QSAR Study of Antimicrobial 3-Hydroxypyridine-4-one and 3-Hydroxypyran-4-one Derivatives Using Different Chemometric Tools
Abstract
:1. Introduction
2. Experimental Section
2.1. Software
2.2. Data set and descriptor generation
2.3. Data screening and model building
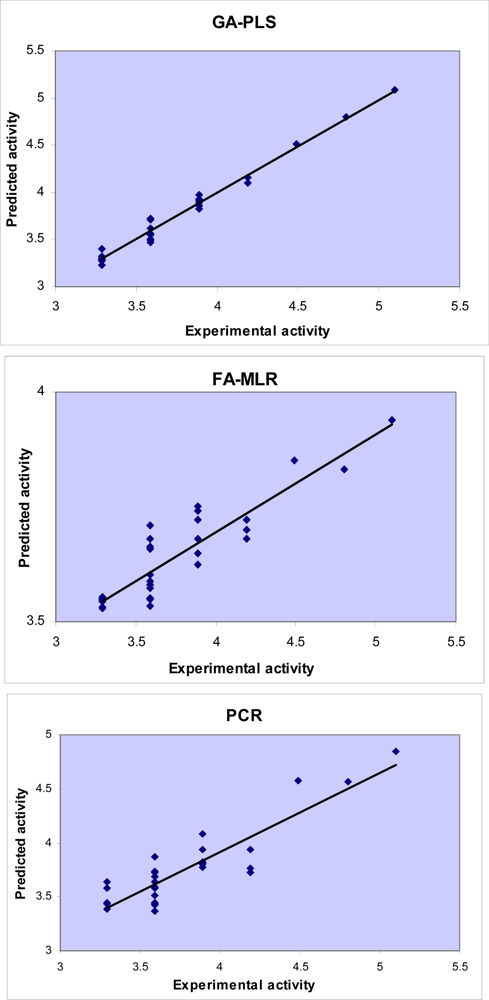
3. Results and Discussion
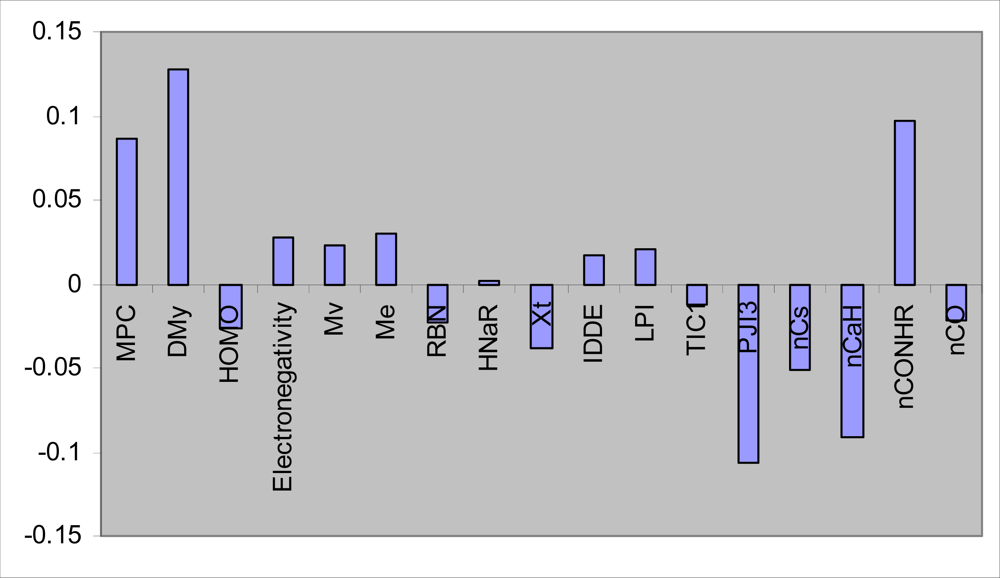
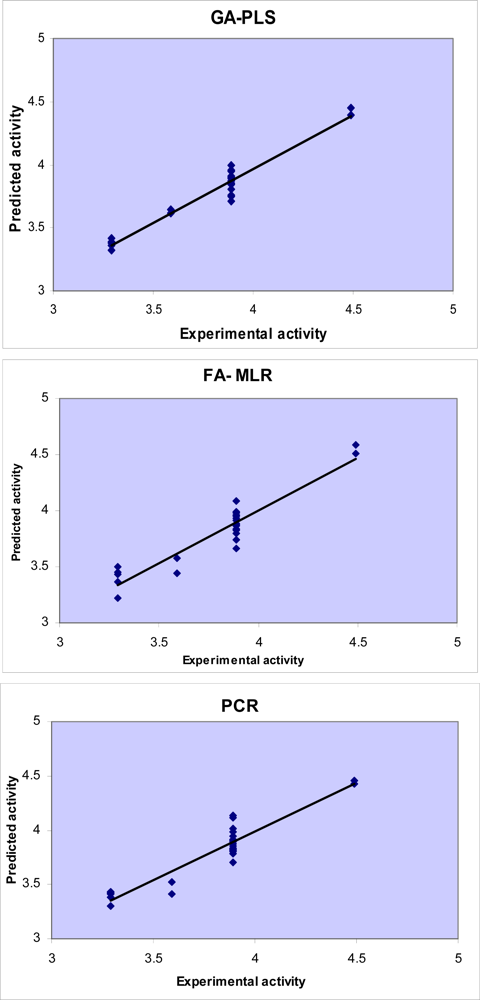
3.1. GA-PLS
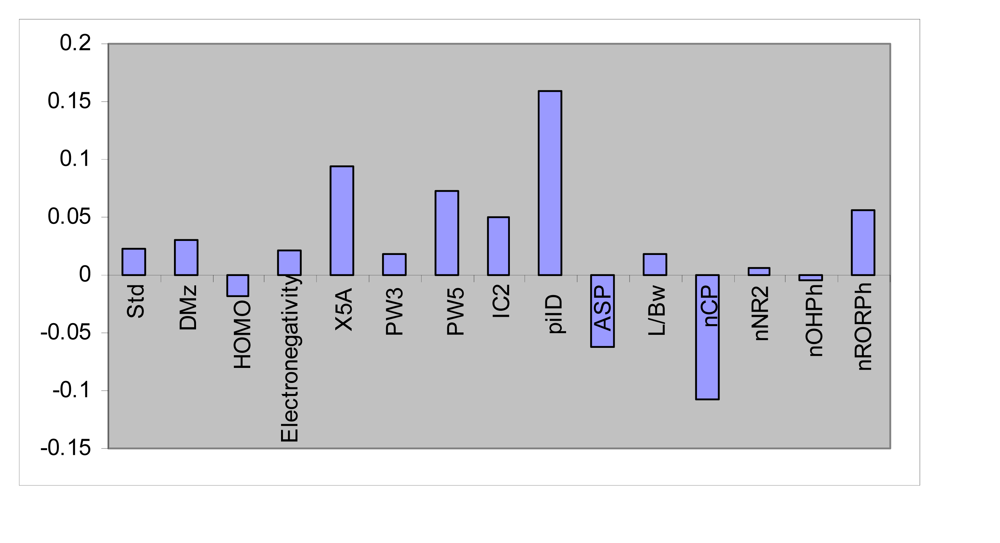
3.2. FA-MLR and PCRA
4. Conclusions
Acknowledgments
References and notes
- Schmidi, H. Multivariate Prediction for QSAR. Chemom. Intell. Lab. Sys 1997, 37, 125–134. [Google Scholar]
- Hansch, C; Kurup, A; Garg, R; Gao, H. Chem-bioinformatics and QSAR. A Review of QSAR Lacking Positive Hydrophobic Terms. Chem. Rev 2001, 101, 619–672. [Google Scholar]
- Wold, S; Trygg, J; Berglund, A; Antii, H. Some Recent Developments in the PLS Modeling. Chemom. Intell. Lab. Syst 2001, 58, 131–150. [Google Scholar]
- Hemmateenejad, B; Miri, R; Akhond, M; Shamsipur, M. QSAR Study of the Calcium Channel Antagonist Activity of some Recently Synthesized Dihydropyridine Derivatives. An Application of Genetic Algorithm for Variable Selection in MLR and PLS Methods. Chemom. Intell. Lab. Syst 2002, 64, 91–99. [Google Scholar]
- Hemmateenejad, B; Miri, R; Akhond, M; Shamsipur, M. Quantitative Structure Activity Relationship Study of Recently Synthesized 1,4- Dihydropyridine Calcium Channel Antagonists. Application of Hansch Analysis Methods. Arch. Pharm. Pharm. Med. Chem 2002, 10, 472–480. [Google Scholar]
- Horvath, D; Mao, B. Neighborhood Behavior. Fuzzy Molecular Descriptors and Their Influence on the Relationship between Structural Similarity and Property Similarity. QSAR. Comb. Sci 2003, 22, 498–509. [Google Scholar]
