Insights into Genomic Dynamics and Plasticity in the Monkeypox Virus from the 2022 Outbreak
Abstract
1. Introduction
2. Results
2.1. QC of Sequencing Data, Read Alignment, and Hybrid Assembly
2.1.1. Single Nucleotide Polymorphisms
2.1.2. Characterization of STRs in MPXV Genomes
Intra-Host STRs Variation Across Matrices and Timepoints
STRs and In Silico Protein Implications
2.1.3. Phylogenetic Analysis and STR-Based Clustering
3. Discussion
4. Materials and Methods
4.1. DNA Extraction, Library Preparation, and Sequencing
4.2. Bioinformatic Analysis
4.2.1. Hybrid Assembly
4.2.2. STR Analysis
4.2.3. Global Dataset, Phylogeny, and STR-Based Clustering Analysis
4.2.4. Protein Translation and In Silico Structural Prediction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reynolds, M.G.; Carroll, D.S.; Karem, K.L. Factors Affecting the Likelihood of Monkeypox’s Emergence and Spread in the Post-Smallpox Era. Curr. Opin. Virol. 2012, 2, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Smith, J.O. Vacated Niches, Competitive Release and the Community Ecology of Pathogen Eradication. Philos. Trans. R. Soc. B Biol. Sci. 2013, 24, 20120150. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization Global Commission for the Certification of Smallpox Eradication. The Achievement of Global Eradication of Smallpox: Final Report of the Global Commission for the Certification of Smallpox Eradication, Geneva, December 1979; WHO: Geneva, Switzerland, 1979. [Google Scholar]
- Naga, N.G.; Nawar, E.A.; Mobarak, A.A.; Faramawy, A.G.; Al-Kordy, H.M.H. Monkeypox: A Re-Emergent Virus with Global Health Implications—A Comprehensive Review. Trop. Dis. Travel. Med. Vaccines 2025, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- 2022-25 Mpox Outbreak: Global Trends. Available online: https://Worldhealthorg.Shinyapps.Io/Mpx_global/ (accessed on 1 September 2025).
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic Characterization and Signs of Microevolution in the 2022 Multi-Country Outbreak of Monkeypox Virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Totmenin, A.V.; Safronov, P.F.; Mikheev, M.V.; Gutorov, V.V.; Ryazankina, O.I.; Petrov, N.A.; Babkin, I.V.; Uvarova, E.A.; Sandakhchiev, L.S.; et al. Moss B Analysis of the Monkeypox Virus Genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef]
- Alcami, A.; Koszinowski, U.H. Viral Mechanisms of Immune Evasion. Trends Microbiol. 2000, 8, 410–418. [Google Scholar] [CrossRef]
- Moss, B.; Shisler, J.L. Immunology 101 at Poxvirus U: Immune Evasion Genes. Semin. Immunol. 2001, 13, 59–66. [Google Scholar] [CrossRef]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, Y.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A Tale of Two Clades: Monkeypox Viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef]
- Doty, J.B.; Malekani, J.M.; Kalemba, L.S.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Tobit Liyandja, L.D.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo. Viruses 2017, 9, 283. [Google Scholar] [CrossRef]
- Nakazawa, Y.; Mauldin, M.R.; Emerson, G.L.; Reynolds, M.G.; Lash, R.R.; Gao, J.; Zhao, H.; Li, Y.; Muyembe, J.-J.; Kingebeni, P.M.; et al. A Phylogeographic Investigation of African Monkeypox. Viruses 2015, 7, 2168–2184. [Google Scholar] [CrossRef]
- WHO Monkeypox Fact Sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/monkeypox (accessed on 1 September 2025).
