From Data to Decision: Integrating Bioinformatics into Glioma Patient Stratification and Immunotherapy Selection
Abstract
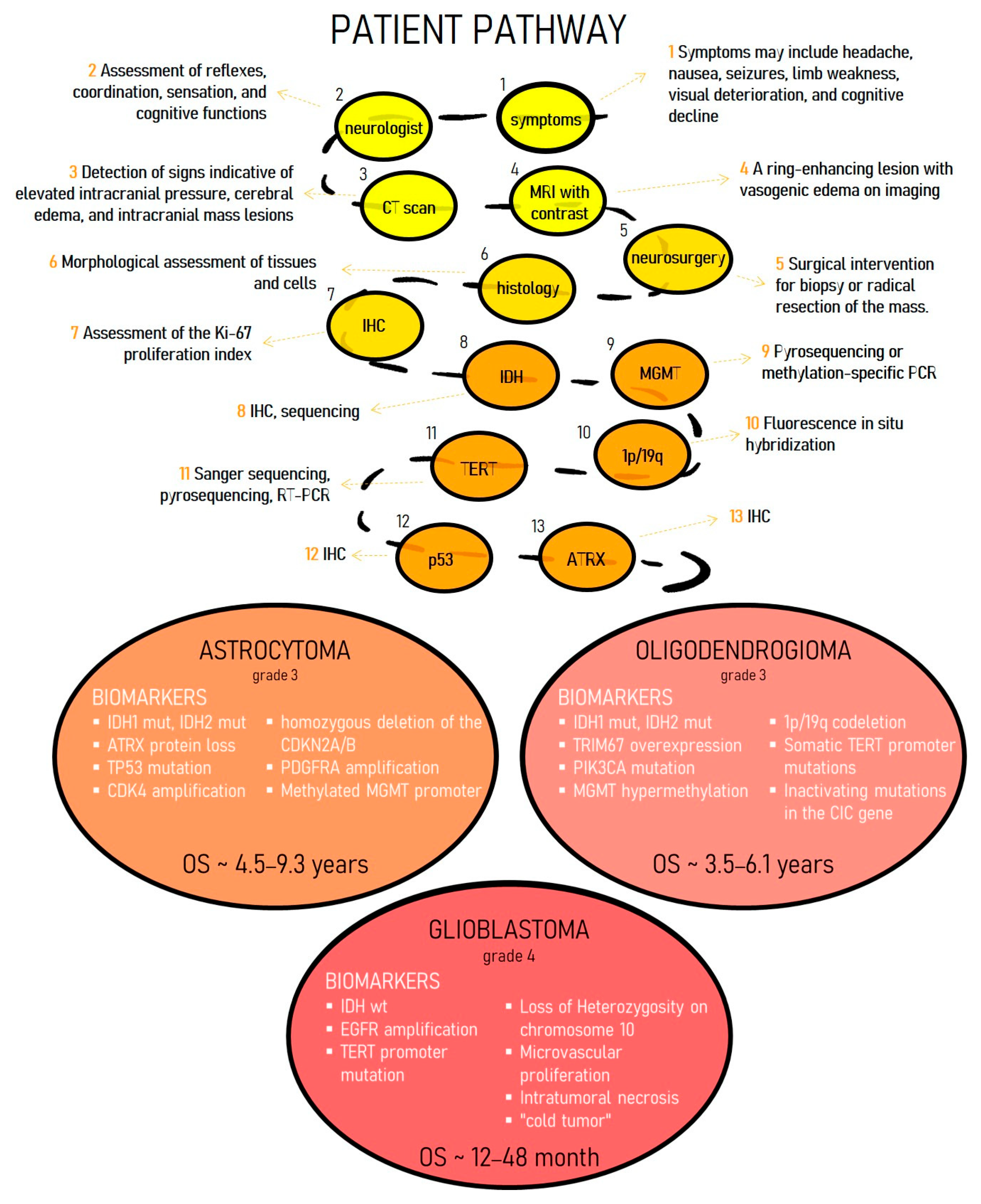
1. Introduction
2. Insufficiency of Standard Diagnostic Criteria for Glioma: Clinical Case Examples
3. Diverse Data Sources for Constructing Classifiers
4. Leveraging NGS Data for Glioma Classification
5. A Methylation-Based Glioma Classifier
6. The Application of Transcriptome Data for Glioma Classifier Development
7. A Multi-Dimensional View: Classifying Glioma Through an Integrative Omics Lens
8. Explainable Artificial Intelligence in Glioma Classification
9. Machine Learning Approaches for Predicting Immunotherapy Response in Glioma
10. Discussion
11. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | artificial intelligence |
| AIPS | artificial intelligence prognostic signature |
| APC | antigen-presenting cell |
| CGGA | Chinese Glioma Genome Atlas |
| CDKN | Cyclin-Dependent Kinase Inhibitor |
| CHGB | Chromogranin B |
| CLEC7A | C-type lectin domain family 7 member A |
| CNN | convolutional neural network |
| CNS | central nervous system |
| CSF | cerebrospinal fluid |
| CT | computed tomography |
| CTLA4 | Cytotoxic T-Lymphocyte-Associated protein 4 |
| CXCL | C-X-C motif chemokine ligand |
| ctDNA | circulating tumor DNA |
| ddPCR | droplet digital PCR |
| DEG | differential gene expression |
| DE-IRGs | differentially expressed invasion-related genes |
| DMRs | differentially methylated regions |
| DNA | deoxyribonucleic acid |
| EGFR | Epidermal Growth Factor Receptor |
| FLT3 | fms-like tyrosine kinase 3 |
| GBM | glioblastoma |
| GEO | Gene Expression Omnibus |
| GO | Gene ontology |
| HGAP | high-grade astrocytoma with piloid features |
| HGG | high-grade glioma |
| ICD | immunogenic cell death |
| ICG | immune checkpoint gene |
| IDH | Isocitrate Dehydrogenase |
| IGRPS | Immune-Gene-Related Prognostic Score |
| IL | Interleukin |
| IRGs | immune-related genes |
| LAG-3 | Lymphocyte-Activation Gene 3 |
| LASSO | least absolute shrinkage and selection operato |
| LGG | low-grade glioma |
| MINGLE | Multi-omics Integrated Network for Graphical Exploration |
| ML | machine learning |
| MRI | magnetic resonance imaging |
| mRNA | messenger ribonucleic acid |
| NGS | next-generation sequencing |
| NMF | nonnegative matrix factorization |
| PAM | prediction analysis of microarray |
| PCR | polymerase chain reaction |
| PD-1 | Programmed Cell Death protein 1 |
| PD-L1 | Programmed Death-Ligand 1 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PRM | prognostic risk model |
| SHAP | Shapley Additive Explanations |
| ssGSVA | single-sample gene set variation analysis |
| SVM | support vector machine |
| OS | overall survival |
| TAM | tumor-associated macrophages |
| TCGA | The Cancer Genome Atlas |
| TIDE | tumor immune dysfunction and exclusion |
| TLR | Toll-like receptors |
| TMB | tumor mutation burden |
| VEGFA | vascular endothelial growth factor A |
| VTN | Vitronectin |
| XAI | explainable artificial intelligence |
References
- Anjum, K.; Shagufta, B.I.; Abbas, S.Q.; Patel, S.; Khan, I.; Shah, S.A.A.; Akhter, N.; Hassan, S.S. Current Status and Future Therapeutic Perspectives of Glioblastoma Multiforme (GBM) Therapy: A Review. Biomed. Pharmacother. 2017, 92, 681–689, Erratum in Biomed. Pharmacother. 2018, 101, 820. [Google Scholar] [CrossRef]
- Lu, C.; Kang, T.; Zhang, J.; Yang, K.; Liu, Y.; Song, K.; Lin, Q.; Dixit, D.; Gimple, R.C.; Zhang, Q.; et al. Combined Targeting of Glioblastoma Stem Cells of Different Cellular States Disrupts Malignant Progression. Nat. Commun. 2025, 16, 2974. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yan, X.; Hu, S. Glioma Stem Cells: Drivers of Tumor Progression and Recurrence. Stem Cell Res. Ther. 2025, 16, 293. [Google Scholar] [CrossRef] [PubMed]
- Yalamarty, S.S.K.; Filipczak, N.; Li, X.; Subhan, M.A.; Parveen, F.; Ataide, J.A.; Rajmalani, B.A.; Torchilin, V.P. Mechanisms of Resistance and Current Treatment Options for Glioblastoma Multiforme (GBM). Cancers 2023, 15, 2116. [Google Scholar] [CrossRef]
- Auffinger, B.; Spencer, D.; Pytel, P.; Ahmed, A.U.; Lesniak, M.S. The Role of Glioma Stem Cells in Chemotherapy Resistance and Glioblastoma Multiforme Recurrence. Expert Rev. Neurother. 2015, 15, 741–752. [Google Scholar] [CrossRef]
- Celiku, O.; Gilbert, M.R.; Lavi, O. Computational Modeling Demonstrates That Glioblastoma Cells Can Survive Spatial Environmental Challenges through Exploratory Adaptation. Nat. Commun. 2019, 10, 5704. [Google Scholar] [CrossRef]
- Berger, F.; Gay, E.; Pelletier, L.; Tropel, P.; Wion, D. Development of Gliomas: Potential Role of Asymmetrical Cell Division of Neural Stem Cells. Lancet Oncol. 2004, 5, 511–514. [Google Scholar] [CrossRef]
- Ling, A.L.; Solomon, I.H.; Landivar, A.M.; Nakashima, H.; Woods, J.K.; Santos, A.; Masud, N.; Fell, G.; Mo, X.; Yilmaz, A.S.; et al. Clinical Trial Links Oncolytic Immunoactivation to Survival in Glioblastoma. Nature 2023, 623, 157–166. [Google Scholar] [CrossRef]
- Chen, L.; Ma, J.; Zou, Z.; Liu, H.; Liu, C.; Gong, S.; Gao, X.; Liang, G. Clinical Characteristics and Prognosis of Patients with Glioblastoma: A Review of Survival Analysis of 1674 Patients Based on SEER Database. Medicine 2022, 101, e32042. [Google Scholar] [CrossRef] [PubMed]
