Abstract
QSAR analysis of a series of previously synthesized N1-aryl-amino/methyl/ethyl-1,2,4-triazole and N1-heteroarylamino/methyl/ethyl-1,2,4-triazole derivatives tested for growth inhibitory activity against Bacillus subtilis, was performed using the computer-assisted multiple regression procedure. Using the Hansch and Free Wilson approaches the activity contribution for either the aminomethyl/aminoethyl unit or the aromatic/heteroaromatic ring was determined from the correlation equation.
1. Introduction
Our society is faced with several challenges that may have a chemical solution. Examples include bacterial drug resistance, new diseases like AIDS, and agricultural pest control. Characterizing the biological activity and properties of all the known compounds is impossible so it is necessary to develop predictive tools for molecular properties and environmental behaviour. Quantitative structure activity relationships (QSAR) and quantitative structure properties relationships (QSPR) play a central role in this effort, so these methods are unquestionably of great importance in modern chemistry and biochemistry [1,2,3]. The concept of QSAR/QSPR is to transform searches for compounds with desired properties using chemical intuition and experience into a mathematically quantified and computerized form. Once a correlation between structure and activity/property is found, any number of compounds, including those not yet synthesized, can be readily screened on the computer in order to select structure with the properties desired. It is then possible to select the most promising compounds to be synthesized and tested in the laboratory.
1,2,4-Triazole and its derivatives represent one of the most biologically active classes of compounds, possessing a wide spectrum of activities. The 1,2,4-triazole nucleus is associated with diverse pharmacological activities such as antibacterial, antifungal, hypoglycemic, antihypertensive and analgesic properties [4,5,6,7]. Consequently, spurred by the need for new antimicrobial agents and the fact that many new effective antimicrobial drugs possess heterocyclic rings in their structure, such as a 1,2,4-triazole ring, over the last few years we have synthesized some novel 1,2,4-triazole derivatives and tested them for antibacterial and antifungal effects against Escherichia coli, Bacillus subtilis, Salmonella enteritidis, Staphylococcus aureus, Aspergillus niger and Candida albicans [8,9,10,11]. The antibacterial activity of synthesized N1-aryl/heteroarylamino/methyl/ethyl-1,2,4-triazole derivatives against Bacillus subtilis were used in this QSAR analysis.
2. Results and Discussion
In our work, the chosen model is based on the in vitro antimicrobial activity of certain N1-aryl- and N1-heteroarylaminoaminomethyl/ethyl-1,2,4-triazole derivatives 1-18, (Table 1 and Table 2; Figure 1 and Figure 2), against Bacillus subtilis.
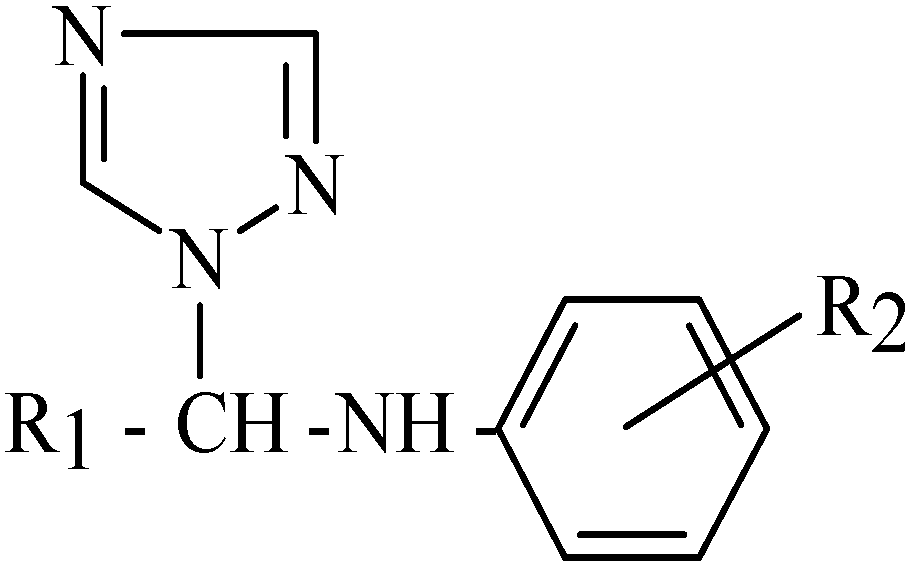
Figure 1.