- Putta, S; Eksterowicz, J; Lemmen, C; Stanton, R. A Novel Subshape Molecular Descriptor. J. Chem. Inf. Comput. Sci 2003, 43, 1623–1635. [Google Scholar]
- Gupta, S; Singh, M; Madan, AK. Superpendentic Index: A Novel Topological Descriptor for Predicting Biological Activity. J. Chem. Inf. Comput. Sci 1999, 39, 272–277. [Google Scholar]
- Consonni, V; Todeschini, R; Pavan, M. Structure/Response Correlations and Similarity/Diversity Analysis by GETAWAY Descriptors. 2. Application of the Novel 3D Molecular Descriptors to QSAR/QSPR Studies. J. Chem. Inf. Comput. Sci 2002, 42, 693–705. [Google Scholar]
- Deeb, O; Hemmateenejad, B; Jaber, A; Garduno-Juarez, R; Miri, R. Effects of the Electronic and Physicochemical Parameters on the Carcinogenecis Activity of Some Sulfa Drug Using QSAR Analysis Based on Genetic-MLR & Genetic-PLS. Chemosphere 2007, 67, 2122–2130. [Google Scholar]
- Eaton, JW; Brandt, P; Mahoney, JR; Lee, JT, Jr. Haptoglobin: A Natural Bacteriostat. Science 1982, 215, 691–693. [Google Scholar]
- Jones, RL; Peterson, CM; Grady, RW; Kumbaraci, T; Cerami, A. Effects of Iron Chelators and Iron Overload on Salmonella Infection. Nature 1977, 267, 63–65. [Google Scholar]
- Weinberg, ED. Cellular Iron Metabolism in Health and Diseased. Drug Metab. Rev 1990, 22, 531–579. [Google Scholar]
- Weinberg, ED. Iron and Infection. Microbiol. Rev 1978, 42, 45–66. [Google Scholar]
- Weinberg, ED. Iron Withholding: A Defense Against Infection and Neoplasia. Physiol. Rev 1984, 64, 65–102. [Google Scholar]
- Skaar, EP; Gaspar, AH; Schneewind, O. Bacillus anthracis IsdG, a Heme-Degrading Monooxygenase. J. Bacteriol 2006, 188, 1071–1080. [Google Scholar]
- Neilands, JB. Siderophores: Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem 1995, 270, 26723–26726. [Google Scholar]