- AlSamhori, J.F.; Khan, M.; Hamed, B.M.; Saleem, M.A.; Shahid, F.; Rath, S.; Nabi, W.; Aziz, N.; Shahnawaz, M.; Basharat, A.; et al. Understanding Glioblastoma Survival: A SEER-Based Study of Demographics and Treatment. J. Clin. Oncol. 2025, 43, e14027. [Google Scholar] [CrossRef]
- Bertolini, G.; Trombini, T.; Zenesini, C.; Mazza, S.; Dascola, I.; Pavarani, A.; Michiara, M.; Ceccon, G.; Peluso, A.; Calamo Specchia, F.M.; et al. Prognostic Impact of Second Surgical Resection in IDH Wildtype Recurrent Glioblastoma Following Chemo-Radiation Therapy: A Propensity Score Analysis Cohort Study. J. Neuro-Oncol. 2025, 174, 789–798. [Google Scholar] [CrossRef]
- Martin-Villalba, A.; Okuducu, A.F.; Von Deimling, A. The Evolution of Our Understanding on Glioma. Brain Pathol. 2008, 18, 455–463. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Bao, J.; Pan, Z.; Wei, S. Unlocking New Horizons: Advances in Treating IDH-Mutant, 1p/19q-Codeleted Oligodendrogliomas. Discov. Oncol. 2025, 16, 971. [Google Scholar] [CrossRef]
- Tang, X.; Wei, W.; Huang, Y.; Zheng, Y.; Xu, Y.; Li, F. A Case Report on IDH-Mutant Astrocytoma, CNS WHO Grade 4: Multi-Omic Characterization of Untreated Clinical Progression. Front. Oncol. 2025, 15, 1557245. [Google Scholar] [CrossRef]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186, Erratum in Nat. Rev. Clin. Oncol. 2022, 19, 357–358. [Google Scholar] [CrossRef]
- Teng, Y.; Chen, C.; Zhang, Y.; Xu, J. The Feasibility of MRI Texture Analysis in Distinguishing Glioblastoma, Anaplastic Astrocytoma and Anaplastic Oligodendroglioma. Transl. Cancer Res. 2022, 11, 4079–4088. [Google Scholar] [CrossRef]
- van Lent, D.I.; van Baarsen, K.M.; Snijders, T.J.; Robe, P.A.J.T. Radiological Differences between Subtypes of WHO 2016 Grade II–III Gliomas: A Systematic Review and Meta-Analysis. Neuro-Oncol. Adv. 2020, 2, vdaa044. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, S.; Ma, C.; Fang, W.; Jing, X.; Yang, C.; Li, H.; Zhang, X.; Ge, C.; Liu, B.; et al. Glioma Subtype Prediction Based on Radiomics of Tumor and Peritumoral Edema under Automatic Segmentation. Sci. Rep. 2024, 14, 27471. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, V.E.; Solheim, O.; Salvesen, Ø.; Torp, S.H. The Histological Representativeness of Glioblastoma Tissue Samples. Acta Neurochir. 2021, 163, 1911–1920. [Google Scholar] [CrossRef]
- Anwer, M.S.; Abdel-Rasol, M.A.; El-Sayed, W.M. Emerging Therapeutic Strategies in Glioblastsoma: Drug Repurposing, Mechanisms of Resistance, Precision Medicine, and Technological Innovations. Clin. Exp. Med. 2025, 25, 117. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial Intelligence in Cancer Diagnosis and Prognosis: Opportunities and Challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hou, P.; Li, X.; Xiao, M.; Zhang, Z.; Li, Z.; Xu, J.; Liu, G.; Tan, Y.; Fang, C. Comprehensive Understanding of Glioblastoma Molecular Phenotypes: Classification, Characteristics, and Transition. Cancer Biol. Med. 2024, 21, 20230510. [Google Scholar] [CrossRef]
- Al Ghafari, M.; El Jaafari, N.; Mouallem, M.; Maassarani, T.; El Sibai, M.; Abi-Habib, R. Key Genes Altered in Glioblastoma Based on Bioinformatics (Review). Oncol. Lett. 2025, 29, 243. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Qiu, J.; Xu, J.; Yang, X.; Zhang, Y.; Tian, X.; Guan, X.; Cen, X.; Zhao, Y. Tumor Biomarkers for Diagnosis, Prognosis and Targeted Therapy. Signal Transduct. Target. Ther. 2024, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.D.; Madhavi, K. Enhancing Bone Cancer Detection through Optimized Pre Trained Deep Learning Models and Explainable AI Using the Osteosarcoma Tumor Assessment Dataset. Sci. Rep. 2025, 15, 39104. [Google Scholar] [CrossRef] [PubMed]
- Mehedi, H.K.; Khandaker, M.; Ara, S.; Alam, A.; Mridha, M.F.; Aung, Z. A Lightweight Deep Learning Method to Identify Different Types of Cervical Cancer. Sci. Rep. 2024, 14, 29446. [Google Scholar] [CrossRef]
- Munquad, S.; Si, T.; Mallik, S.; Li, A.; Das, A.B. Subtyping and Grading of Lower-Grade Gliomas Using Integrated Feature Selection and Support Vector Machine. Brief. Funct. Genom. 2022, 21, 408–421. [Google Scholar] [CrossRef]
- Niu, B.; Liang, C.; Lu, Y.; Zhao, M.; Chen, Q.; Zhang, Y.; Zheng, L.; Chou, K.-C. Glioma Stages Prediction Based on Machine Learning Algorithm Combined with Protein-Protein Interaction Networks. Genomics 2020, 112, 837–847. [Google Scholar] [CrossRef]
- Cai, Y.-D.; Zhang, S.; Zhang, Y.-H.; Pan, X.; Feng, K.; Chen, L.; Huang, T.; Kong, X. Identification of the Gene Expression Rules That Define the Subtypes in Glioma. J. Clin. Med. 2018, 7, 350. [Google Scholar] [CrossRef]
- Vieira, F.G.; Bispo, R.; Lopes, M.B. Integration of Multi-Omics Data for the Classification of Glioma Types and Identification of Novel Biomarkers. Bioinform. Biol. Insights 2024, 18, 11779322241249563. [Google Scholar] [CrossRef]
- Lin, Z.; Wang, R.; Huang, C.; He, H.; Ouyang, C.; Li, H.; Zhong, Z.; Guo, J.; Chen, X.; Yang, C.; et al. Identification of an Immune-Related Prognostic Risk Model in Glioblastoma. Front. Genet. 2022, 13, 926122. [Google Scholar] [CrossRef]