- Esteban, D.J.; Hutchinson, A.P. Genes in the Terminal Regions of Orthopoxvirus Genomes Experience Adaptive Molecular Evolution. BMC Genom. 2011, 12, 261. [Google Scholar] [CrossRef]
- Hughes, A.L.; Friedman, R. Poxvirus Genome Evolution by Gene Gain and Loss. Mol. Phylogenetics Evol. 2005, 35, 186–195. [Google Scholar] [CrossRef]
- Kugelman, J.R.; Johnston, S.C.; Mulembakani, P.M.; Kisalu, N.; Lee, M.S.; Koroleva, G.; McCarthy, S.E.; Gestole, M.C.; Wolfe, N.D.; Fair, J.N.; et al. Genomic Variability and Tandem Repeats in Variola Virus and Monkeypox Virus. Emerg. Infect. Dis. 2014, 20, 232–239. [Google Scholar]
- Elde, N.C.; Child, S.J.; Eickbush, M.T.; Kitzman, J.O.; Rogers, K.S.; Shendure, J.; Geballe, A.P.; Malik, H.S. Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses. Cell 2012, 150, 831–841. [Google Scholar] [CrossRef]
- Yeh, T.Y.; Hsieh, Z.Y.; Feehley, M.C.; Feehley, P.J.; Contreras, G.P.; Su, Y.C.; Hsieh, S.-L.; Lewis, D.A. Recombination Shapes the 2022 Monkeypox (Mpox) Outbreak. Med 2022, 3, 824–826. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Yadav, V.K.; Singh, H. Identification and Analysis of Microsatellites in Coronaviridae. Virus Res. 2024, 336, 199328. [Google Scholar] [CrossRef]
- Siddiqe, R.; Ghosh, A. Genome-wide in silico identification and characterization of Simple Sequence Repeats in diverse completed SARS-CoV-2 genomes. Gene Rep. 2021, 23, 101020. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Tenorio, F.J.; Rojas-López, A.; Arias, C.F. Microsatellite Repeats in the Genome of Caliciviruses. J. Virol. 2002, 76, 10638–10643. [Google Scholar]
- Burrel, S.; Ait-Arkoub, Z.; Voujon, D.; Deback, C.; Abrao, E.P.; Agut, H.; Boutolleau, D. Molecular characterization of herpes simplex virus 2 strains by analysis of microsatellite polymorphism. J. Clin. Microbiol. 2013, 51, 3616–3623. [Google Scholar] [CrossRef]
- Sahu, B.P.; Majee, P.; Singh, R.R.; Nayak, D. Comparative analysis, distribution, and characterization of microsatellites in Orf virus genome. Sci. Rep. 2020, 10, 13852. [Google Scholar] [CrossRef]
- Monzón, S.; Varona, S.; Negredo, A.; Vidal-Freire, S.; Patiño-Galindo, J.A.; Ferressini-Gerpe, N.; Zaballos, A.; Orviz, E.; Ayerdi, O.; Muñoz-Gómez, A.; et al. Monkeypox Virus Genomic Accordion Strategies. Nat. Commun. 2024, 15, 3059. [Google Scholar] [CrossRef] [PubMed]
- Laliberte, J.-P.; Moss, B. Membrane Fusion by Poxvirus Entry/Fusion Complex. Proc. Natl. Acad. Sci. USA 2014, 111, E2834–E2843. [Google Scholar]
- Satheshkumar, P.S.; Chavre, J.; Moss, B. Role of the vaccinia virus O3 protein in cell entry can be fulfilled by its Sequence flexible transmembrane domain. Virology 2013, 444, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Deiana, M.; Lavezzari, D.; Mori, A.; Accordini, S.; Pomari, E.; Piubelli, C.; Malagò, S.; Cordioli, M.; Ronzoni, N.; Angheben, A.; et al. Exploring Viral Genome Profile in Mpox Patients during the 2022 Outbreak, in a North-Eastern Centre of Italy. Viruses 2024, 16, 726. [Google Scholar] [CrossRef]
- Luna, N.; Ramírez, A.L.; Muñoz, M.; Ballesteros, N.; Patiño, L.H.; Castañeda, S.A.; Bonilla-Aldana, D.K.; Paniz-Mondolfi, A.; Ramírez, J.D. Phylogenomic analysis of the monkeypox virus (MPXV) 2022 outbreak: Emergence of a novel viral lineage? Travel Med. Infect Dis. 2022, 49, 102402. [Google Scholar] [CrossRef]
- Alakunle, E.; Kolawole, D.; Diaz-Cánova, D.; Alele, F.; Adegboye, O.; Moens, U.; Okeke, M.I. A Comprehensive Review of Monkeypox Virus and Mpox Characteristics. Front. Cell. Infect. Microbiol. 2024, 14, 1360586. [Google Scholar] [CrossRef]
- GISAID EpiPox GISAID EpiPox. Available online: https://gisaid.org// (accessed on 21 August 2025).