- Sebat, JL; Paszczynski, AJ; Cortese, MS; Crawford, RL. Antimicrobial Properties of Pyridine-2,6-Dithiocarboxylic Acid, a Metal Chelator Produced by Pseudomonas spp. Appl. Envir. Microbiol 2001, 67, 3934–3942. [Google Scholar]
- Weinberg, GA. Iron Chelators as Therapeutic Agents against Pneumocystis carinii. Antimicrob. Agents Chemother 1994, 38, 997–1003. [Google Scholar]
- van Asbeck, BS; Marcelis, JH; Marx, JJM; Struyvenberg, A; van Kats, JH; Verhoef, J. Inhibition of Bacterial Multiplication by the Iron Chelator Deferoxamine: Potentiating Effect of Ascorbic Acid. Eur. J. Clin. Microbiol. Infect. Dis 1983, 2, 426–431. [Google Scholar]
- Erol, DD; Yulug, N. Synthesis and Antimicrobial Investigation of Thiazolinoalkyl-4(1H)-pyridones. Eur. J. Med. Chem 1994, 29, 893–897. [Google Scholar]
- Min-Hua, F; van der Does, L; Bantjes, A. Iron (III)-Chelating Resins. 3. Synthesis, Iron (III)-Chelating Properties, and in vitro Antibacterial Activity of Compounds Containing 3-hydroxy-2-methyl-4(1H)-pyridinone Ligands. J. Med. Chem 1993, 36, 2822–2827. [Google Scholar]
- Aytemir, MD; Erol, DD; Hider, RC; Ozalp, M. Synthesis and Evaluation of Antimicrobial Activity of New 3-Hydroxy-6-methyl-4-oxo-4H-pyran-2- carboxamide Derivatives. Turk. J. Chem 2003, 27, 757–764. [Google Scholar]
- Aytemir, MD; Hider, RC; Erol, DD; Ozalp, M; Ekizoglu, M. Synthesis of New Antimicrobial Agents; Amide Derivatives of Pyranones and Pyridinones. Turk. J. Chem 2003, 27, 445–452. [Google Scholar]
- Fassihi, A; Abedi, D; Saghaie, L; Sabet, R; Fazeli, H; Bostaki, Gh; Deilami, O; Sadinpour, H. Synthesis, Antimicrobial Evaluation and QSAR Study of Some 3-hydroxypyridine-4- one and 3-hydroxypyran-4-one Derivatives. In Eur. J. Med. Chem; 2008; DOI:10.1016/j.emech.2008.10.022. [Google Scholar]
- Todeschini, R. Milano Chemometrics and QSPR Group, http://michem.disat.unimib.it/, accessed 9 September, 2008.