- Luo, X.; Wang, Q.; Tang, H.; Chen, Y.; Li, X.; Chen, J.; Zhang, X.; Li, Y.; Sun, J.; Han, S. A Novel Immune Gene-Related Prognostic Score Predicts Survival and Immunotherapy Response in Glioma. Medicina 2022, 59, 23. [Google Scholar] [CrossRef]
- Guo, K.; Yang, J.; Jiang, R.; Ren, X.; Liu, P.; Wang, W.; Zhou, S.; Wang, X.; Ma, L.; Hu, Y. Identification of Key Immune and Cell Cycle Modules and Prognostic Genes for Glioma Patients through Transcriptome Analysis. Pharmaceuticals 2024, 17, 1295. [Google Scholar] [CrossRef]
- Chen, G.; He, Z.; Jiang, W.; Li, L.; Luo, B.; Wang, X.; Zheng, X. Construction of a Machine Learning-Based Artificial Neural Network for Discriminating PANoptosis Related Subgroups to Predict Prognosis in Low-Grade Gliomas. Sci. Rep. 2022, 12, 22119. [Google Scholar] [CrossRef]
- Li, H.; He, J.; Li, M.; Li, K.; Pu, X.; Guo, Y. Immune Landscape-Based Machine-Learning–Assisted Subclassification, Prognosis, and Immunotherapy Prediction for Glioblastoma. Front. Immunol. 2022, 13, 1027631. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Luo, A.; Hu, S.; Ye, G.; Li, D.; Zhao, H.; Peng, B. Genomics and Proteomics to Determine Novel Molecular Subtypes and Predict the Response to Immunotherapy and the Effect of Bevacizumab in Glioblastoma. Sci. Rep. 2024, 14, 17630. [Google Scholar] [CrossRef] [PubMed]
- Romo, C.G.; Palsgrove, D.N.; Sivakumar, A.; Elledge, C.R.; Kleinberg, L.R.; Chaichana, K.L.; Gocke, C.D.; Rodriguez, F.J.; Holdhoff, M. Widely Metastatic IDH1-Mutant Glioblastoma with Oligodendroglial Features and Atypical Molecular Findings: A Case Report and Review of Current Challenges in Molecular Diagnostics. Diagn. Pathol. 2019, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.A.; Bao, S.; Kong, X.-T.; Terry, M.; Himstead, A.; Zheng, M.; Pekmezci, M. A High-Grade Glioma, Not Elsewhere Classified in an Older Adult with Discordant Genetic and Epigenetic Analyses. Biomedicines 2024, 12, 2042. [Google Scholar] [CrossRef]
- Ballester, L.Y.; Huse, J.T.; Tang, G.; Fuller, G.N. Molecular Classification of Adult Diffuse Gliomas: Conflicting IDH1/IDH2, ATRX, and 1p/19q Results. Hum. Pathol. 2017, 69, 15–22. [Google Scholar] [CrossRef]
- Karimi, S.; Zuccato, J.A.; Mamatjan, Y.; Mansouri, S.; Suppiah, S.; Nassiri, F.; Diamandis, P.; Munoz, D.G.; Aldape, K.D.; Zadeh, G. The Central Nervous System Tumor Methylation Classifier Changes Neuro-Oncology Practice for Challenging Brain Tumor Diagnoses and Directly Impacts Patient Care. Clin. Epigenetics 2019, 11, 185. [Google Scholar] [CrossRef]
- Penson, A.; Camacho, N.; Zheng, Y.; Varghese, A.M.; Al-Ahmadie, H.; Razavi, P.; Chandarlapaty, S.; Vallejo, C.E.; Vakiani, E.; Gilewski, T.; et al. Development of Genome-Derived Tumor Type Prediction to Inform Clinical Cancer Care. JAMA Oncol. 2020, 6, 84–91. [Google Scholar] [CrossRef]
- Raufaste-Cazavieille, V.; Santiago, R.; Droit, A. Multi-Omics Analysis: Paving the Path toward Achieving Precision Medicine in Cancer Treatment and Immuno-Oncology. Front. Mol. Biosci. 2022, 9, 962743. [Google Scholar] [CrossRef]
- Park, Y.; Heider, D.; Hauschild, A.-C. Integrative Analysis of Next-Generation Sequencing for Next-Generation Cancer Research toward Artificial Intelligence. Cancers 2021, 13, 3148. [Google Scholar] [CrossRef] [PubMed]
- Nicora, G.; Vitali, F.; Dagliati, A.; Geifman, N.; Bellazzi, R. Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools. Front. Oncol. 2020, 10, 1030. [Google Scholar] [CrossRef]
- Pan, Y.-T.; Lin, Y.-P.; Yen, H.-K.; Yen, H.-H.; Huang, C.-C.; Hsieh, H.-C.; Janssen, S.; Hu, M.-H.; Lin, W.-H.; Groot, O.Q. Are Current Survival Prediction Tools Useful When Treating Subsequent Skeletal-Related Events from Bone Metastases? Clin. Orthop. Relat. Res. 2024, 482, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R.; et al. Clinical Applications of Artificial Intelligence and Machine Learning in Cancer Diagnosis: Looking into the Future. Cancer Cell Int. 2021, 21, 270. [Google Scholar] [CrossRef] [PubMed]
- Badillo, S.; Banfai, B.; Birzele, F.; Davydov, I.I.; Hutchinson, L.; Kam-Thong, T.; Siebourg-Polster, J.; Steiert, B.; Zhang, J.D. An Introduction to Machine Learning. Clin. Pharmacol. Ther. 2020, 107, 871–885. [Google Scholar] [CrossRef]
- Rai, H.M.; Yoo, J. A Comprehensive Analysis of Recent Advancements in Cancer Detection Using Machine Learning and Deep Learning Models for Improved Diagnostics. J. Cancer Res. Clin. Oncol. 2023, 149, 14365–14408. [Google Scholar] [CrossRef]
- Taye, M.M. Understanding of Machine Learning with Deep Learning: Architectures, Workflow, Applications and Future Directions. Computers 2023, 12, 91. [Google Scholar] [CrossRef]
- Wei, L.; Niraula, D.; Gates, E.D.H.; Fu, J.; Luo, Y.; Nyflot, M.J.; Bowen, S.R.; El Naqa, I.M.; Cui, S. Artificial Intelligence (AI) and Machine Learning (ML) in Precision Oncology: A Review on Enhancing Discoverability through Multiomics Integration. Br. J. Radiol. 2023, 96, 20230211. [Google Scholar] [CrossRef]
- Gui, Y.; He, X.; Yu, J.; Jing, J. Artificial Intelligence-Assisted Transcriptomic Analysis to Advance Cancer Immunotherapy. J. Clin. Med. 2023, 12, 1279. [Google Scholar] [CrossRef]