- Yadav, P.; Devasurmutt, Y.; Tatu, U. Phylogenomic and Structural Analysis of the Monkeypox Virus Shows Evolution towards Increased Stability. Viruses 2022, 31, 127. [Google Scholar] [CrossRef]
- Delamonica, B.; Davalos, L.; Larijani, M.; Anthony, S.J.; Liu, J.; MacCarthy, T. Evolutionary potential of the monkeypox genome arising from interactions with human APOBEC3 enzymes. Virus Evol. 2023, 2, vead047. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lum, F.-M.; Torres-Ruesta, A.; Tay, M.Z.; Lin, R.T.P.; Lye, D.C.; Rénia, L.; Ng, L.F.P. Monkeypox: Disease Epidemiology, Host Immunity and Clinical Interventions. Nat. Rev. Immunol. 2022, 22, 597–613. [Google Scholar] [CrossRef]
- Molteni, C.; Forni, D.; Cagliani, R.; Arrigoni, F.; Pozzoli, U.; De Gioia, L.; Sironi, M. Selective Events at Individual Sites Underlie the Evolution of Monkeypox Virus Clades. Virus Evol. 2023, 9, vead031. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus Entry and Membrane Fusion. Virology 2006, 344, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, X.; Liu, J.; Xiang, L.; Huang, S.; Xie, X.; Fang, L.; Lin, Y.; Zhang, M.; Wang, L.; et al. Phylogeny and molecular evolution of the first local monkeypox virus cluster in Guangdong Province, China. Nat. Commun. 2023, 14, 8241. [Google Scholar] [CrossRef] [PubMed]
- Paciello, I.; Rundlet, E.J.; Zhou, L.; Realini, G.; Perrone, F.; Pierleoni, G.; Troisi, M.; Postal, J.; Guivel-Benhassine, F.; Porrot, F.; et al. Antigen-agnostic identification of poxvirus broadly neutralizing antibodies targeting OPG153. Sci Transl Med. 2025, 10, eaeb3840. [Google Scholar] [CrossRef] [PubMed]
- Akther, S.; Su, M.; Wang, J.C.; Amin, H.; Taki, F.; De La Cruz, N.; Chowdhury, M.; Clabby, T.; Kopping, E.; Ruiz, V.E.; et al. Genomic Epidemiology of Mpox Virus during the 2022 Outbreak in New York City. Nat. Commun. 2025, 16, 8354. [Google Scholar] [CrossRef]