- Frisch, MJ; Trucks, MJ; Schlegel, HB; Scuseria, GE; Robb, MA; Cheeseman, JR; Zakrzewski, VG; Montgomery, JA; Stratmann, JR; Burant, JC; et al. Gaussian 98, Revision A.7; Gaussian, Inc: Pittsburgh PA, 1998. [Google Scholar]
- Roy, K. QSAR of Adenosine Receptor Antagonists II: Exploring Physicochemical Requirements for Selective Binding of 2-arylpyrazolo [3,4-c] quinoline Derivatives with Adenosine A1 and A3 Receptor Subtypes. QSAR. Comb. Sci 2003, 22, 614–621. [Google Scholar]
- Siedlecki, W; Sklansky, J. On Automatic Feature Selection. Int. J. Pattern Recog. Artif. Intell 1988, 2, 197–220. [Google Scholar]
- Leardi, R. Application of Genetic Algorithm-PLS for Feature Selection in Spectral Data Sets. J. Chemomtr 2000, 14, 643–655. [Google Scholar]
- Leardi, R; Gonzalez, AL. Genetic Algorithm Applied to Feature Selection in PLS Regression: How and When to Use Them. Chemom. Intell. Lab. Syst 1998, 41, 195–207. [Google Scholar]
- Fassihi, A; Sabet, R. QSAR Study of p56lck Protein Tyrosine Kinase Inhibitory Activity of Flavonoid Derivatives Using MLR and GA-PLS. Int. J. Mol. Sci 2008, 9, 1876–1892. [Google Scholar]
- Leardi, R. Genetic Algorithms in Chemometrics and Chemistry: A Review. J. Chemometrics 2001, 15, 559–569. [Google Scholar]
- Hemmateenejad, B. Optimal QSAR Analysis of the Carcinogenic Activity of Drugs by Correlation Ranking and Genetic Algorithm-Based. J. Chemometrics 2004, 18, 475–485. [Google Scholar]
- Franke, R; Gruska, A. Chemometrics Methods in Molecular Design. In Methods and Principles in Medicinal Chemistry; van Waterbeemd, H, Ed.; VCH: Weinheim, Germany, 1995; Volume 2, pp. 113–119. [Google Scholar]
- Kubinyi, H. The Quantitative Analysis of Structure-Activity Relationships. In Burger’s Medicinal Chemistry and Drug Discovery, 5th Ed.; Wolff, ME, Ed.; Wiley: New York, USA, 1995; Volume 1, pp. 506–509. [Google Scholar]