- Wu, M.; Luan, J.; Zhang, D.; Fan, H.; Qiao, L.; Zhang, C. Development and Validation of a Clinical Prediction Model for Glioma Grade Using Machine Learning. Technol. Health Care 2024, 32, 1977–1990. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA Methylation-Based Classification of Central Nervous System Tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Arteaga-Arteaga, H.B.; Candamil-Cortés, M.S.; Breaux, B.; Guillen-Rondon, P.; Orozco-Arias, S.; Tabares-Soto, R. Machine Learning Applications on Intratumoral Heterogeneity in Glioblastoma Using Single-Cell RNA Sequencing Data. Brief. Funct. Genom. 2023, 22, 428–441, Erratum in Brief. Funct. Genom. 2024, 23, 679. [Google Scholar] [CrossRef]
- Takahashi, S.; Takahashi, M.; Tanaka, S.; Takayanagi, S.; Takami, H.; Yamazawa, E.; Nambu, S.; Miyake, M.; Satomi, K.; Ichimura, K.; et al. A New Era of Neuro-Oncology Research Pioneered by Multi-Omics Analysis and Machine Learning. Biomolecules 2021, 11, 565. [Google Scholar] [CrossRef] [PubMed]
- Tambi, R.; Zehra, B.; Vijayakumar, A.; Satsangi, D.; Uddin, M.; Berdiev, B.K. Artificial Intelligence and Omics in Malignant Gliomas. Physiol. Genom. 2024, 56, 876–895. [Google Scholar] [CrossRef] [PubMed]
- Kleppe, A.; Skrede, O.-J.; De Raedt, S.; Liestøl, K.; Kerr, D.J.; Danielsen, H.E. Designing Deep Learning Studies in Cancer Diagnostics. Nat. Rev. Cancer 2021, 21, 199–211. [Google Scholar] [CrossRef]
- Wang, F.; Kaushal, R.; Khullar, D. Should Health Care Demand Interpretable Artificial Intelligence or Accept “Black Box” Medicine? Ann. Intern. Med. 2020, 172, 59–60. [Google Scholar] [CrossRef]
- McCoy, L.G.; Brenna, C.T.A.; Chen, S.S.; Vold, K.; Das, S. Believing in Black Boxes: Machine Learning for Healthcare Does Not Need Explainability to Be Evidence-Based. J. Clin. Epidemiol. 2022, 142, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Banegas-Luna, A.J.; Peña-García, J.; Iftene, A.; Guadagni, F.; Ferroni, P.; Scarpato, N.; Zanzotto, F.M.; Bueno-Crespo, A.; Pérez-Sánchez, H. Towards the Interpretability of Machine Learning Predictions for Medical Applications Targeting Personalised Therapies: A Cancer Case Survey. Int. J. Mol. Sci. 2021, 22, 4394. [Google Scholar] [CrossRef]
- Li, A.; Walling, J.; Ahn, S.; Kotliarov, Y.; Su, Q.; Quezado, M.; Oberholtzer, J.C.; Park, J.; Zenklusen, J.C.; Fine, H.A. Unsupervised Analysis of Transcriptomic Profiles Reveals Six Glioma Subtypes. Cancer Res. 2009, 69, 2091–2099. [Google Scholar] [CrossRef]
- Zacher, A.; Kaulich, K.; Stepanow, S.; Wolter, M.; Köhrer, K.; Felsberg, J.; Malzkorn, B.; Reifenberger, G. Molecular Diagnostics of Gliomas Using Next Generation Sequencing of a Glioma-Tailored Gene Panel. Brain Pathol. 2017, 27, 146–159. [Google Scholar] [CrossRef]
- Petersen, J.K.; Boldt, H.B.; Sorensen, M.; Dahlrot, R.H.; Hansen, S.; Burton, M.; Thomassen, M.; Kruse, T.; Poulsen, F.R.; Andreasen, L.; et al. GENE-33. INTEGRATED GLIOMA DIAGNOSTICS USING TARGETED NEXT-GENERATION SEQUENCING. Neuro Oncol. 2019, 21, vi104–vi105. [Google Scholar] [CrossRef]
- Gashi, M.; Vuković, M.; Jekic, N.; Thalmann, S.; Holzinger, A.; Jean-Quartier, C.; Jeanquartier, F. State-of-the-Art Explainability Methods with Focus on Visual Analytics Showcased by Glioma Classification. BioMedInformatics 2022, 2, 139–158. [Google Scholar] [CrossRef]
- Coletti, R.; Carrilho, J.F.; Martins, E.P.; Gonçalves, C.S.; Costa, B.M.; Lopes, M.B. A Novel Tool for Multi-Omics Network Integration and Visualization: A Study of Glioma Heterogeneity. Comput. Biol. Med. 2025, 188, 109811. [Google Scholar] [CrossRef] [PubMed]
- Vershinina, O.; Turubanova, V.; Krivonosov, M.; Trukhanov, A.; Ivanchenko, M. Explainable Machine Learning Models for Glioma Subtype Classification and Survival Prediction. Cancers 2025, 17, 2614. [Google Scholar] [CrossRef]
- Palkar, A.; Dias, C.C.; Chadaga, K.; Sampathila, N. Empowering Glioma Prognosis with Transparent Machine Learning and Interpretative Insights Using Explainable AI. IEEE Access 2024, 12, 31697–31718. [Google Scholar] [CrossRef]
- Akpinar, E.; Oduncuoglu, M. Hybrid Classical and Quantum Computing for Enhanced Glioma Tumor Classification Using TCGA Data. Sci. Rep. 2025, 15, 25935. [Google Scholar] [CrossRef]
- Yang, P.; Feng, P.; Tian, G.; Zhao, G.; Yuan, G.; Pan, Y. Integrative Machine Learning and Bioinformatics Analysis Unveil Key Genes for Precise Glioma Classification and Prognosis Evaluation. Comput. Biol. Chem. 2025, 119, 108510. [Google Scholar] [CrossRef]
- Li, Y.; Sun, H. Multi-Omics Analysis Identifies Novels Genes Involved in Glioma Prognosis. Sci. Rep. 2025, 15, 5806. [Google Scholar] [CrossRef] [PubMed]
- Noviandy, T.R.; Idroes, G.M.; Hardi, I. Integrating Explainable Artificial Intelligence and Light Gradient Boosting Machine for Glioma Grading. Inform. Health 2025, 2, 1–8. [Google Scholar] [CrossRef]
- Lin, T.-H.; Lin, H.-Y. Genetic Feature Selection Algorithm as an Efficient Glioma Grade Classifier. Sci. Rep. 2025, 15, 15497. [Google Scholar] [CrossRef]
