Genome-Wide Analysis of the JAZ Family in Brassica rapa and the Roles of BrJAZ1a and 6b in Response to Stresses
Abstract
1. Introduction
2. Results
2.1. The Isolation and Identification of the JAZ Family Genes in B. rapa
- A total of 31 candidate JAZ family members were initially identified in B. rapa via annotation and homology searches in the BRAD database. The presence of the characteristic ZIM domain in these candidate proteins was then validated using the InterPro (http://www.ebi.ac.uk/interpro/, accessed on 20 February 2025), SMART (http://smart.embl-heidelberg.de/, accessed on 20 February 2025), and ScanProsite (https://prosite.expasy.org/, accessed on 20 February 2025) tools. This analysis confirmed that only 25 genes contained a typical TIFY domain, and these were retained for further study. Based on their homology to A. thaliana JAZ genes, these 25 genes were designated as BrJAZ1 to BrJAZ12 (Table S1). Notably, no homologous genes for AtJAZ11 and AtJAZ12 were found in the Chinese cabbage genome. The key characteristics of the 25 BrJAZ genes are summarized in Table 1. The CDS lengths ranged from 342 bp to 1071 bp, while the genomic sequences were considerably longer (501 bp to 3273 bp), a difference attributable to the variation in intron number and size of the introns. The encoded proteins ranged from 113 to 356 amino acids in length, with molecular weights between approximately 13.0 kDa and 39.8 kDa. The theoretical isoelectric points (pI) of most BrJAZ proteins were basic, ranging from 7.9 to 10.07, except for BrJAZ12a, which had an acidic pI of 4.97.
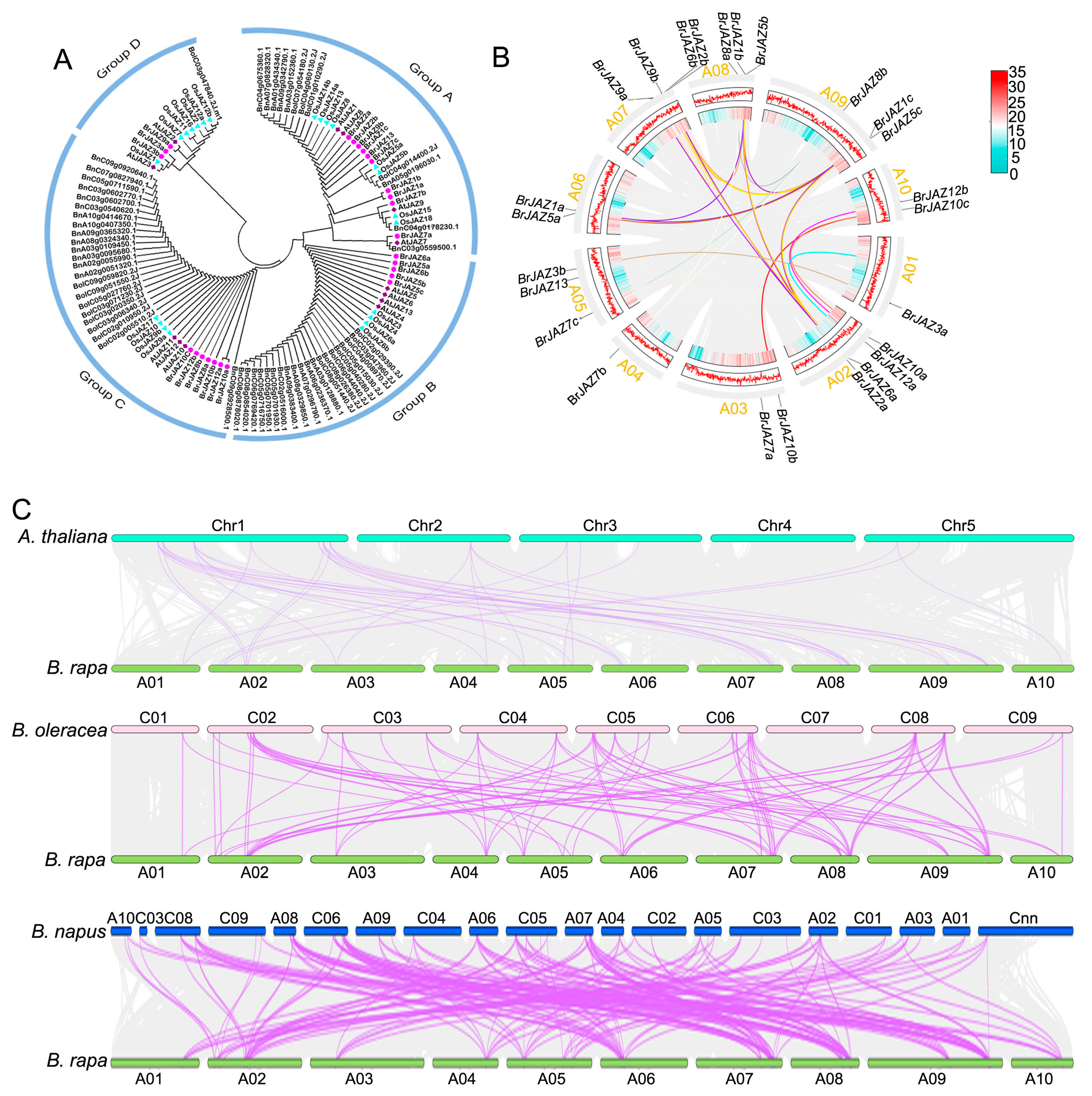
2.2. Phylogenetic Relationships and Synteny of JAZ Members Among B. rapa, A. thaliana, B. napus, and B. oleracea
2.3. The Gene Localization, Structures, and Conserved Motifs of BrJAZ Members
2.4. The Cis-Acting Elements on the BrJAZ Promoters
2.5. The Expression Levels of BrJAZs in Different Tissues and in Response to Stresses
2.6. The Transcriptional Levels of BrJAZs in Response to Stresses
2.7. The Phenotypic Characteristics of Over-Expressed BrJAZ1a and BrJAZ6b Transgenic A. thaliana Lines Under Abiotic Stresses
2.8. The Subcellular Localization of BrJAZ1a-GFP and BrJAZ6b-GFP
3. Discussion
Limitations and Future Perspectives
4. Materials and Methods
4.1. Plant Materials
4.2. Phylogenetic Tree and Collinearity Analysis
4.3. Gene Location, Structure, and Protein Motifs
4.4. Total RNA Isolation and Quantitative Real-Time PCR
4.5. Gene Clone and Vector Constructions
4.6. Plant Transformations
4.7. Enzyme Activities
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, Y.; Meng, Y.; Huang, D.; Qi, Y.; Chen, M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 128–136. [Google Scholar] [CrossRef]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Meng, Y.; Hu, J.; Ding, M.; Bian, J.; Yan, M.; Han, J.; Zhou, M. Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA59 transcription factor. Plant J. 2018, 95, 444–457. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Pandey, B.K.; Mehra, P.; Heitz, T.; Giri, J. OsJAZ9 overexpression modulates jasmonic acid biosynthesis and potassium deficiency responses in rice. Plant Mol. Biol. 2020, 104, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.L.; Zhang, J.X.; Jiang, S.; Lu, Y.; Huang, Y.S.; Dong, X.M.; Hu, Q.; Yao, W.; Zhang, M.Q.; Xiao, S.H. Genome-wide identification of JAZ gene family in sugarcane and function analysis of ScJAZ1/2 in drought stress response and flowering regulation. Plant Physiol. Biochem. 2024, 210, 108577. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, C.; Liu, Z.; Zhao, T.; Jiang, J.; Li, J.; Xu, X.; Yang, H. Genome-Wide Identification, Characterization and Expression Analysis of the JAZ Gene Family in Resistance to Gray Leaf Spots in Tomato. Int. J. Mol. Sci. 2021, 22, 9974. [Google Scholar] [CrossRef]
- Yu, X.; Yang, J.; Liu, Y.; Li, Z.; Yan, H.; Cao, Y.; Ma, X.; Zhang, X.; Xu, W.; Su, Z. Chromatin state analysis for regulatory features of JAZ family genes in Arabidopsis thaliana. Plant Sci. 2025, 359, 112652. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Yan, C.; Zhang, J.; Cheng, Y.; Li, W.; Yan, H.; Gao, J. Genome-wide identification and expression analysis of the JAZ gene family in turnip. Sci. Rep. 2021, 11, 21330. [Google Scholar] [CrossRef]
- Zhao, P.; Zheng, J.; Xu, Q.; Zhao, S.; Li, J.; Zhu, Y.; Yan, J.; Chen, Q.; Yang, B.; Li, C.; et al. Genome-Wide Identification and Analysis of the JAZ Gene Family in Rapeseed Reveal JAZ2 and JAZ3 Roles in Drought and Salt Stress Tolerance. J. Agric. Food Chem. 2025, 73, 20955–20971, Correction in J. Agric. Food Chem. 2025, 73, 25689. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, C.; Yang, L.; Zhang, Y.; Wang, Y.; Fang, Z.; Lv, H. Genome-Wide Identification, Expression Profile of the TIFY Gene Family in Brassica oleracea var. capitata, and Their Divergent Response to Various Pathogen Infections and Phytohormone Treatments. Genes 2020, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Xiang, S.; Chen, Y.; Zhang, H.; Yu, D. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 2021, 72, 1473–1489. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhang, T.; Geng, S.; Scott, P.B.; Li, H.; Chen, S. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteom. 2019, 196, 81–91. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef]
- Chen, L.; Shen, Y.; Yang, W.; Pan, Q.; Li, C.; Sun, Q.; Zeng, Q.; Li, B.; Zhang, L. Comparative Metabolic Study of Two Contrasting Chinese Cabbage Genotypes under Mild and Severe Drought Stress. Int. J. Mol. Sci. 2022, 23, 5947. [Google Scholar] [CrossRef]
- Pavlović, I.; Mlinarić, S.; Tarkowská, D.; Oklestkova, J.; Novák, O.; Lepeduš, H.; Bok, V.V.; Brkanac, S.R.; Strnad, M.; Salopek-Sondi, B. Early Brassica Crops Responses to Salinity Stress: A Comparative Analysis Between Chinese Cabbage, White Cabbage, and Kale. Front. Plant Sci. 2019, 10, 450. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef]