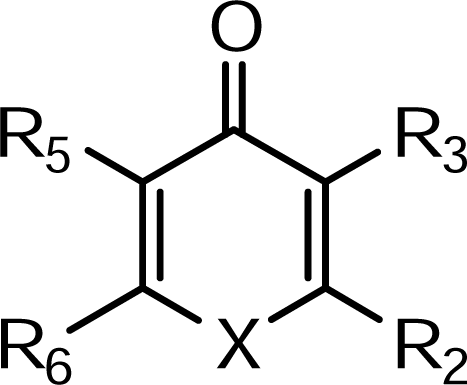
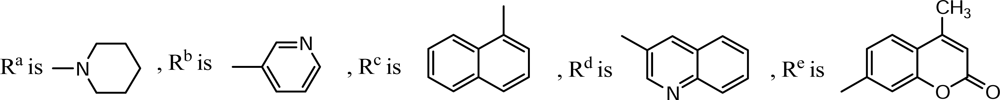
| Compound | X | R2 | R3 | R5 | R6 |
|---|---|---|---|---|---|
| 1 | NH | CH3 | OH | CH2-Ra | H |
| 2 | NH | C2H5 | OH | CH2-Ra | H |
| 3 | NH | CH3 | OH | CH2-N(CH3)2 | H |
| 4 | NH | C2H5 | OH | CH2-N(CH3)2 | H |
| 5 | NH | CH3 | OH | CH2-N(C2H5)2 | H |
| 6 | NH | C2H5 | OH | CH2-N(C2H5)2 | H |
| 7 | N-Ph | CH3 | OH | H | H |
| 8 | N-m-OH-Ph | CH3 | OH | H | H |
| 9 | N-C3H7 | CH3 | OH | H | H |
| 10 | N-C4H9 | CH3 | OH | H | H |
| 11 | O | CH2Cl | H | OH | H |
| 12 | O | CH3 | H | OH | H |
| 13 | O | CH2OH | OH | H | CH3 |
| 14 | O | CH2OH | OCH2Ph | H | CH3 |
| 15 | O | CHO | OCH2Ph | H | CH3 |
| 16 | O | COOH | OCH2Ph | H | CH3 |
| 17 | O | CONHRb | OCH2Ph | H | CH3 |
| 18 | O | CONHRc | OCH2Ph | H | CH3 |
| 19 | O | CONHRd | OCH2Ph | H | CH3 |
| 20 | O | CONHRb | OH | H | CH3 |
| 21 | O | CONHRc | OH | H | CH3 |
| 22 | O | CONHRd | OH | H | CH3 |
| 23 | O | CH2OH | H | OCH2Ph | H |
| 24 | O | COOH | H | OCH2Ph | H |
| 25 | O | CONHPh | H | OCH2Ph | H |
| 26 | N-CH3 | CONHPh | H | OCH2Ph | H |
| 27 | N-CH3 | CONHPh | H | OH | H |
| 28 | O | CONH-Re | H | OCH2Ph | H |
| 29 | N-CH3 | CONH-Re | H | OCH2Ph | H |
| 30 | N-CH3 | CONH-Re | H | OH | H |
| 31 | O | CH2OH | H | OH | H |
| Descriptor Type | Molecular Description |
|---|---|
| Constitutional | Mean atomic van der Waals volume (Mv) (scaled on Carbon atom), no. of heteroatoms, no. of multiple bonds (nBM), no. of rings, no. of circuits, no of H-bond donors, no of H-bond acceptors, no. of Nitrogen atoms (nN), chemical composition, sum of Kier-Hall electrotopological states (Ss), mean atomic polarizability (Mp), number of rotable bonds (RBN), mean atomic Sanderson electronegativity (Me), etc.