- Yan, W.; Zhang, W.; You, G.; Zhang, J.; Han, L.; Bao, Z.; Wang, Y.; Liu, Y.; Jiang, C.; Kang, C.; et al. Molecular Classification of Gliomas Based on Whole Genome Gene Expression: A Systematic Report of 225 Samples from the Chinese Glioma Cooperative Group. Neuro Oncol. 2012, 14, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.M.H.; Tran, L.K.H.; Nechtman, J.; dos Santos, B.; Purohit, S.; Bin Satter, K.; Dun, B.; Kolhe, R.; Sharma, S.; Bollag, R.; et al. Comparative Analysis of Transcriptomic Profile, Histology, and IDH Mutation for Classification of Gliomas. Sci. Rep. 2020, 10, 20651. [Google Scholar] [CrossRef]
- Han, H.; Feng, P.; Yuan, G. Molecular Typing of Gliomas on the Basis of Integrin Family Genes and a Functional Study of ITGA7. Sci. Rep. 2025, 15, 12306. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular Subclasses of High-Grade Glioma Predict Prognosis, Delineate a Pattern of Disease Progression, and Resemble Stages in Neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Paul, Y.; Mondal, B.; Patil, V.; Somasundaram, K. DNA Methylation Signatures for 2016 WHO Classification Subtypes of Diffuse Gliomas. Clin. Epigenetics 2017, 9, 32. [Google Scholar] [CrossRef]
- Teo, W.-Y.; Sekar, K.; Seshachalam, P.; Shen, J.; Chow, W.-Y.; Lau, C.C.; Yang, H.; Park, J.; Kang, S.-G.; Li, X.; et al. Relevance of a TCGA-Derived Glioblastoma Subtype Gene-Classifier among Patient Populations. Sci. Rep. 2019, 9, 7442. [Google Scholar] [CrossRef]
- Tang, Y.; Qazi, M.A.; Brown, K.R.; Mikolajewicz, N.; Moffat, J.; Singh, S.K.; McNicholas, P.D. Identification of Five Important Genes to Predict Glioblastoma Subtypes. Neuro-Oncol. Adv. 2021, 3, vdab144. [Google Scholar] [CrossRef]
- Madurga, R.; García-Romero, N.; Jiménez, B.; Collazo, A.; Pérez-Rodríguez, F.; Hernández-Laín, A.; Fernández-Carballal, C.; Prat-Acín, R.; Zanin, M.; Menasalvas, E.; et al. Normal Tissue Content Impact on the GBM Molecular Classification. Brief. Bioinform. 2021, 22, bbaa129. [Google Scholar] [CrossRef]
- Steponaitis, G.; Kucinskas, V.; Golubickaite, I.; Skauminas, K.; Saudargiene, A. Glioblastoma Molecular Classification Tool Based on MRNA Analysis: From Wet-Lab to Subtype. Int. J. Mol. Sci. 2022, 23, 15875. [Google Scholar] [CrossRef]
- Munquad, S.; Si, T.; Mallik, S.; Das, A.B.; Zhao, Z. A Deep Learning–Based Framework for Supporting Clinical Diagnosis of Glioblastoma Subtypes. Front. Genet. 2022, 13, 855420. [Google Scholar] [CrossRef]
- Munquad, S.; Das, A.B. DeepAutoGlioma: A Deep Learning Autoencoder-Based Multi-Omics Data Integration and Classification Tools for Glioma Subtyping. BioData Min. 2023, 16, 32. [Google Scholar] [CrossRef]
- Wang, D.; Shah, M.; Arjuna, S.; Dono, A.; Patel, C.; Huse, J.; Kerrigan, B.P.; Nguyen, S.; Lang, F.; Esquenazi, Y.; et al. BIOM-67. Differential DNA methylation patterns in the CSF of patients with diffuse gliomas. Neuro Oncol. 2024, 26, viii35. [Google Scholar] [CrossRef]
- Lin, H.; Wang, K.; Xiong, Y.; Zhou, L.; Yang, Y.; Chen, S.; Xu, P.; Zhou, Y.; Mao, R.; Lv, G.; et al. Identification of Tumor Antigens and Immune Subtypes of Glioblastoma for MRNA Vaccine Development. Front. Immunol. 2022, 13, 773264. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Li, Z.; Li, Y.; Zhang, Y.; Miao, X. Characterization of Different Subtypes of Immune Cell Infiltration in Glioblastoma to Aid Immunotherapy. Front. Immunol. 2022, 13, 799509. [Google Scholar] [CrossRef]
- Zhu, Y.; Feng, S.; Song, Z.; Wang, Z.; Chen, G. Identification of Immunological Characteristics and Immune Subtypes Based on Single-Sample Gene Set Enrichment Analysis Algorithm in Lower-Grade Glioma. Front. Genet. 2022, 13, 894865. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yang, X.; Deng, T.; Yan, J.; Guo, F.; Mo, L.; An, S.; Huang, Q. Comprehensive Machine Learning-Based Integration Develops a Novel Prognostic Model for Glioblastoma. Mol. Ther. Oncol. 2024, 32, 200838. [Google Scholar] [CrossRef]
- Yuan, F.; Wang, Y.; Yuan, L.; Ye, L.; Hu, Y.; Cheng, H.; Li, Y. Machine Learning–Based New Classification for Immune Infiltration of Gliomas. PLoS ONE 2024, 19, e0312071. [Google Scholar] [CrossRef]
- Tong, M.; Xu, Z.; Wang, L.; Chen, H.; Wan, X.; Xu, H.; Yang, S.; Tu, Q. An Analysis of Prognostic Risk and Immunotherapy Response of Glioblastoma Patients Based on Single-Cell Landscape and Nitrogen Metabolism. Neurobiol. Dis. 2025, 211, 106935. [Google Scholar] [CrossRef]
- Tian, J.; Zhao, J.; Xu, Z.; Liu, B.; Pu, J.; Li, H.; Lei, Q.; Zhao, Y.; Zhou, W.; Li, X.; et al. Bioinformatics Analysis to Identify Key Invasion Related Genes and Construct a Prognostic Model for Glioblastoma. Sci. Rep. 2025, 15, 10773. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Zhang, S.; Zhou, L. Expression Profile and Prognostic Relevance of Immune Infiltration-Related RBMS1 in Gliomas: A Multidimensional Integrative Analysis. J. Cancer Res. Clin. Oncol. 2025, 151, 205. [Google Scholar] [CrossRef]
- Li, G.; Zhao, Y.; He, Y.; Qian, Z.; Liu, Y.; Li, X.; Li, L.; Liu, Z. Machine Learning-Based Construction of Immunogenic Cell Death-Related Score for Improving Prognosis and Personalized Treatment in Glioma. Sci. Rep. 2025, 15, 30417. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, L.A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D.J.; Agrawal, N.; Ahn, K.S.; Ahn, S.-M.; Aikata, H.; Akbani, R.; et al. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93, Correction in Nature 2023, 614, E39. [Google Scholar] [CrossRef] [PubMed]