- O’Toole, Á.; Neher, R.A.; Ndodo, N.; Borges, V.; Gannon, B.; Gomes, J.P.; Groves, N.; King, D.J.; Maloney, D.; Lemey, P.; et al. Putative APOBEC3 deaminase editing in MPXV as evidence for sustained human transmission since at least 2016. bioRxiv 2023. [Google Scholar] [CrossRef]
- Brito, A.F.; Baele, G.; Nahata, K.D.; Grubaugh, N.D.; Pinney, J.W. Intrahost speciations and host switches played an important role in the evolution of herpesviruses. Virus Evol. 2021, 7, veab025. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Jung, Y.; Han, D. BWA-MEME: BWA-MEM Emulated with a Machine Learning Approach. Bioinformatics 2022, 38, 2404–2413. [Google Scholar] [CrossRef]
- Genovese, G.; Rockweiler, N.B.; Gorman, B.R.; Bigdeli, T.B.; Pato, M.T.; Pato, C.N.; Ichihara, K.; McCarroll, S.A. BCFtools/Liftover: An Accurate and Comprehensive Tool to Convert Genetic Variants across Genome Assemblies. Bioinformatics 2024, 40, btae038. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS. Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Marçais, G.; Delcher, A.L.; Phillippy, A.M.; Coston, R.; Salzberg, S.L.; Zimin, A. MUMmer4: A Fast and Versatile Genome Alignment System. PLoS. Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids. Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT Version 5: Improvement in Accuracy of Multiple Sequence Alignment. Nucleic Acids. Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids. Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R Language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Galili, T. Dendextend: An R Package for Visualizing, Adjusting and Comparing Trees of Hierarchical Clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- The EMBOSS Project. The EMBOSS Website 2024. Available online: https://www.ebi.ac.uk/jdispatcher/emboss (accessed on 21 August 2025).