|
| Topological | Narumi harmonic topological index (HNar), Total structure connectivity index (Xt), information content index (IC), mean information content on the distance degree equality (IDDE), total walk count, path/walk-Randic shape indices (PW3, PW4, PW5, Zagreb indices, Schultz indices, Balaban J index (such as MSD) Wiener indices, Information content index (neighborhood symmetry of 2-order) (IC2), Ratio of multiple path count to path counts (PCR), Lovasz-Pelikan index (leading eigenvalue) (LP1), total information content index (neighborhood symmetry of 1-order) (TIC1), reciprocal hyper-detour index (Rww), Average connectivity index chi-5 (X5A), piID (conventional bond-order ID number), etc.
|
| Geometrical | 3D Petijean shape index (PJI3), Asphericity (ASP), Gravitational index, Balaban index, Wiener index, Length-to-breadth ratio by WHIM (L/Bw), etc.
|
| Quantum | Highest occupied Molecular Orbital Energy (HOMO), Lowest Unoccupied Molecular Orbital Energy (LUMO), Most positive charge (MPC), Sum of square of positive charges (SSPC), Sum of square of negative charges (SSNC), Sum of positive charges (SUMPC), Sum of negative charges (SUMNC), Sum of absolute of charges (SAC), Standard deviation (Std), Total dipole moment (DMt), Molecular dipole moment at X-direction (DMX), Molecular dipole moment at Y-direction (DMY), Molecular dipole moment at Z-direction (DMZ), Electronegativity (χ= −0.5 (HOMO-LUMO)), Electrophilicity (ω= χ2/2 η), Hardness (η = 0.5 (HOMO+LUMO)), Softness (S=1/ η).
|
| Functional group | Number of total secondary C(sp3) (nCs), Number of total tertiary carbons (nCt), Number of H-bond acceptor atoms (nHAcc), Number of secondary amides (aliphatic) (nCONHR), Number of unsubstituted aromatic C (nCaH), Number of ethers (aromatic) (nRORPh), Number of ketones (aliphatic) (nCO), Number of tertiary amines (aliphatic) (nNR2), Number of phenols (nOHPh), Number of total primary C(sp3) (nCp), etc.
|
| Chemical | LogP (Octanol-water partition coefficient), Hydration Energy (HE), Polarizability (Pol), Molar refractivity (MR), Molecular volume (V), Molecular surface area (SA).
|
| Compound | Experimental pMICa | Predicted pMIC | REP b (%) |
|---|---|---|---|
| 1 | 3.29 | 3.3205 | 0.9173 |
| 2 | 3.29 | 3.3007 | 0.3242 |
| 3 | 3.29 | 3.2266 | −1.9664 |
| 4* | 3.29 | 3.3976 | 3.1675 |
| 5 | 4.19 | 3.7498 | −11.740 |
| 6 | 3.29 | 3.3205 | 0.9173 |
| 7 | 3.89 | 3.8255 | −1.6850 |
| 8 | 3.29 | 3.2698 | −0.6172 |
| 9 | 3.29 | 3.2886 | −0.0440 |