- Mardis, E.R.; Wilson, R.K. Cancer Genome Sequencing: A Review. Hum. Mol. Genet. 2009, 18, R163–R168. [Google Scholar] [CrossRef]
- Martínez-Ricarte, F.; Mayor, R.; Martínez-Sáez, E.; Rubio-Pérez, C.; Pineda, E.; Cordero, E.; Cicuéndez, M.; Poca, M.A.; López-Bigas, N.; Cajal, S.R.Y.; et al. Molecular Diagnosis of Diffuse Gliomas through Sequencing of Cell-Free Circulating Tumor DNA from Cerebrospinal Fluid. Clin. Cancer Res. 2018, 24, 2812–2819. [Google Scholar] [CrossRef] [PubMed]
- Kinnersley, B.; Jung, J.; Cornish, A.J.; Chubb, D.; Laxton, R.; Frangou, A.; Gruber, A.J.; Sud, A.; Caravagna, G.; Sottoriva, A.; et al. Genomic Landscape of Diffuse Glioma Revealed by Whole Genome Sequencing. Nat. Commun. 2025, 16, 4233. [Google Scholar] [CrossRef]
- Na, K.; Kim, H.-S.; Shim, H.S.; Chang, J.H.; Kang, S.-G.; Kim, S.H. Targeted Next-Generation Sequencing Panel (TruSight Tumor 170) in Diffuse Glioma: A Single Institutional Experience of 135 Cases. J. Neuro-Oncol. 2019, 142, 445–454. [Google Scholar] [CrossRef]
- Petersen, J.K.; Boldt, H.B.; Sørensen, M.D.; Blach, S.; Dahlrot, R.H.; Hansen, S.; Burton, M.; Thomassen, M.; Kruse, T.; Poulsen, F.R.; et al. Targeted Next-generation Sequencing of Adult Gliomas for Retrospective Prognostic Evaluation and Up-front Diagnostics. Neuropathol. Appl. Neurobiol. 2021, 47, 108–126. [Google Scholar] [CrossRef]
- Buchwald, Z.S.; Tian, S.; Rossi, M.; Smith, G.H.; Switchenko, J.; Hauenstein, J.E.; Moreno, C.S.; Press, R.H.; Prabhu, R.S.; Zhong, J.; et al. Genomic Copy Number Variation Correlates with Survival Outcomes in WHO Grade IV Glioma. Sci. Rep. 2020, 10, 7355. [Google Scholar] [CrossRef]
- Aran, D.; Sabato, S.; Hellman, A. DNA Methylation of Distal Regulatory Sites Characterizes Dysregulation of Cancer Genes. Genome Biol. 2013, 14, R21. [Google Scholar] [CrossRef]
- Jurmeister, P.; Bockmayr, M.; Seegerer, P.; Bockmayr, T.; Treue, D.; Montavon, G.; Vollbrecht, C.; Arnold, A.; Teichmann, D.; Bressem, K.; et al. Machine Learning Analysis of DNA Methylation Profiles Distinguishes Primary Lung Squamous Cell Carcinomas from Head and Neck Metastases. Sci. Transl. Med. 2019, 11, eaaw8513. [Google Scholar] [CrossRef]
- Yoon, J.; Kim, M.; Posadas, E.M.; Freedland, S.J.; Liu, Y.; Davicioni, E.; Den, R.B.; Trock, B.J.; Karnes, R.J.; Klein, E.A.; et al. A Comparative Study of PCS and PAM50 Prostate Cancer Classification Schemes. Prostate Cancer Prostatic Dis. 2021, 24, 733–742. [Google Scholar] [CrossRef]
- Sabedot, T.S.; Malta, T.M.; Snyder, J.; Nelson, K.; Wells, M.; deCarvalho, A.C.; Mukherjee, A.; Chitale, D.A.; Mosella, M.S.; Sokolov, A.; et al. A Serum-Based DNA Methylation Assay Provides Accurate Detection of Glioma. Neuro Oncol. 2021, 23, 1494–1508. [Google Scholar] [CrossRef]
- Huse, J.T.; Phillips, H.S.; Brennan, C.W. Molecular Subclassification of Diffuse Gliomas: Seeing Order in the Chaos. Glia 2011, 59, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cui, B.; Zhou, Y.; Wang, X.; Wu, W.; Wang, Z.; Dai, Z.; Cheng, Q.; Yang, K. B2M Overexpression Correlates with Malignancy and Immune Signatures in Human Gliomas. Sci. Rep. 2021, 11, 5045. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal Differentiation Mediated by NF-ΚB Promotes Radiation Resistance in Glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef]
- Zorniak, M.; Clark, P.A.; Leeper, H.E.; Tipping, M.D.; Francis, D.M.; Kozak, K.R.; Salamat, M.S.; Kuo, J.S. Differential Expression of 2′,3′-Cyclic-Nucleotide 3′-Phosphodiesterase and Neural Lineage Markers Correlate with Glioblastoma Xenograft Infiltration and Patient Survival. Clin. Cancer Res. 2012, 18, 3628–3636. [Google Scholar] [CrossRef] [PubMed]
- Crisman, T.J.; Zelaya, I.; Laks, D.R.; Zhao, Y.; Kawaguchi, R.; Gao, F.; Kornblum, H.I.; Coppola, G. Identification of an Efficient Gene Expression Panel for Glioblastoma Classification. PLoS ONE 2016, 11, e0164649. [Google Scholar] [CrossRef]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e6. [Google Scholar] [CrossRef]
- Park, J.; Shim, J.-K.; Yoon, S.-J.; Kim, S.H.; Chang, J.H.; Kang, S.-G. Transcriptome Profiling-Based Identification of Prognostic Subtypes and Multi-Omics Signatures of Glioblastoma. Sci. Rep. 2019, 9, 10555. [Google Scholar] [CrossRef]
- Segato, A.; Marzullo, A.; Calimeri, F.; De Momi, E. Artificial Intelligence for Brain Diseases: A Systematic Review. APL Bioeng. 2020, 4, 041503. [Google Scholar] [CrossRef] [PubMed]
- Chin-Yee, B.; Upshur, R. Three Problems with Big Data and Artificial Intelligence in Medicine. Perspect. Biol. Med. 2019, 62, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Gilvary, C.; Madhukar, N.; Elkhader, J.; Elemento, O. The Missing Pieces of Artificial Intelligence in Medicine. Trends Pharmacol. Sci. 2019, 40, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of Explainable Artificial Intelligence for Healthcare: A Systematic Review of the Last Decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef]
- Frasca, M.; La Torre, D.; Pravettoni, G.; Cutica, I. Explainable and Interpretable Artificial Intelligence in Medicine: A Systematic Bibliometric Review. Discov. Artif. Intell. 2024, 4, 15. [Google Scholar] [CrossRef]