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids. Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Bertoni, D.; Tsenkov, M.; Magana, P.; Nair, S.; Pidruchna, I.; Querino, L.A.M.; Midlik, A.; Paramval, U.; Lawal, D.; Tanweer, A.; et al. AlphaFold Protein Structure Database 2025: A redesigned interface and updated structural coverage. Nucleic Acids Res. 2026, 54, D358–D362. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

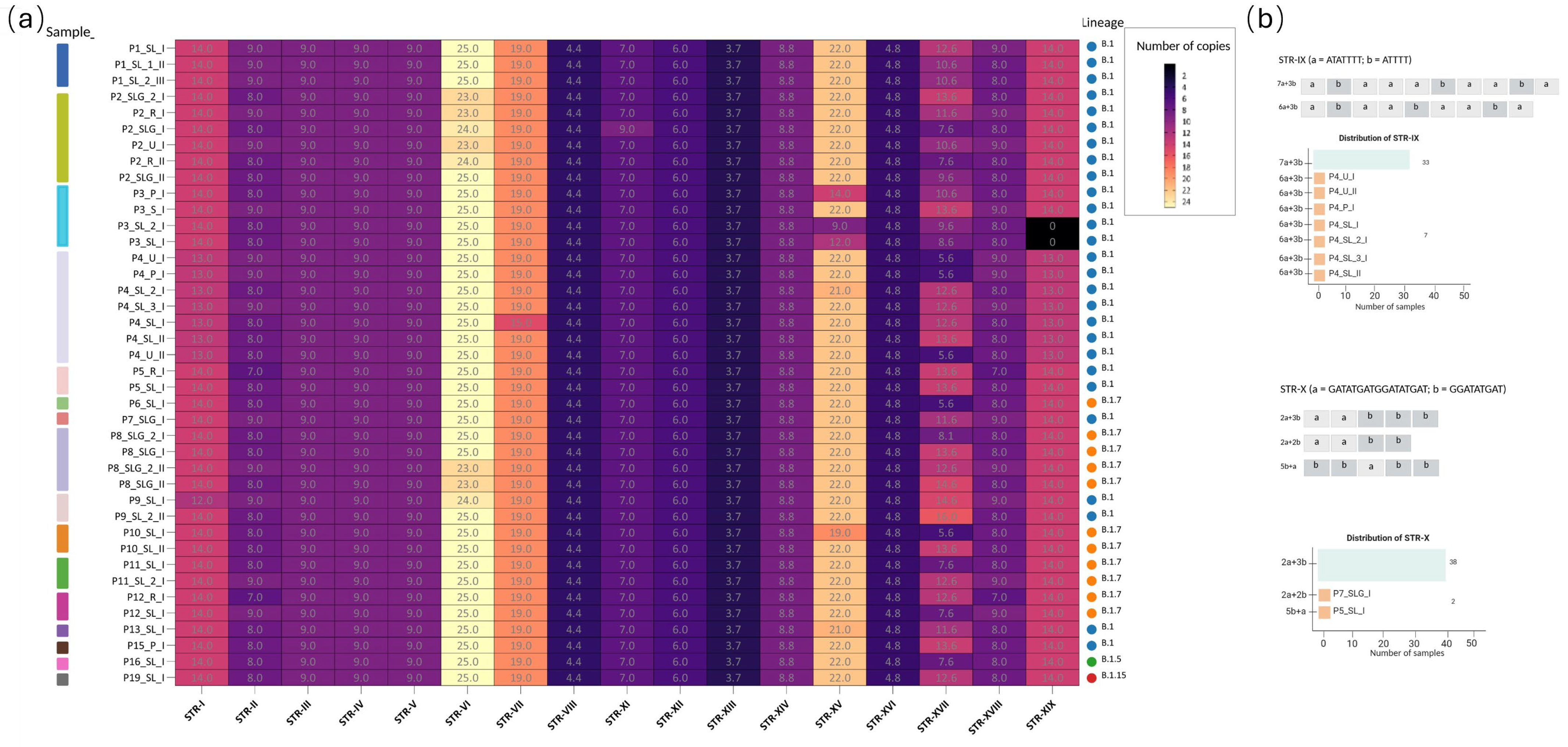
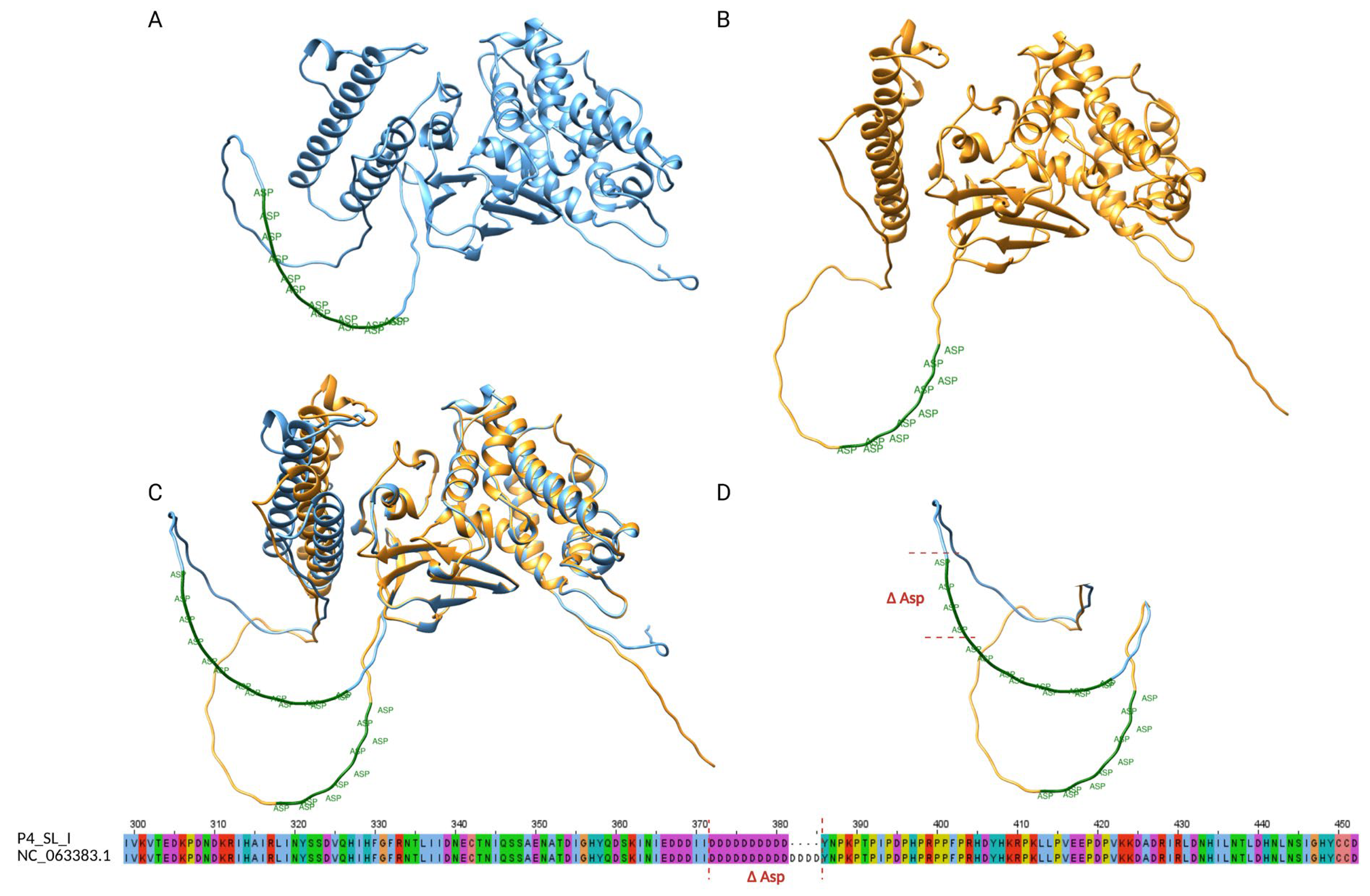
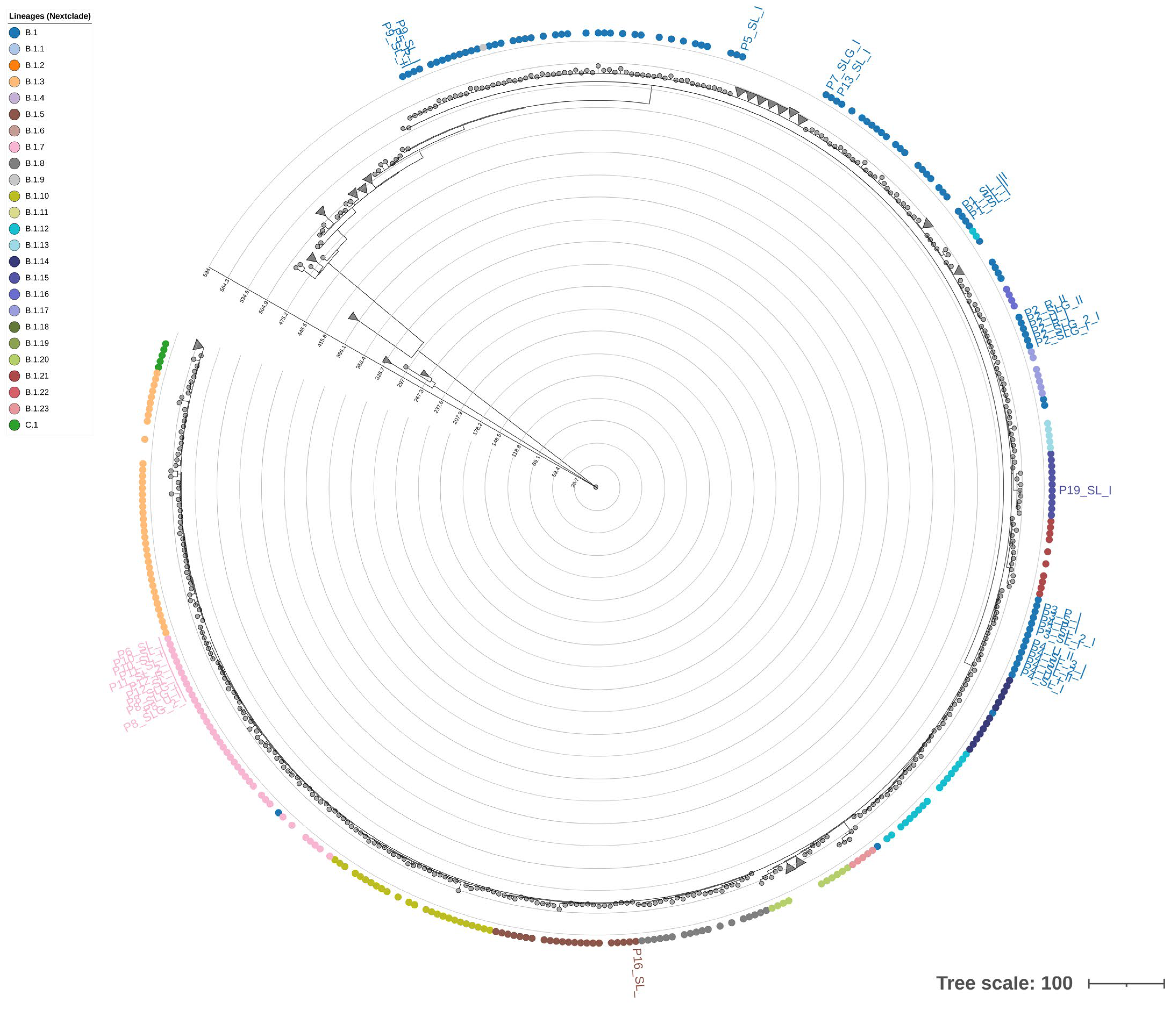
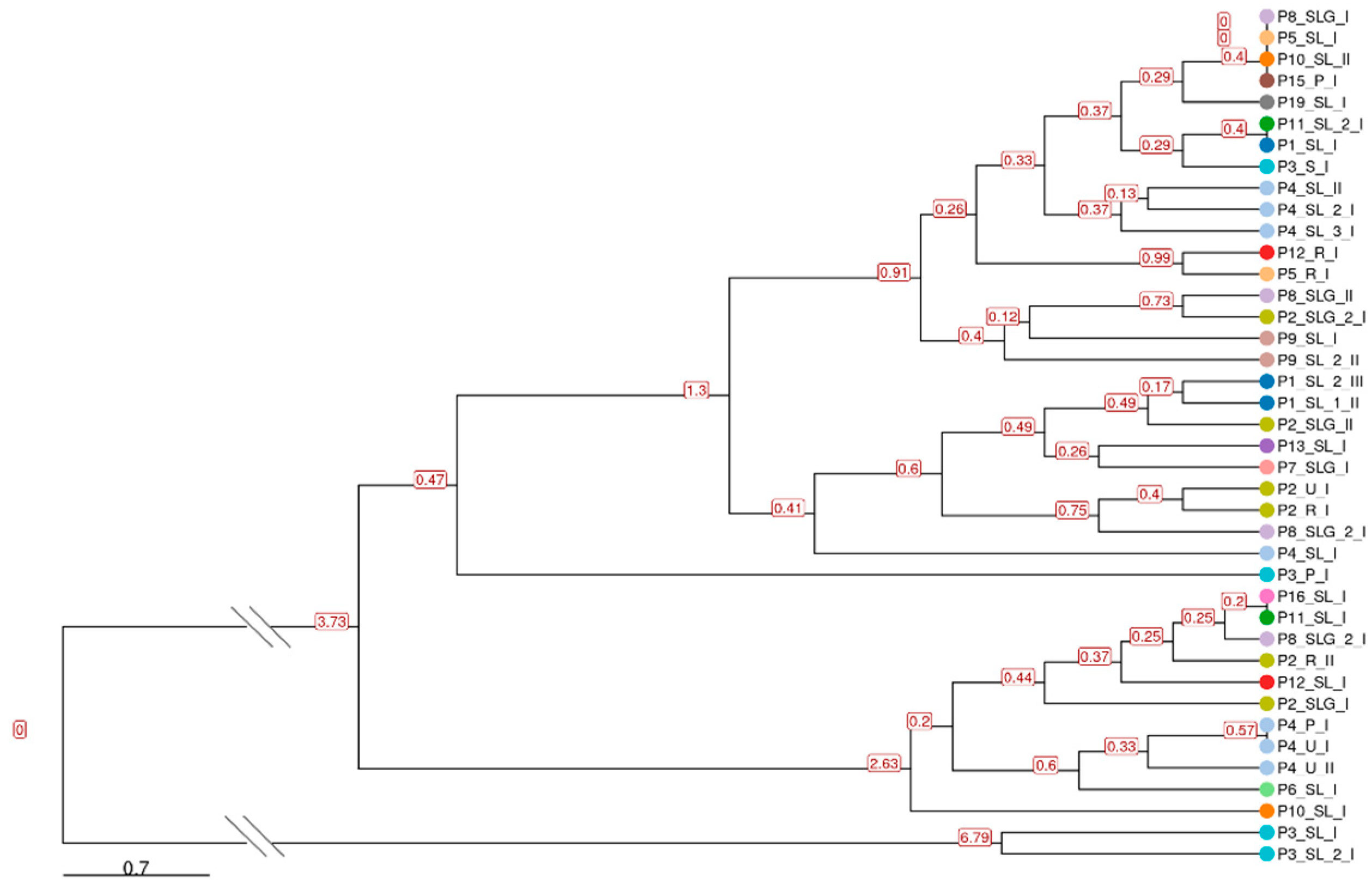
| Name | Correspondence with Monzòn et al. [24] | Start | End | Pattern | # Max Rep | Str length (bp) | # Samples Sharing the STR | Nearest Gene | Relative Position to the Gene | Distance in bp | Functional Insight |
|---|---|---|---|---|---|---|---|---|---|---|---|
| STR-I | LCR10 | 595 | 611 | [T] | 14 | 14 | 40/40 | OPG001 | Upstream | −224 | Chemokine-binding protein |
| STR-II | LCR1 * | 4677 | 4801 | [TAACTAACTTATGACT] | 9 | 144 | 40/40 | OPG015 | Downstream | 37 | Ankyrin domains and immune evasion (TNF mimic) |
| STR-III | LCR12 | 28,527 | 28,537 | [A] | 9 | 9 | 40/40 | OPG044 | Intragene | NA | Bcl-2-like protein |
| STR-IV | LCR13 | 76,082 | 76,090 | [T] | 9 | 9 | 40/40 | OPG097 | Intragene | NA | Unknown |
| STR-V | LCR14 | 80,844 | 80,825 | [T] | 9 | 9 | 40/40 | OPG104 | Intragene | NA | Unknown |
| STR-VI | LCR5 | 133,074 | 133,098 | [T] | 25 | 25 | 40/40 | OPG151 | Upstream | −657 | Unknown |
| STR-VII | LCR7 | 136,498 | 136,555 | [ATC]n + TATGAT + [ATC]n | 19 | 57 | 40/40 | OPG153 | Intragene | NA | Key attachment/egress protein |
| STR-VIII | LCR15 | 140,092 | 140,131 | [ATAACAATT] | 4.4 | 39 | 40/40 | OPG159 | Intragene | NA | Unknown |
| STR-IX | LCR8 | 146,835 | 146,920 | [ATATTTT]n + [ATTTT]n | 10 | 64 | 40/40 | OPG170 | Upstream | −57 | Unknown |
| STR-X | LCR9 * | 150,544 | 150,621 | [GATATGATGGATATGAT]n + [GGATATGAT]n | 5 | 62 | 40/40 | OPG176 | Intragene | NA | Bcl-2-like protein |