| 10* | 3.89 | 3.9283 | 0.9738 |
| 11 | 3.59 | 3.6207 | 0.8470 |
| 12 | 3.59 | 3.7254 | 3.6340 |
| 13 | 3.59 | 3.5063 | −2.3883 |
| 14 | 3.59 | 3.6212 | 0.8627 |
| 15* | 4.19 | 4.1563 | −0.8119 |
| 16 | 3.59 | 3.5611 | −0.8123 |
| 17 | 3.59 | 3.6177 | 0.7647 |
| 18 | 3.59 | 3.5548 | −0.9915 |
| 19* | 3.89 | 3.8950 | 0.1293 |
| 20 | 4.19 | 4.0995 | −2.2079 |
| 21 | 3.59 | 3.7117 | 3.2787 |
| 22 | 5.10 | 5.0840 | −0.3141 |
| 23 | 3.59 | 3.5533 | −1.0318 |
| 24* | 3.59 | 3.7223 | 3.5534 |
| 25 | 3.89 | 3.9222 | 0.8214 |
| 26 | 3.89 | 3.9779 | 2.2092 |
| 27 | 4.80 | 4.8022 | 0.0453 |
| 28 | 3.89 | 3.8591 | −0.8011 |
| 29 | 3.59 | 3.4907 | −2.8470 |
| 30* | 4.49 | 4.5105 | 0.4549 |
| 31 | 3.59 | 3.4728 | −3.3746 |
| Compd. | Experimental pMIC | Predicted pMIC | REP(%) |
|---|---|---|---|
| 2 | 3.29 | 3.4139 | 3.6304 |
| 4* | 3.29 | 3.3893 | 2.9303 |
| 5 | 3.89 | 3.8920 | 0.0514 |
| 6 | 3.29 | 3.3591 | 2.0577 |
| 7 | 3.29 | 3.3835 | 2.7631 |
| 8 | 3.59 | 3.6477 | 1.5813 |
| 9 | 3.29 | 3.3208 | 0.9272 |
| 10* | 3.59 | 3.6196 | 0.8175 |
| 11 | 3.89 | 3.9567 | 1.6857 |
| 12 | 3.89 | 3.7481 | −3.7870 |
| 13 | 3.89 | 3.9092 | 0.4922 |
| 14 | 3.89 | 3.7076 | −4.9191 |
| 15 | 3.89 | 3.8892 | −0.0203 |
| 16 | 3.89 | 3.8422 | −1.2433 |
| 17* | 4.49 | 4.3961 | −2.1360 |
| 18 | 4.49 | 4.4476 | −0.9524 |
| 19 | 3.89 | 3.7076 | −4.9191 |
| 20 | 3.89 | 3.8014 | −2.3296 |
| 21 | 3.89 | 3.9525 | 1.5813 |
| 23 | 3.89 | 3.7450 | −3.8727 |
| 24* | 3.89 | 3.9056 | 0.3994 |
| 25 | 3.89 | 3.9969 | 2.6755 |
| 26 | 3.89 | 3.8489 | −1.0691 |
| 27 | 3.89 | 3.7573 | −3.5304 |
| 28 | 3.89 | 3.9503 | 1.5262 |
| 29* | 3.89 | 3.9964 | 2.6619 |
| 30 | 3.89 | 3.8978 | 0.2006 |
| 31 | 3.89 | 3.8732 | −0.4333 |
| 1 | 2 | 3 | 4 | Commonality | |
|---|---|---|---|---|---|
| MPC | 0.588 | −0.105 | 0.587 | −0.313 | 0.799 |
| DMy | 0.195 | −0.054 | 0.762 | 0.071 | 0.627 |
| HOMO | 0.059 | 0.637 | −0.013 | 0.620 | 0.794 |
| Electonegativity | −0.643 | −0.206 | −0.199 | −0.496 | 0.741 |
| Mv | 0.751 | −0.413 | 0.362 | −0.259 | 0.934 |
| Me | 0.001 | −0.781 | 0.097 | −0.298 | 0.708 |
| RBN | 0.087 | 0.902 | 0.068 | 0.003 | 0.826 |
| HNar | 0.866 | 0.051 | 0.217 | −0.252 | 0.863 |
| Xt | −0.645 | −0.505 | −0.307 | 0.081 | 0.772 |
| IDDE | 0.746 | 0.359 | 0.215 | 0.324 | 0.837 |
| LP1 | 0.667 | 0.460 | 0.368 | 0.292 | 0.877 |
| TIC1 | 0.714 | 0.413 | 0.175 | 0.127 | 0.726 |
| PJI3 | 0.375 | 0.611 | −0.315 | −0.276 | 0.689 |
| nCS | −0.559 | 0.578 | −0.411 | 0.199 | 0.855 |