- Rodríguez Mallma, M.J.; Zuloaga-Rotta, L.; Borja-Rosales, R.; Rodríguez Mallma, J.R.; Vilca-Aguilar, M.; Salas-Ojeda, M.; Mauricio, D. Explainable Machine Learning Models for Brain Diseases: Insights from a Systematic Review. Neurol. Int. 2024, 16, 1285–1307. [Google Scholar] [CrossRef]
- Ruzevick, J.; Jackson, C.; Phallen, J.; Lim, M. Clinical Trials with Immunotherapy for High-Grade Glioma. Neurosurg. Clin. N. Am. 2012, 23, 459–470. [Google Scholar] [CrossRef]
- Franson, A.; McClellan, B.L.; Varela, M.L.; Comba, A.; Syed, M.F.; Banerjee, K.; Zhu, Z.; Gonzalez, N.; Candolfi, M.; Lowenstein, P.; et al. Development of Immunotherapy for High-Grade Gliomas: Overcoming the Immunosuppressive Tumor Microenvironment. Front. Med. 2022, 9, 966458. [Google Scholar] [CrossRef]
- Keskin, D.B.; Anandappa, A.J.; Sun, J.; Tirosh, I.; Mathewson, N.D.; Li, S.; Oliveira, G.; Giobbie-Hurder, A.; Felt, K.; Gjini, E.; et al. Neoantigen Vaccine Generates Intratumoral T Cell Responses in Phase Ib Glioblastoma Trial. Nature 2019, 565, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Pyonteck, S.M.; Akkari, L.; Schuhmacher, A.J.; Bowman, R.L.; Sevenich, L.; Quail, D.F.; Olson, O.C.; Quick, M.L.; Huse, J.T.; Teijeiro, V.; et al. CSF-1R Inhibition Alters Macrophage Polarization and Blocks Glioma Progression. Nat. Med. 2013, 19, 1264–1272. [Google Scholar] [CrossRef]
- Hilf, N.; Kuttruff-Coqui, S.; Frenzel, K.; Bukur, V.; Stevanović, S.; Gouttefangeas, C.; Platten, M.; Tabatabai, G.; Dutoit, V.; van der Burg, S.H.; et al. Actively Personalized Vaccination Trial for Newly Diagnosed Glioblastoma. Nature 2019, 565, 240–245, Correction in Nature 2018, 533, E13. [Google Scholar] [CrossRef]
- Johanns, T.M.; Miller, C.A.; Liu, C.J.; Perrin, R.J.; Bender, D.; Kobayashi, D.K.; Campian, J.L.; Chicoine, M.R.; Dacey, R.G.; Huang, J.; et al. Detection of Neoantigen-Specific T Cells Following a Personalized Vaccine in a Patient with Glioblastoma. Oncoimmunology 2019, 8, e1561106. [Google Scholar] [CrossRef]
- O’Rourke, D.M.; Nasrallah, M.P.; Desai, A.; Melenhorst, J.J.; Mansfield, K.; Morrissette, J.J.D.; Martinez-Lage, M.; Brem, S.; Maloney, E.; Shen, A.; et al. A Single Dose of Peripherally Infused EGFRvIII-Directed CAR T Cells Mediates Antigen Loss and Induces Adaptive Resistance in Patients with Recurrent Glioblastoma. Sci. Transl. Med. 2017, 9, eaaa0984. [Google Scholar] [CrossRef]
- Miller, T.E.; El Farran, C.A.; Couturier, C.P.; Chen, Z.; D’Antonio, J.P.; Verga, J.; Villanueva, M.A.; Gonzalez Castro, L.N.; Tong, Y.E.; Al Saadi, T.; et al. Programs, Origins and Immunomodulatory Functions of Myeloid Cells in Glioma. Nature 2025, 640, 1072–1082. [Google Scholar] [CrossRef]
- Jackson, C.M.; Lim, M.; Drake, C.G. Immunotherapy for Brain Cancer: Recent Progress and Future Promise. Clin. Cancer Res. 2014, 20, 3651–3659. [Google Scholar] [CrossRef]
- Young, J.S.; Dayani, F.; Morshed, R.A.; Okada, H.; Aghi, M.K. Immunotherapy for High-Grade Gliomas: A Clinical Update and Practical Considerations for Neurosurgeons. World Neurosurg. 2019, 124, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Shklovskaya, E.; Satgunaseelan, L.; Mao, Y.; da Silva, I.P.; Perry, K.A.; Diefenbach, R.J.; Gide, T.N.; Shivalingam, B.; Buckland, M.E.; et al. Neoadjuvant Triplet Immune Checkpoint Blockade in Newly Diagnosed Glioblastoma. Nat. Med. 2025, 31, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Savage, W.M.; Yeary, M.D.; Tang, A.J.; Sperring, C.P.; Argenziano, M.G.; Adapa, A.R.; Yoh, N.; Canoll, P.; Bruce, J.N. Biomarkers of Immunotherapy in Glioblastoma. Neuro-Oncol. Pract. 2024, 11, 383–394. [Google Scholar] [CrossRef]
- Lynes, J.P.; Nwankwo, A.K.; Sur, H.P.; Sanchez, V.E.; Sarpong, K.A.; Ariyo, O.I.; Dominah, G.A.; Nduom, E.K. Biomarkers for Immunotherapy for Treatment of Glioblastoma. J. Immunother. Cancer 2020, 8, e000348. [Google Scholar] [CrossRef]
- Liu, X.-P.; Jin, X.; Seyed Ahmadian, S.; Yang, X.; Tian, S.-F.; Cai, Y.-X.; Chawla, K.; Snijders, A.M.; Xia, Y.; van Diest, P.J.; et al. Clinical Significance and Molecular Annotation of Cellular Morphometric Subtypes in Lower-Grade Gliomas Discovered by Machine Learning. Neuro Oncol. 2023, 25, 68–81. [Google Scholar] [CrossRef]
- Doucette, T.; Rao, G.; Rao, A.; Shen, L.; Aldape, K.; Wei, J.; Dziurzynski, K.; Gilbert, M.; Heimberger, A.B. Immune Heterogeneity of Glioblastoma Subtypes: Extrapolation from the Cancer Genome Atlas. Cancer Immunol. Res. 2013, 1, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Vedunova, M.; Turubanova, V.; Vershinina, O.; Savyuk, M.; Efimova, I.; Mishchenko, T.; Raedt, R.; Vral, A.; Vanhove, C.; Korsakova, D.; et al. DC Vaccines Loaded with Glioma Cells Killed by Photodynamic Therapy Induce Th17 Anti-Tumor Immunity and Provide a Four-Gene Signature for Glioma Prognosis. Cell Death Dis. 2022, 13, 1062. [Google Scholar] [CrossRef] [PubMed]
- de Mendonça, M.L.; Coletti, R.; Gonçalves, C.S.; Martins, E.P.; Costa, B.M.; Vinga, S.; Lopes, M.B. Updating TCGA Glioma Classification Through Integration of Molecular Profiling Data Following the 2016 and 2021 WHO Guidelines; University of Palermo: Palermo, Sicilia, Italy, 2023. [Google Scholar]