| STR-XI | LCR16 | 152,661 | 152,566 | [A] | 7 | 9 | 40/40 | OPG180 | Upstream | −31 | Unknown |
| STR-XII | LCR17 | 1631,69 | 163,207 | [TAAC] | 6 | 24 | 40/40 | OPG188 | Upstream | −63 | Unknown |
| STR-XIII | LCR18 | 166,070 | 166,109 | [AATAATT] | 3.7 | 39 | 40/40 | OPG190 | Intragene | NA | IFNα/β binding protein homolog |
| STR-XIV | LCR19 * | 169,709 | 169,761 | [CAGATA] | 8.8 | 52 | 40/40 | OPG197 | Intragene | NA | Unknown |
| STR-XV | LCR2 * | 173,252 | 173,295 | [AT] | 22 | 43 | 40/40 | OPG200 | Upstream | −654 | Unknown |
| STR-XVI | LCR21 | 174,506 | 174,538 | [GATGAA] | 4.8 | 32 | 40/40 | OPG204 | Intragene | NA | Interferon decoy receptor; immune modulation |
| STR-XVII | LCR3 * | 179,055 | 179,245 | [ATATACATT] | 16 | 144 | 40/40 | OPG208 | Downstream | 34 | Serine protease inhibitor (anti-apoptotic, potential virulence) |
| STR-XVIII | LCR4 | 192,382 | 192,534 | [AGTCATAAGTTAGTTA] | 9 | 144 | 40/40 | OPG015 | Upstream | −16 | Ankyrin domains and immune evasion (TNF mimic) |
| STR-XIX | LCR11 * | 196,608 | 196,624 | [A] | 14 | 14 | 38/40 | OPG001 | Downstream | 233 | Chemokine-binding protein |
| P. | Matrix | Timepoint | Sample_Id | Collection_Date | Travel_History | qPCR |
|---|---|---|---|---|---|---|
| P1 | Skin lesion swab | I | P1_SL_I | 25 May2022 | Mallorca | 21.00 |
| P1 | Skin lesion swab | II | P1_SL_1_II | 03 June 2022 | Mallorca | 21.51 |
| P1 | Skin lesion swab | III | P1_SL_2_III | 07 June 2022 | Mallorca | 21.00 |
| P2 | Skin lesion swab (genital) | I | P2_SLG_I | 18 July 2022 | Spain | 18.96 |
| P2 | urine | I | P2_U_I | 18 July 2022 | Spain | 25.91 |
| P2 | Rectal swab | I | P2_R_I | 18 July 2022 | Spain | 18.52 |
| P2 | Skin lesion swab (genital) | I | P2_SLG_2_I | 18 July 2022 | Spain | 17.07 |
| P2 | Skin lesion swab (genital) | II | P2_SLG_II | 21 July 2022 | Spain | 20.96 |
| P2 | Rectal swab | II | P2_R_II | 21 July 2022 | Spain | 20.63 |