Table 1.
N1-arylaminomethyl/ethyl-1,2,4-triazole derivatives (1-10) used in the present study.
| Comp. No: | R1 | R2 |
|---|---|---|
| 1 | H | p-COOC2H5 |
| 2 | H | p-COOH |
| 3 | H | o-COOH |
| 4 | H | p-Cl |
| 5 | H | p-Br |
| 6 | H | p-CH3 |
| 7 | H | p-C6H5 |
| 8 | H | p-C6H4-CH2-NH-  |
| 9 | CH3 | p-COOC2H5 |
| 10 | CH3 | p-NO2 |
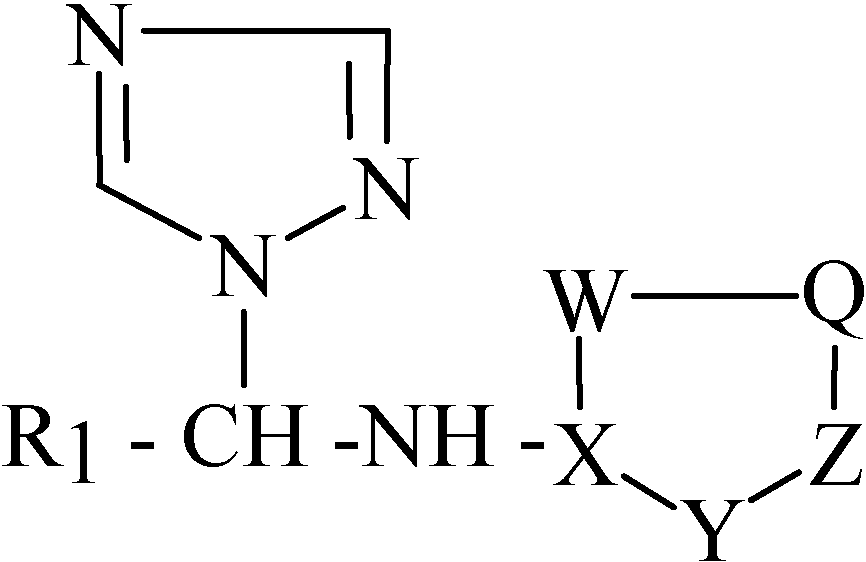
Figure 2.

Table 2.
N1-heteroarylaminomethyl/ethyl-1,2,4-triazole derivatives (11-18) used in the present study.
| Comp. No: | R1 | X | Y | Z | Q | W |
|---|---|---|---|---|---|---|
| 11 | H | -C= | =N- | -CH= | =CH- | -CH=CH- |
| 12 | H | -C= | =N- | -CH= | =CH- | -C(CH3)=CH- |
| 13 | H | -C= | =N- | =C(CH3)- | =CH- | -CH=CH- |
| 14 | H | -C= | =N- | -CH= | =C(Cl)- | -CH=CH- |
| 15 | H | -C= | =N- | -CH= | =N- | -CH=CH- |
| 16 | H | -N- | -CH= | =N- | -N= | =CH- |
| 17 | CH3 | -C= | =N- | =C(CH3)- | =CH- | -CH=CH- |
| 18 | CH3 | -C= | =N- | -CH= | =CH- | -S- |
The results of antimicrobial tests indicated that not all compounds exhibited antibacterial and antifungal activities. It must also be noted that compounds 17 and 18 do not inhibit the growth of the selected microorganisms. The inhibitory effects of compounds 1-18, against B. subtilis, expressed as minimum inhibition concentration (MIC), are given in Table 3, where C is the MIC value expressed in molar concentration units (Table 3).

Table 3.