| nCaH | 0.894 | −0.140 | −0.143 | −0.079 | 0.845 |
| nCONHR | 0.261 | 0.220 | 0.695 | −0.906 | 0.765 |
| nCO | −0.082 | 0.081 | −0.214 | 0.853 | 0.787 |
| pMIC S. aureus | 0.041 | −0.116 | 0.898 | −0.051 | 0.824 |
| %variance | 29.87 | 20.10 | 17.15 | 12.12 | 79.24 |
| 1 | 2 | 3 | 4 | 5 | Commonality | |
|---|---|---|---|---|---|---|
| Std | −0.491 | −0.431 | −0.459 | −0.107 | 0.095 | 0.657 |
| DMz | −0.007 | 0.102 | −0.209 | 0.860 | 0.322 | 0.898 |
| HOMO | 0.240 | 0.811 | −0.156 | −0.349 | 0.014 | 0.861 |
| Electonegativity | −0.706 | −0.389 | 0.142 | 0.323 | −0.310 | 0.871 |
| X5A | −0.627 | −0.664 | −0.134 | −0.102 | 0.129 | 0.879 |
| PW3 | −0.166 | 0.594 | −0.377 | −0.158 | 0.893 | 0.584 |
| PW5 | 0.913 | −0.079 | 0.055 | 0.135 | −0.132 | 0.879 |
| IC2 | 0.579 | 0.272 | −0.164 | 0.210 | 0.584 | 0.820 |
| piID | 0.750 | −0.070 | −0.333 | −0.190 | −0.208 | 0.758 |
| ASP | −0.075 | 0.087 | 0.866 | −0.198 | 0.322 | 0.905 |
| L/Bw | 0.064 | 0.117 | 0.926 | −0.023 | 0.164 | 0.902 |
| nCp | −0.206 | 0.754 | −0.224 | −0.097 | −0.325 | 0.777 |
| nNR2 | −0.366 | 0.722 | 0.148 | 0.287 | −0.234 | 0.814 |
| nOHPh | −0.191 | −0.415 | −0.165 | −0.447 | 0.356 | 0.562 |
| nRORPh | 0.571 | −0.522 | 0.379 | 0.002 | −0.341 | 0.858 |
| pMIC C. albicans | 0.628 | −0.627 | −0.277 | −0.107 | 0.602 | 0.872 |
| %variance | 22.58 | 20.58 | 14.71 | 14.02 | 8.71 | 80.60 |
© 2008 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/). This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sabet, R.; Fassihi, A. QSAR Study of Antimicrobial 3-Hydroxypyridine-4-one and 3-Hydroxypyran-4-one Derivatives Using Different Chemometric Tools. Int. J. Mol. Sci. 2008, 9, 2407-2423. https://doi.org/10.3390/ijms9122407
Sabet R, Fassihi A. QSAR Study of Antimicrobial 3-Hydroxypyridine-4-one and 3-Hydroxypyran-4-one Derivatives Using Different Chemometric Tools. International Journal of Molecular Sciences. 2008; 9(12):2407-2423. https://doi.org/10.3390/ijms9122407
Chicago/Turabian StyleSabet, Razieh, and Afshin Fassihi. 2008. "QSAR Study of Antimicrobial 3-Hydroxypyridine-4-one and 3-Hydroxypyran-4-one Derivatives Using Different Chemometric Tools" International Journal of Molecular Sciences 9, no. 12: 2407-2423. https://doi.org/10.3390/ijms9122407
APA StyleSabet, R., & Fassihi, A. (2008). QSAR Study of Antimicrobial 3-Hydroxypyridine-4-one and 3-Hydroxypyran-4-one Derivatives Using Different Chemometric Tools. International Journal of Molecular Sciences, 9(12), 2407-2423. https://doi.org/10.3390/ijms9122407