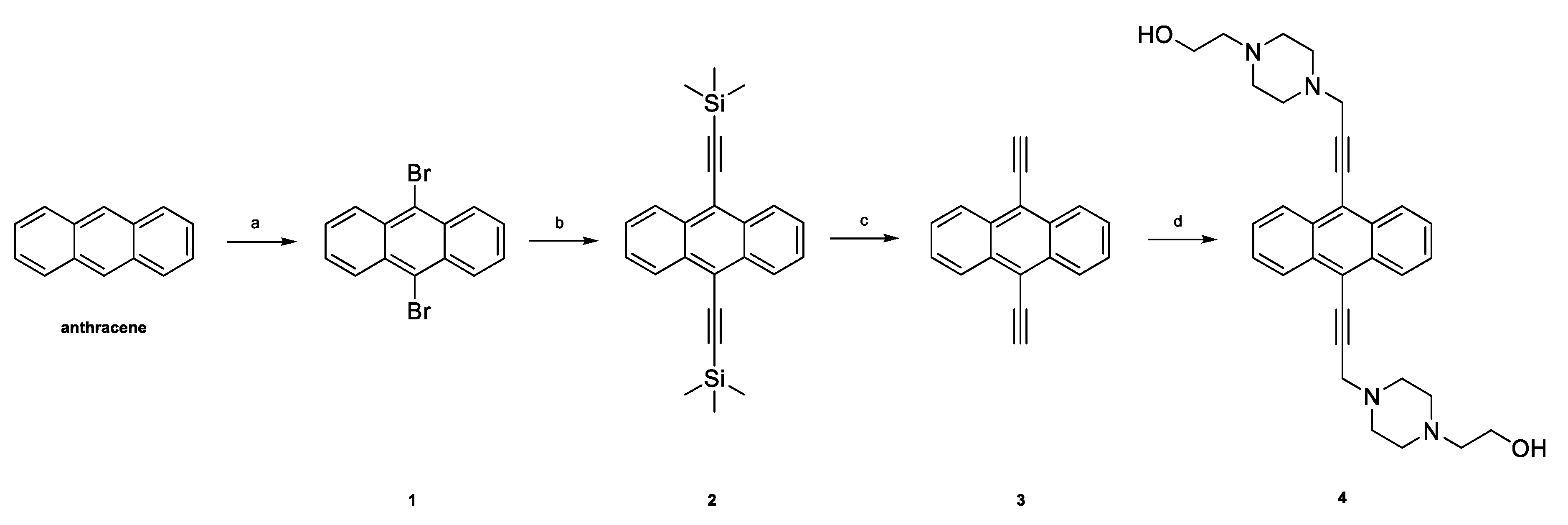
2.1. Chemistry
2.2. DNA Binding Studies
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. Synthesis of 9,10-Dibromoanthracene (1)
3.1.3. Synthesis of 9,10-Bis((trimethylsilyl)ethynyl)anthracene (2)
3.1.4. Synthesis of 9,10-Diethynylanthracene (3)
3.1.5. Synthesis of 9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene (4)
3.2. ESI-MS Binding Studies
3.3. Molecular Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Altieri, A.; Alvino, A.; Ohnmacht, S.; Ortaggi, G.; Neidle, S.; Nocioni, D.; Franceschin, M.; Bianco, A. Xanthene and Xanthone Derivatives as G-Quadruplex Stabilizing Ligands. Molecules 2013, 18, 13446–13470. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Evidence on selective binding to G-quadruplex DNA of isoflavones from Maclura pomifera by mass spectrometry and molecular docking. Nat. Prod. Res. 2019, 1–5. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Photoactivated semi-synthetic derivative of osajin selectively interacts with G-quadruplex DNA. Nat. Prod. Res. 2020, 1–6. [Google Scholar] [CrossRef]
- Asamitsu, S.; Bando, T.; Sugiyama, H. Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures. Chem. A Eur. J. 2019, 25, 417–430. [Google Scholar] [CrossRef]
- Rigo, R.; Palumbo, M.; Sissi, C. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1399–1413. [Google Scholar] [CrossRef]
- Chang, Y.-S.; Ko, B.-H.; Ju, J.-C.; Chang, H.-H.; Huang, S.-H.; Lin, C.-W. SARS Unique Domain (SUD) of Severe Acute Respiratory Syndrome Coronavirus Induces NLRP3 Inflammasome-Dependent CXCL10-Mediated Pulmonary Inflammation. Int. J. Mol. Sci. 2020, 21, 3179. [Google Scholar] [CrossRef]
- Shen, L.-W.; Qian, M.-Q.; Yu, K.; Narva, S.; Yu, F.; Wu, Y.-L.; Zhang, W. Inhibition of Influenza A virus propagation by benzoselenoxanthenes stabilizing TMPRSS2 Gene G-quadruplex and hence down-regulating TMPRSS2 expression. Sci. Rep. 2020, 10, 7635. [Google Scholar] [CrossRef]
- Di Fonzo, S.; Amato, J.; D’Aria, F.; Caterino, M.; D’Amico, F.; Gessini, A.; Brady, J.W.; Cesàro, A.; Pagano, B.; Giancola, C. Ligand binding to G-quadruplex DNA: New insights from ultraviolet resonance Raman spectroscopy. Phys. Chem. Chem. Phys. 2020, 22, 8128–8140. [Google Scholar] [CrossRef]
- Sengupta, A.; Ganguly, A.; Chowdhury, S. Promise of G-Quadruplex Structure Binding Ligands as Epigenetic Modifiers with Anti-Cancer Effects. Molecules 2019, 24, 582. [Google Scholar] [CrossRef]
- Pirota, V.; Nadai, M.; Doria, F.; Richter, S. Naphthalene Diimides as Multimodal G-Quadruplex-Selective Ligands. Molecules 2019, 24, 426. [Google Scholar] [CrossRef]