Experimentally obtained MIC values, calculated log 1/C and log P.
| Compound No: | MICa | log 1/ C | log Pb | MRc |
|---|---|---|---|---|
| 1 | 4.061 x 10-6 | 5.3914 | 1.2981 | 17.47 |
| 2 | 2.291 x 10-5 | 4.6399 | 0.4579 | 6.93 |
| 3 | - | - | 1.4690 | 6.93 |
| 4 | 4.793 x 10-5 | 4.3194 | 1.2210 | 6.03 |
| 5 | 1.975 x 10-5 | 4.7044 | 1.4665 | 8.88 |
| 6 | 2.656 x 10-5 | 4.5757 | 1.1238 | 5.65 |
| 7 | 3.995 x 10-6 | 5.3984 | 2.3405 | 25.36 |
| 8 | - | - | -1.6900 | - |
| 9 | 3.842 x 10-5 | 4.4154 | 1.7157 | 17.47 |
| 10 | 4.287 x 10-5 | 4.3678 | 1.3895 | 0.67 |
| 11 | 5.708 x 10-5 | 4.2435 | 0.0301 | - |
| 12 | - | - | 0.5774 | - |
| 13 | 5.399 x 10-5 | 4.2676 | 0.5774 | - |
| 16 | 6.055 x 10-5 | 4.2178 | -2.1800 | - |
a minimum inhibition concentration expressed in molar concentration; b Ref. [12]; c Ref. [1].
Using the filter paper disc method [13] compounds 3, 8, 12, 14 and 15 do not inhibit the growth of the chosen microorganism [10]. From the data obtained, first MIC values were calculated and then the log 1/C values (Table 3). It was important for further analysis to find the correlation matrix for the descriptors used (Table 4).

Table 4.
Correlation matrix for the chosen electronic, steric and hydrophobic parameters.
| σ | 1.0000 | ||||||
| π | -0.8861 | 1.0000 | |||||
| logP | -0.4792 | 0.8180 | 1.0000 | ||||
| MR | -0.4751 | 0.7235 | 0.7732 | 1.0000 | |||
| F | 0.2547 | -0.1186 | 0.6125 | -0.2499 | 1.0000 | ||
| R | 0.6327 | -0.7356 | 0.0348 | -0.5815 | 0.3997 | 1.0000 | |
In our work, attempts were made to establish a correlation between selected physicochemical properties and experimental values for antimicrobial activities against B. subtilis, in three ways:
- ➢
- applying the general Hansch equation for structurally identical compounds (1-8);
- ➢
- using the Free Wilson approach which included derivatives with some structural changes (aminomethyl unit has been replaced with aminoethyl group), compounds (1-10);
- ➢
- extend the Free Wilson equation, for determination of the influence of heterocyclic ring, substituted on the amino group, compounds (1-18).
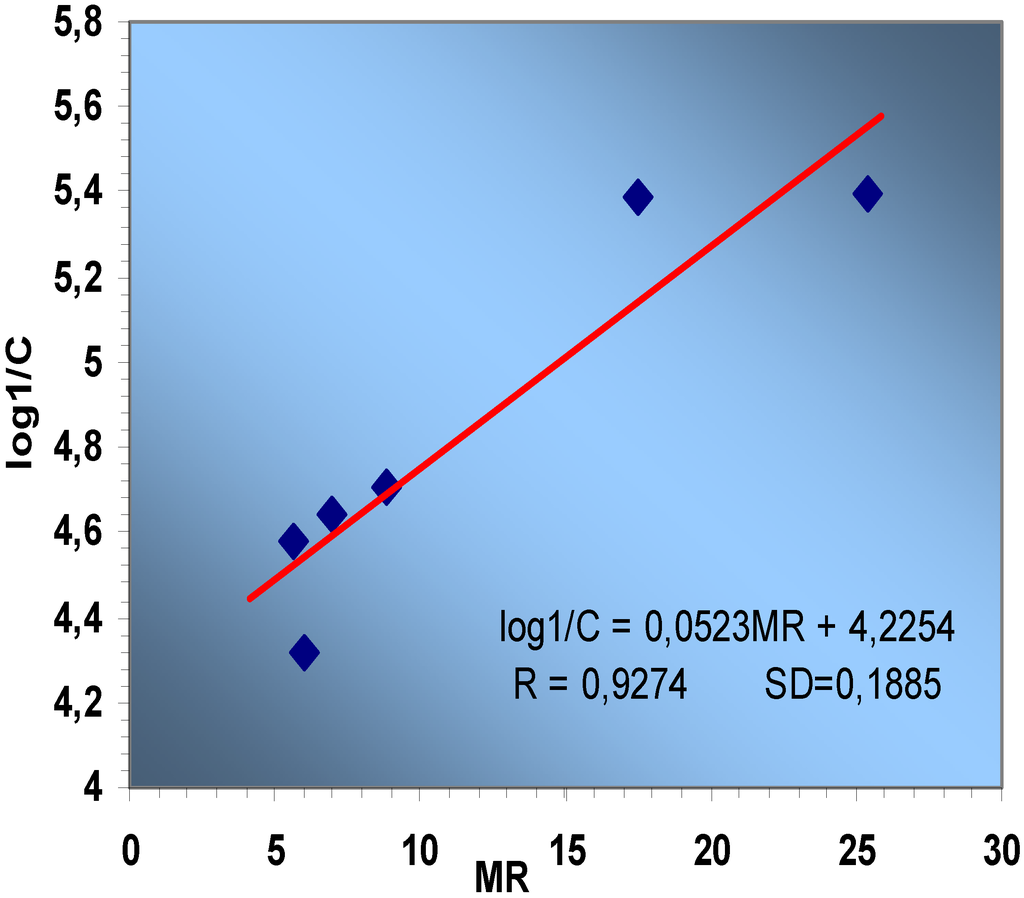
Figure 3.
Linear correlation between log 1/C and MR
When the data in Table 3 and Table 7 were submitted to linear regression (Figure 3) the resulting QSAR equation is:
where R is correlation coefficients and SD is the standard deviation.
log1/C = 0.0523MR + 4.2254
R = 0.9274 SD=0.1885
R = 0.9274 SD=0.1885
The data for the chosen compounds were reasonably well correlated with a regression coefficient of 0.9274, indicating a relatively good fit.
However, moderate linear collinearities exist between log1/C and other selected descriptors (σ, π, logP, F, R and L) where R is below 0.7. This weak correlation is unable to describe the biological activities of the selected set of compounds. Addition of some other groups to the parent triazole ring would certainly improve those linear fittings.
An attempt was also made to find the parabolic correlation. Parabolic relationships between biological response (log1/C) and logP term, can be explained by the fact that many membranes must be traversed for compounds to get to the target site, and those with greatest hydrophobicity will become localized in the membranes they encounter initially. Thus, an optimum hydrophobicity may be found in some test systems.