| P3 | Skin lesion swab | I | P3_SL_I | 18 October 2022 | None | 14.70 |
| P3 | Pharyngeal swab | I | P3_P_I | 18 October 2022 | None | 16.01 |
| P3 | Saliva | I | P3_S_I | 18 October 2022 | None | 20.06 |
| P3 | Skin lesion swab (head) | I | P3_SL_2_I | 18 October2022 | None | 18.99 |
| P4 | Skin lesion swab (abdomen) | I | P4_SL_I | 02 November 2022 | Bosnia-Herzegovina | 16.70 |
| P4 | Skin lesion swab (hand) | I | P4_SL_2_I | 02 November 2022 | Bosnia-Herzegovina | 14.40 |
| P4 | Skin lesion swab (head) | I | P4_SL_3_I | 02 November 2022 | Bosnia-Herzegovina | 14.00 |
| P4 | Pharyngeal swab | I | P4_P_I | 02 November 2022 | Bosnia-Herzegovina | 23.10 |
| P4 | Urine | I | P4_U_I | 02 November 2022 | Bosnia-Herzegovina | 25.80 |
| P4 | Skin lesion swab (leg) | II | P4_SL_II | 02 November 2022 | Bosnia-Herzegovina | 25.30 |
| P4 | Urine | II | P4_U_II | 02 November 2022 | Bosnia-Herzegovina | 25.76 |
| P5 | Skin lesion swab | I | P5_SL_I | 23 June 2022 | Spain | 22.19 |
| P5 | Rectal swab | I | P5_R_I | 23 June 2022 | Spain | 18.78 |
| P6 | Skin lesion swab | I | P6_SL_I | 21 July 2022 | None | 26.65 |
| P7 | Skin lesion swab (genital) | I | P7_SLG_I | 01 July 2022 | None | 21.02 |
| P8 | Skin lesion swab (genital) | I | P8_SLG_I | 15 July 2022 | None | 19.83 |
| P8 | Skin lesion swab (genital) | I | P8_SLG_2_I | 15 July 2022 | None | 15.05 |
| P8 | Skin lesion swab (genital) | II | P8_SLG_II | 21 July 2022 | None | 19.07 |
| P8 | Urine | II | P8_SLG_2_II | 21 July 2022 | None | 25.10 |
| P9 | Skin lesion swab | I | P9_SL_I | 01 July 2022 | USA | 26.67 |
| P9 | Skin lesion swab | II | P9_SL_2_II | 01 July 2022 | USA | 22.74 |
| P10 | Skin lesion swab | I | P10_SL_I | 26 July2022 | None | 24.05 |
| P10 | Skin lesion swab | II | P10_SL_II | 29 July 2022 | None | 19.05 |
| P11 | Skin lesion swab (abdomen) | I | P11_SL_I | 05 August 2022 | None | 20.17 |
| P11 | Skin lesion swab (arm) | I | P11_SL_2_I | 05 August 2022 | None | 19.32 |