- Bortolus, M.; Ribaudo, G.; Toffoletti, A.; Carbonera, D.; Zagotto, G. Photo-induced spin switching in a modified anthraquinone modulated by DNA binding. Photochem. Photobiol. Sci. 2019, 18, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.; Ribaudo, G.; Braud, E.; Ethève-Quelquejeu, M.; De Franco, M.; Garbay, C.; Demange, L.; Gresh, N.; Zagotto, G. Design and synthesis of a peptide derivative of ametantrone targeting the major groove of the d(GGCGCC) 2 palindromic sequence. New J. Chem. 2020, 44, 3624–3631. [Google Scholar] [CrossRef]
- Ribaudo, G.; Scalabrin, M.; Pavan, V.; Fabris, D.; Zagotto, G. Constrained bisantrene derivatives as G-quadruplex binders. ARKIVOC 2016, 2016, 145. [Google Scholar]
- Chilka, P.; Desai, N.; Datta, B. Small Molecule Fluorescent Probes for G- Quadruplex Visualization as Potential Cancer Theranostic Agents. Molecules 2019, 24, 752. [Google Scholar] [CrossRef]
- Ongaro, A.; Ribaudo, G.; Zagotto, G.; Memo, M.; Gianoncelli, A. Synthesis via A3 Coupling Reaction of Anthracene-Propargylamine as a New Scaffold for the Interaction with DNA. ChemistrySelect 2019, 4, 13138–13142. [Google Scholar] [CrossRef]
- Zhu, J.; Zhong, K.; Liang, Y.; Wang, Z.; Chen, T.; Jin, L.Y. Synthesis and self-assembly of oligomers containing cruciform 9,10-bis(arylethynyl)anthracene unit: formation of supramolecular nanostructures based on rod-length-dependent organization. Tetrahedron 2014, 70, 1230–1235. [Google Scholar] [CrossRef]
- Zhang, C.-H.; Yu, P.-P.; Tan, W.-Y.; Luo, D.; Wang, L.-P.; Xia, Y.; Liu, C.-C.; Cao, Y. An easily and environmentally friendly accessible small-molecule acetylenic donor for organic solar cells. Dye. Pigment. 2019, 160, 983–988. [Google Scholar] [CrossRef]
- Ferreira, R.; Marchand, A.; Gabelica, V. Mass spectrometry and ion mobility spectrometry of G-quadruplexes. A study of solvent effects on dimer formation and structural transitions in the telomeric DNA sequence d(TAGGGTTAGGGT). Methods 2012, 57, 56–63. [Google Scholar] [CrossRef]
- Mazzitelli, C.L.; Brodbelt, J.S.; Kern, J.T.; Rodriguez, M.; Kerwin, S.M. Evaluation of binding of perylene diimide and benzannulated perylene diimide ligands to dna by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2006, 17, 593–604. [Google Scholar] [CrossRef]
- Xu, N.; Yang, H.; Cui, M.; Song, F.; Liu, Z.; Liu, S. Evaluation of alkaloids binding to the parallel quadruplex structure [d(TGGGGT)]4 by electrospray ionization mass spectrometry. J. Mass Spectrom. 2012, 47, 694–700. [Google Scholar] [CrossRef]
- Rosu, F. Determination of affinity, stoichiometry and sequence selectivity of minor groove binder complexes with double-stranded oligodeoxynucleotides by electrospray ionization mass spectrometry. Nucleic Acids Res. 2002, 30, e82. [Google Scholar] [CrossRef]