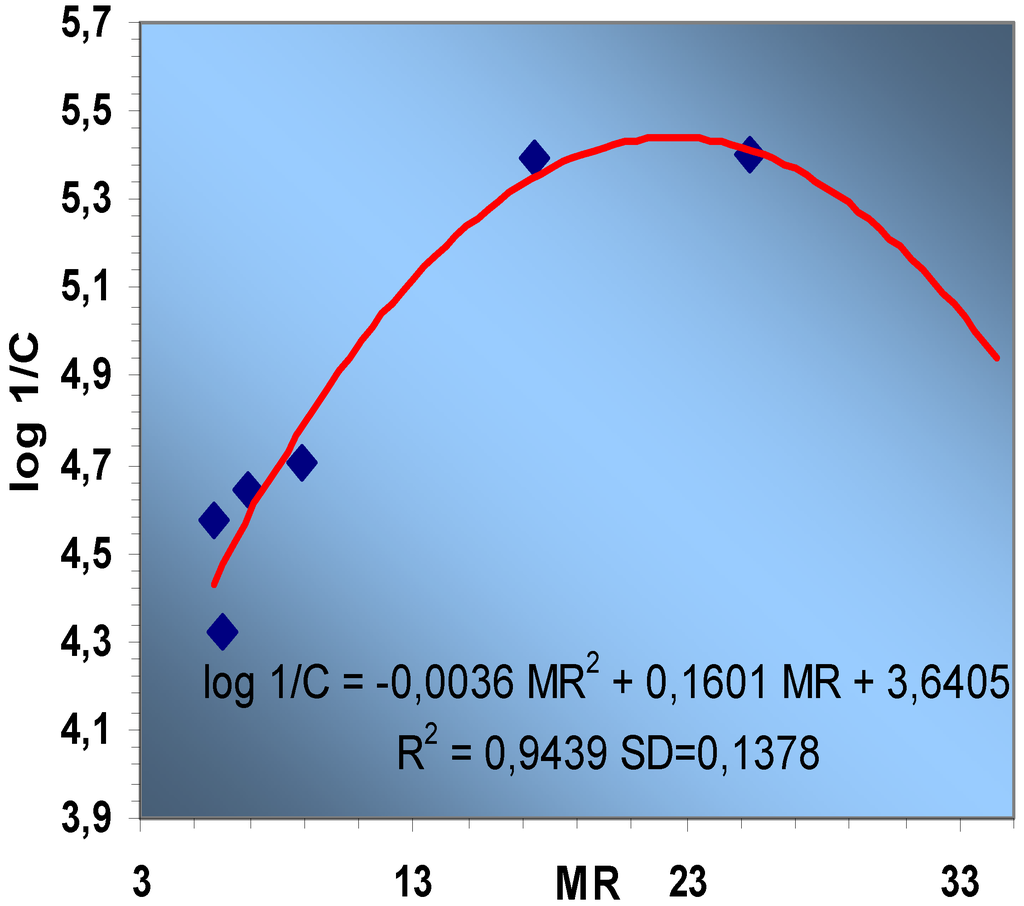
Figure 4.
Parabolic correlation between log 1/C and MR.
Knowing this fact, we tried to find parabolic correlations and thus an excellent correlation is obtained between log1/C and MR, (Figure 4):
log 1/C = - 0.0036 MR2 + 0.1601 MR + 3.6405
R2 = 0.9439 SD=0.1378
R2 = 0.9439 SD=0.1378
For many QSAR studies many descriptors are needed. Addition of more then one descriptor would certainly improve the QSAR model.
Several multivariate correlations between the structural parameters mentioned above and log1/C are presented in Table 5, together with correlation coefficients (R) and standard deviations (SD).
In bivariate correlation analysis also, the correlations involving π, logP and σ are found to be good (Table 5, correlation No 1-13; R=0.91-0.96). Excellent correlation (0.9679) is obtained when in bivariate correlation logP and R were used. The correlation is expressed as:
log1/C = 0.7156logP + 2.5765R + 3.9939
R=0.9679 SD=0.1462
R=0.9679 SD=0.1462
It is interesting to note that an excellent correlation is also obtained in tervariate correlation involving the same parameters (correlation No 14-18, Table 5). The correlation coefficients in all the cases were found to be approximately the same (0.97-0.98), and the standard deviation below 0.16.

Table 5.
Regression parameters and the quality of correlation of log 1/C with σ, π, log P, MR, F, R and L, multivariate regressions for substituted 1,2,4-triazoles.