| P12 | Skin lesion swab (head) | I | P12_SL_I | 18 August 2022 | None | 18.06 |
| P12 | Rectal swab | I | P12_R_I | 18 August 2022 | None | 25.25 |
| P13 | Skin lesion swab | I | P13_SL_I | 19September 2022 | None | 21.31 |
| P15 | Pharyngeal swab | I | P15_P_I | 08 July 2022 | None | 27.03 |
| P16 | Skin lesion swab | I | P16_SL_I | NA | NA | NA |
| P19 | Skin lesion swab | I | P19_SL_I | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Deiana, M.; Locatelli, E.; Veschetti, L.; Malagò, S.; Mori, A.; Lavezzari, D.; Accordini, S.; Ronzoni, N.; Angheben, A.; Malerba, G.; et al. Insights into Genomic Dynamics and Plasticity in the Monkeypox Virus from the 2022 Outbreak. Int. J. Mol. Sci. 2026, 27, 1371. https://doi.org/10.3390/ijms27031371
Deiana M, Locatelli E, Veschetti L, Malagò S, Mori A, Lavezzari D, Accordini S, Ronzoni N, Angheben A, Malerba G, et al. Insights into Genomic Dynamics and Plasticity in the Monkeypox Virus from the 2022 Outbreak. International Journal of Molecular Sciences. 2026; 27(3):1371. https://doi.org/10.3390/ijms27031371
Chicago/Turabian StyleDeiana, Michela, Elena Locatelli, Laura Veschetti, Simone Malagò, Antonio Mori, Denise Lavezzari, Silvia Accordini, Niccolò Ronzoni, Andrea Angheben, Giovanni Malerba, and et al. 2026. "Insights into Genomic Dynamics and Plasticity in the Monkeypox Virus from the 2022 Outbreak" International Journal of Molecular Sciences 27, no. 3: 1371. https://doi.org/10.3390/ijms27031371
APA StyleDeiana, M., Locatelli, E., Veschetti, L., Malagò, S., Mori, A., Lavezzari, D., Accordini, S., Ronzoni, N., Angheben, A., Malerba, G., Tacconelli, E., Cusi, M. G., Gobbi, F. G., Piubelli, C., & Castilletti, C. (2026). Insights into Genomic Dynamics and Plasticity in the Monkeypox Virus from the 2022 Outbreak. International Journal of Molecular Sciences, 27(3), 1371. https://doi.org/10.3390/ijms27031371