- Tjernberg, A.; Carnö, S.; Oliv, F.; Benkestock, K.; Edlund, P.-O.; Griffiths, W.J.; Hallén, D. Determination of Dissociation Constants for Protein−Ligand Complexes by Electrospray Ionization Mass Spectrometry. Anal. Chem. 2004, 76, 4325–4331. [Google Scholar] [CrossRef]
- Erba, E.B.; Zenobi, R. Mass spectrometric studies of dissociation constants of noncovalent complexes. Annu. Reports Sect. “C” (Physical Chem.) 2011, 107, 199–228. [Google Scholar] [CrossRef]
- Ishii, K.; Noda, M.; Uchiyama, S. Mass spectrometric analysis of protein–ligand interactions. Biophys. Physicobiology 2016, 13, 87–95. [Google Scholar] [CrossRef]
- Tawani, A.; Kumar, A. Structural Insight into the interaction of Flavonoids with Human Telomeric Sequence. Sci. Rep. 2015, 5, 17574. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zorzan, M.; Pezzani, R.; Redaelli, M.; Zagotto, G.; Memo, M.; Gianoncelli, A. Investigation of the molecular reactivity of bioactive oxiranylmethyloxy anthraquinones. Arch. Pharm. (Weinheim) 2019, 352, 1900030. [Google Scholar] [CrossRef]
- Torvinen, M.; Kalenius, E.; Sansone, F.; Casnati, A.; Jänis, J. Noncovalent Complexation of Monoamine Neurotransmitters and Related Ammonium Ions by Tetramethoxy Tetraglucosylcalix [4]arene. J. Am. Soc. Mass Spectrom. 2012, 23, 359–365. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

| Compound | Binding Affinity G-quadruplex | Binding Affinity dsDNA | Selectivity Ratio (G-quadruplex/dsDNA) | ECOM50% 2:1 ligand/G-quadruplex | ECOM50% 1:1 ligand/G-quadruplex | ECOM50% 1:1 ligand/dsDNA |
|---|---|---|---|---|---|---|
| 4 | 80.3 | 41.8 | 1.92 | 43.58 | 34.39 | 33.42 |
| Ant4b | 68.4 | 79.8 | 0.86 | 40.07 | 34.57 | 39.43 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribaudo, G.; Ongaro, A.; Oselladore, E.; Zagotto, G.; Memo, M.; Gianoncelli, A. 9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene: Synthesis and G-quadruplex Selectivity. Molbank 2020, 2020, M1138. https://doi.org/10.3390/M1138
Ribaudo G, Ongaro A, Oselladore E, Zagotto G, Memo M, Gianoncelli A. 9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene: Synthesis and G-quadruplex Selectivity. Molbank. 2020; 2020(2):M1138. https://doi.org/10.3390/M1138
Chicago/Turabian StyleRibaudo, Giovanni, Alberto Ongaro, Erika Oselladore, Giuseppe Zagotto, Maurizio Memo, and Alessandra Gianoncelli. 2020. "9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene: Synthesis and G-quadruplex Selectivity" Molbank 2020, no. 2: M1138. https://doi.org/10.3390/M1138
APA StyleRibaudo, G., Ongaro, A., Oselladore, E., Zagotto, G., Memo, M., & Gianoncelli, A. (2020). 9,10-Bis[(4-(2-hydroxyethyl)piperazine-1-yl)prop-2-yne-1-yl]anthracene: Synthesis and G-quadruplex Selectivity. Molbank, 2020(2), M1138. https://doi.org/10.3390/M1138