| Correlation No | Correlation parameters used a | Slope Ai i=1-2 | Intercept B | R | SD | |
|---|---|---|---|---|---|---|
| 1 | σ | A1= | 0.2668 | 4.1667 | 0.9389 | 0.2002 |
| MR | A2= | 0.0528 | ||||
| 2 | σ | A1= | -0.2059 | 3.4557 | 0.9565 | 0.1697 |
| L | A2= | 0.3222 | ||||
| 3 | π | A1= | -3.0467 | 1.6286 | 0.9130 | 0.2373 |
| logP | A2= | 4.0843 | ||||
| 4 | π | A1= | -0.2347 | 4.2092 | 0.9647 | 0.1542 |
| MR | A2= | 0.0679 | ||||
| 5 | π | A1= | 0.6084 | 4.5146 | 0.9452 | 0.1899 |
| R | A2= | 2.8805 | ||||
| 6 | π | A1= | -0.0131 | 3.4419 | 0.9502 | 0.1812 |
| L | A2= | 0.3182 | ||||
| 7 | logP | A1= | -0.2789 | 4.3958 | 0.9556 | 0.1714 |
| MR | A2= | 0.0691 | ||||
| 8 | logP | A1= | 0.7156 | 3.9939 | 0.9679 | 0.1462 |
| R | A2= | 2.5765 | ||||
| 9 | logP | A1= | 0.0007 | 3.4492 | 0.9500 | 0.1815 |
| L | A2= | 003143 | ||||
| 10 | MR | A1= | 0.0495 | 4.2878 | 0.9599 | 0.1630 |
| R | A2= | 0.7650 | ||||
| 11 | MR | A1= | 0.0120 | 3.6068 | 0.9519 | 0.1780 |
| L | A2= | 0.2469 | ||||
| 12 | F | A1= | -0.4005 | 3.5676 | 0.9654 | 0.1517 |
| L | A2= | 0.3111 | ||||
| 13 | R | A1= | 0.1041 | 3.4743 | 0.9505 | 0.1807 |
| L | A2= | 0.3097 | ||||
| 14 | σ | A1= | -0.6667 | 3.3456 | 0.9736 | 0.1625 |
| π | A2= | -0.2226 | ||||
| L | A3= | 0.4036 | ||||
| 15 | π | A1= | -1.4854 | 3.0706 | 0.9866 | 0.1165 |
| logP | A2= | 1.7219 | ||||
| MR | A3= | 0.0476 | ||||
| 16 | π | A1= | -1.2182 | 3.0139 | 0.9853 | 0.1217 |
| logP | A2= | 2.0982 | ||||
| R | A3= | 1.8779 | ||||
| 17 | π | A1= | -1.7249 | 3.6820 | 0.9771 | 0.1513 |
| MR | A2= | 0.1866 | ||||
| R | A3= | -5.1995 | ||||
| 18 | logP | A1= | 1.8755 | 3.5299 | 0.9728 | 0.1649 |
| MR | A2= | -0.0812 | ||||
| R | A3= | 5.5237 | ||||
a x, y, z from the Eqn. 1; R - correlation coefficient; SD - standard deviation
After including the compounds with little structural modification (compounds 9 and 10) the following correlations were obtained (Table 6):

Table 6.
Regression parameters and the quality of correlation of log 1/C with σ, π, log P, MR, F, R and L; (log1/C = ΣAi x Ii + B).
| Correlation No | Correlation parameters useda | Slope Ai i=1-4 | Intercept B | R | SD | |
|---|---|---|---|---|---|---|
| 1. | IH | A1= | 0.4783 | 3.8239 | 0.9034 | 0.2199 |
| MR | A2= | 0.0457 | ||||
| 2. | IH | A1= | 0.6556 | 3.5334 | 0.9289 | 0.2124 |
| σ | A2= | 0.4197 | ||||
| MR | A3= | 0.0483 | ||||
| 3. | IH | A1= | 0.6410 | 3.6390 | 0.9373 | 0.1998 |
| π | A2= | -0.2520 | ||||
| MR | A3= | 0.0629 | ||||
| 4. | IH | A1= | 0.442 | 3.9815 | 0.9138 | 0.2328 |
| logP | A2= | -0.1849 | ||||
| MR | A3= | 0.0562 | ||||
| 5. | IH | A1= | 0.6674 | 3.5784 | 0.9387 | 0.2282 |
| σ | A2 = | 0.1454 | ||||
| π | A3= | -0.1976 | ||||
| MR | A4= | 0.0600 | ||||
| 6. | I H | A1= | 0.6394 | 3.5760 | 0.9290 | 0.2449 |
| σ | A2 = | 0.3951 | ||||
| logP | A3= | -0.0300 | ||||
| MR | A4= | 0.0499 | ||||
| 7. | IH | A1= | 1.5164 | 1.8039 | 0.9885 | 0.0909 |
| π | A2 = | -1.2034 | ||||
| logP | A3= | 1.3356 | ||||
| MR | A4= | 0.0525 | ||||
R - correlation coefficient; SD - standard deviation
As it can be seen from Table 6, an excellent correlation was obtained when π, logP and MR were used:
where IH is structural indicator parameter representing –CH2- group as 1 and CH3-CH- group as 0. From the value of R (0.9885) can be seen that a relatively good model was chosen.