- Sandmann, T.; Bourgon, R.; Garcia, J.; Li, C.; Cloughesy, T.; Chinot, O.L.; Wick, W.; Nishikawa, R.; Mason, W.; Henriksson, R.; et al. Patients With Proneural Glioblastoma May Derive Overall Survival Benefit From the Addition of Bevacizumab to First-Line Radiotherapy and Temozolomide: Retrospective Analysis of the AVAglio Trial. JCO 2015, 33, 2735–2744. [Google Scholar] [CrossRef]
- Wang, L.-B.; Karpova, A.; Gritsenko, M.A.; Kyle, J.E.; Cao, S.; Li, Y.; Rykunov, D.; Colaprico, A.; Rothstein, J.H.; Hong, R.; et al. Proteogenomic and Metabolomic Characterization of Human Glioblastoma. Cancer Cell 2021, 39, 509–528.e20. [Google Scholar] [CrossRef] [PubMed]
| Category | Reference | Data | ML Model | Number of Features | External Validation |
|---|---|---|---|---|---|
| Main glioma subtypes (astrocytoma, oligodendroglioma, and GBM) | Li et al., 2009 [62] | Transcriptome | PAM | 33–352 | Yes |
| Zacher et al., 2017 [63] | Genome | Clustering | 20 | No | |
| Cai et al., 2018 [30] | Transcriptome | SVM | 539 | No | |
| Petersen et al., 2019 [64] | Genome | Clustering | 20 | No | |
| Gashi et al., 2022 [65] | Genome | RF | 252 | No | |
| Vieira et al., 2024 [31] | Multi-omics | DIABLO | 100 | No | |
| Coletti et al., 2025 [66] | Multi-omics | MINGLE | Not reported | No | |
| Vershinina et al., 2025 [67] | Transcriptome | SVM | 13 | Yes | |
| Main glioma subtypes (LGG versus GBM) | Palkar et al., 2024 [68] | Genome | XGBoost | 23 | No |
| Akpinar et al., 2025 [69] | Genome | VQC | 5 | No | |
| Yang et al., 2025 [70] | Transcriptome | SVM | Not reported | Yes | |
| Li et al., 2025 [71] | Multi-omics | Clustering | 178 | Yes | |
| Noviandy et al., 2025 [72] | Genome | LightGBM | 23 | No | |
| Lin et al., 2025 [73] | Transcriptome | Genetic algorithm | Not reported | No | |
| Glioma subtypes identified by the authors | Yan et al., 2012 [74] | Transcriptome | Clustering | 1801 | Yes |
| Tran et al., 2020 [75] | Transcriptome | Ensemble of LSVC | 168 | Yes | |
| Han et al., 2025 [76] | Transcriptome | Clustering | 26 | Yes | |
| GBM subtypes | Phillips et al., 2006 [77] | Transcriptome | Clustering | 35 | Yes |
| Li et al., 2009 [62] | Transcriptome | PAM | 33–352 | Yes | |
| Verhaak et al., 2010 [78] | Transcriptome | ClaNC | 840 | Yes | |
| Paul et al., 2017 [79] | Methylome | SVM | 13 | Yes | |
| Teo et al., 2019 [80] | Transcriptome | Clustering | 500 | Yes | |
| Tang et al., 2021 [81] | Transcriptome | XGBoost | 5 | Yes | |
| Madurga et al., 2021 [82] | Transcriptome | ClaNC | 20 | Yes | |
| Steponaitis et al., 2022 [83] | Transcriptome | LR | 5 or 20 | Yes | |
| Munquad et al., 2022 [84] | Multi-omics | CNN | 75 | Yes | |
| Munquad et al., 2023 [85] | Multi-omics | CNN | 100 | Yes | |
| Wang et al., 2024 [86] | Methylome | RF | 900 | No | |
| LGG subtypes | Li et al., 2009 [62] | Transcriptome | PAM | 33–352 | Yes |
| Paul et al., 2017 [79] | Methylome | SVM | 14 | Yes | |
| Munquad et al., 2022 [28] | Transcriptome | SVM | 178 | Yes | |
| Munquad et al., 2023 [85] | Multi-omics | CNN | 400 | Yes | |
| Vieira et al., 2024 [31] | Multi-omics | DIABLO | 89 | No | |
| Immune-related glioma subtypes or risk groups | Lin et al., 2022 [87] | Transcriptome | Clustering | 1658 | Yes |
| Luo et al., 2022 [33] | Transcriptome | Cox regression | 23 | Yes | |
| Feng et al., 2022 [88] | Transcriptome | Clustering | 34 | Yes | |
| Zhu et al., 2022 [89] | Transcriptome | Clustering | 29 | Yes | |
| Li et al., 2022 [36] | Transcriptome | SVM | 61 | No | |
| Lin et al., 2022 [32] | Transcriptome | Cox regression | 5 | Yes | |
| Guo et al., 2024 [34] | Transcriptome | Cox regression | 11 | Yes | |
| Jiang et al., 2024 [90] | Transcriptome | RSF | 79 | No | |
| Luo et al., 2024 [37] | Transcriptome | LR | 13, 14, or 17 | Yes | |
| Yuan et al., 2024 [91] | Transcriptome | LSTM | 122 | No | |
| Tong et al., 2025 [92] | Transcriptome | Clustering | 285 | No | |
| Tian et al., 2025 [93] | Transcriptome | Cox regression | 5 | Yes | |
| Zhang et al., 2025 [94] | Transcriptome | Cox regression | 1 | Yes | |
| Li et al., 2025 [95] | Transcriptome | Cox regression | 14 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Sleptsova, E.; Vershinina, O.; Ivanchenko, M.; Turubanova, V. From Data to Decision: Integrating Bioinformatics into Glioma Patient Stratification and Immunotherapy Selection. Int. J. Mol. Sci. 2026, 27, 667. https://doi.org/10.3390/ijms27020667
Sleptsova E, Vershinina O, Ivanchenko M, Turubanova V. From Data to Decision: Integrating Bioinformatics into Glioma Patient Stratification and Immunotherapy Selection. International Journal of Molecular Sciences. 2026; 27(2):667. https://doi.org/10.3390/ijms27020667
Chicago/Turabian StyleSleptsova, Ekaterina, Olga Vershinina, Mikhail Ivanchenko, and Victoria Turubanova. 2026. "From Data to Decision: Integrating Bioinformatics into Glioma Patient Stratification and Immunotherapy Selection" International Journal of Molecular Sciences 27, no. 2: 667. https://doi.org/10.3390/ijms27020667
APA StyleSleptsova, E., Vershinina, O., Ivanchenko, M., & Turubanova, V. (2026). From Data to Decision: Integrating Bioinformatics into Glioma Patient Stratification and Immunotherapy Selection. International Journal of Molecular Sciences, 27(2), 667. https://doi.org/10.3390/ijms27020667