log 1/C = 1.5164 IH - 1.2034 π + 1.3356 log P + 0.0525MR + 1.8039
The last step was determination of the influence of heterocyclic ring by using the extend Free – Wilson equation. The following equation was obtained:
log1/C = 0.3094 IH + 0.3712 I=CH- - 0.2104 logP + 4.6375
R = 0.7171 SD = 0.3567
R = 0.7171 SD = 0.3567
Extending the investigation system may lead to developing the better QSAR system when another heterocyclic nucleus, besides triazole, is included in the chosen substituted 1,2,4-triazoles.
3. Conclusions
Spurred by the need for new antimicrobial agents and the fact that many effective drugs, insecticides and fungicides possess heterocyclic systems in their structure, such as the triazole ring, we synthesized some new 1,2,4-triazole derivatives. Analysis of this limited set of substituted 1,2,4-triazole molecules allowed us to build a QSAR model of their antimicrobial activity against B. subtilis in which π, logP and σ are important factors. When we summarized the results derived from this limited set of triazole derivatives we came to the conclusion that the QSAR results don’t really explain anything, but rather the QSAR equations just point to correlations. On the other hand, QSAR is a very important and routine method for many areas of chemistry. QSAR is best appreciated as a guide for our chemical intuition and QSAR models may guide us to what to synthesize next in our search for more effective solutions to our problems. This, in turn, will help medical as well as agriculture chemist in their prediction of increasing activity and thus the synthesis of new triazoles exhibiting better activities then those reported in this paper.
4. Experimental
4.1 Microbiology
The filter paper disc method [13] was performed in Sabouraud dextrose broth and Mueller Hinton broth. These agar media were inoculated with 24 h liquid cultures containing 107 microorganisms/mL (0.5 mL). Filter paper discs (5 mm diameter) saturated with each compound solution (1 mg/mL; 5 mg/mL and 10 mg/mL in DMSO) were placed on the indicated agar mediums. The incubation time was 24 h at 37 ºC for bacterial and 48 h at 30 ºC for Candida sp. Discs with DMSO were used as control. The diameter of zone inhibition (mm) was measured. The tests were repeated 3 times to confirm the findings.
4.2 QSAR analysis
The MVA (multi variable analysis) approach in QSAR analysis has been most widely and effectively used for theoretical drug design due to various physicochemical (electronic, steric and hydrophobic) parameters and structural indicator parameters used together (Hansch and Free Wilson approaches) [1,2].
The assumption can be formulated as given in Eqn. 1, (Hansch approach):
where x, y and z are molecular properties, and log1/C is the desired biological activities. From the values of linear slopes A1, A2, A3 we can see the correlation of the particular molecular properties with the activity of the investigated compounds.
log1/C = A1 x + A2 y + A3 z+ B
Applying the same chosen descriptors in Free Wilson analysis (Eqn. 2) the activity contributions of either methyl- or substituted heterocyclic ring systems were determined from the correlation equation:
where Ii is the structural indicator parameter; xi and log1/C had the same meaning as in Eqn.1.
log1/C = Σai Ii + Σbi xi + B
The variables used as descriptors in the analysis are electronic, steric and structural parameters (Table 3 and Table 7). Physicochemical parameters taken into consideration in QSAR study are σ electronic parameter of substituents, π hydrophobic parameter, F (field effect) as electronic influences, Verloop’s STERIMOL parameter L for the steric interactions of the substituents R2. L is defined as the length of a substituent along the axis of its substitution to the parent skeleton. Electronic effect of the substituents, expressed in term of F, is found to be important in determining the activity, as it is predictive in electrophilic reactions of bimolecules. The classical Hammett σ parameters and MR value were used (Table 3 and Table 7). For each compound the partition coefficient logP has been calculated [12] (Table 3).

Table 7.
Physicochemical parameters of the triazole derivatives studied
| R | σa | π a | F a | R a | L a |
|---|---|---|---|---|---|
| p-COOC2H5 | 0.45 | 0.51 | 0.33 | 0.15 | 5.96 |
| p-COOH | 0.45 | -0.32 | 0.33 | 0.15 | 3.91 |
| o-COOH | 1.2 | -0.32 | 0.33 | 0.15 | 3.91 |
| p-Cl | 0.23 | 0.71 | 0.41 | -0.15 | 3.52 |
| p-Br | 0.23 | 0.86 | 0.44 | -0.17 | 3.83 |
| p-CH3 | -0.17 | 0.56 | -0.04 | -0.13 | 3.00 |
| p-C6H5 | -0.01 | 1.96 | 0.08 | -0.08 | 6.28 |
| p-NO2 | 0.78 | -0.28 | 0.67 | 0.16 | 3.44 |
a Ref. [1]
Applying the Free Wilson analysis, in first step, the structural variable indicator IH expresses the replacement of hydrogen atom by the methyl group in the aminomethyl unit. IH is defined as 1 for the N1-aryl/heteroarylaminomethyl-1,2,4-triazoles (1-8, 11-16), and 0 for N1-aryl/heteroarylaminoethyl-1,2,4-triazole derivatives (9,10,17,18). In second step, the other indicator I=CH- is defined as 1 for compounds with =CH- in the six membered ring (1-10), and 0 for compound with –N= group in the six membered ring (11-18) (Table 8).

Table 8.
Matrix for Free Wilson approach.
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IH | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| I=CH- | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
References
- Available online: http://www.chem.swin.edu.au/modules/mod4/index.html.
- Hansch, C.; Leo, A. Exploring QSAR: Fundamentals and Applications in Chemistry and Biology; American Chemical Society: Washington, DC, 1995. [Google Scholar]
- Karelson, M.; Lobanov, V.; Katritzky, A. Chem. Rev. 1996, 96, 1027–1043. [CrossRef]
- Jones, D.H.; Slack, R.; Squires, S.; Woolridge, K.R.H. J. Med. Chem. 1965, 8, 676–680. [CrossRef]
- Goswami, B.N.; Kataky, J.C.S.; Baruah, J.N. J. Heterocyclic Chem. 1984, 21, 1225–1229. [CrossRef]
- Holla, B.S.; Kalluraya, B.; Sridhar, K.R. Curr. Sci. 1987, 56, 236–239.
- Mishra, R.K.; Tewari, R.K.; Srivastava, S.K.; Bahel, S.C. J. Indian Chem. Soc. 1991, 68, 1.
- Colanceska, K.; Dimova, V.; Kakurinov, V.; Molnar-Gabor, D.; Buzarovska, A. Molecules 2001, 6, 815–824. [CrossRef]
- Perisic-Janjic, N.; Acinski, M.; Janjic, N.; Lazarevic, M.; Dimova, V. J. Planar Chromatogr. 2000, 13, 281–284.
- Lazarevic, M.; Dimova, V.; Molnar Gabor, D.; Kakurinov, V.; Colanceska Ragenovic, K. Heterocycl. Comm. 2001, 7, 577–582.
- Spasovska-Gerasimova, B.; Ilievska, S.; Colanceska Ragenovic, K.; Lazarevic, M.; Dimova, V. Vlákna a textil 2002, 9, 46–49.
- Available online: http://esc.syrres.com/interkow/kowdemo.htm.
- Rollas, S.; Kalyoncuoglu, N.; Sür-Altiner, D.; Yegenoglu, Y. Pharmazie 1993, 48, 308–309.
© 2006 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.