- Wang, Y.; Li, N.; Zhan, J.; Wang, X.; Zhou, X.R.; Shi, J.; Wang, H. Genome-wide analysis of the JAZ subfamily of transcription factors and functional verification of BnC08.JAZ1-1 in Brassica napus. Biotechnol. Biofuels Bioprod. 2022, 15, 93. [Google Scholar] [CrossRef]
- Singh, A.P.; Pandey, B.K.; Deveshwar, P.; Narnoliya, L.; Parida, S.K.; Giri, J. JAZ Repressors: Potential Involvement in Nutrients Deficiency Response in Rice and Chickpea. Front. Plant Sci. 2015, 6, 975. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Yu, H.; Li, J.; Chang, X.; Dong, N.; Chen, B.; Wang, J.; Zha, L.; Gui, S. Genome-wide identification and expression profiling of the WRKY gene family reveals abiotic stress response mechanisms in Platycodon grandiflorus. Int. J. Biol. Macromol. 2024, 257, 128617. [Google Scholar] [CrossRef]
- He, X.; Kang, Y.; Li, W.; Liu, W.; Xie, P.; Liao, L.; Huang, L.; Yao, M.; Qian, L.; Liu, Z.; et al. Genome-wide identification and functional analysis of the TIFY gene family in the response to multiple stresses in Brassica napus L. BMC Genomics 2020, 21, 736. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, Y.; Liao, J.; Wang, D. Genome-wide identification and expression analysis of jasmonate ZIM domain gene family in tuber mustard (Brassica juncea var. tumida). PLoS ONE 2020, 15, e0234738. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Song, Y.; Wang, Q.; Yao, D.; Li, D.; Qin, W.; Ge, X.; Yang, Z.; Xu, W.; Su, Z.; et al. Gossypium hirsutum Salt Tolerance Is Enhanced by Overexpression of G. arboreum JAZ1. Front. Bioeng. Biotechnol. 2020, 8, 157. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]






| Name | ID | Position | Genome (bp) | Exon | CDS (bp) | Protein (aa) | MW (Da) | PI |
|---|---|---|---|---|---|---|---|---|
| BrJAZ1a | BraA06g014970.3.5C | A06 | 2797 | 4 | 765 | 254 | 27,300 | 9.85 |
| BrJAZ1b | BraA08g028950.3.5C | A08 | 2038 | 4 | 780 | 259 | 28,310 | 9.49 |
| BrJAZ1c | BraA09g058470.3.5C | A09 | 1909 | 4 | 969 | 322 | 36,007 | 9.95 |
| BrJAZ2a | BraA02g023330.3.5C | A02 | 1516 | 5 | 723 | 240 | 26,198 | 9.21 |
| BrJAZ2b | BraA07g039730.3.5C | A07 | 1618 | 5 | 738 | 245 | 26,810 | 9.2 |
| BrJAZ3a | BraA01g036030.3.5C | A01 | 3096 | 7 | 1071 | 356 | 37,877 | 9.37 |
| BrJAZ3b | BraA05g030660.3.5C | A05 | 3273 | 7 | 1008 | 335 | 35,868 | 9.51 |
| BrJAZ5a | BraA06g013220.3.5C | A06 | 1466 | 4 | 357 | 118 | 13,379 | 7.9 |
| BrJAZ5b | BraA08g029870.3.5C | A08 | 1573 | 4 | 813 | 270 | 30,032 | 8.32 |
| BrJAZ5c | BraA09g059410.3.5C | A09 | 1762 | 4 | 900 | 299 | 33,021 | 9.12 |
| BrJAZ6a | BraA02g021780.3.5C | A02 | 1710 | 4 | 822 | 292 | 32,429 | 9.1 |
| BrJAZ6b | BraA07g037970.3.5C | A07 | 1812 | 4 | 825 | 274 | 30,596 | 9.08 |
| BrJAZ7a | BraA03g018070.3.5C | A03 | 342 | 1 | 342 | 113 | 13,014 | 8.74 |
| BrJAZ7b | BraA04g025940.3.5C | A04 | 501 | 2 | 360 | 119 | 13,705 | 9.76 |
| BrJAZ7c | BraA05g010440.3.5C | A05 | 556 | 2 | 351 | 116 | 13,144 | 9.64 |
| BrJAZ8a | BraA08g023940.3.5C | A08 | 586 | 3 | 393 | 130 | 14,972 | 9.85 |
| BrJAZ8b | BraA09g036440.3.5C | A09 | 582 | 3 | 402 | 133 | 15,329 | 9.62 |
| BrJAZ9a | BraA07g030900.3.5C | A07 | 1518 | 5 | 639 | 212 | 23,526 | 9.03 |
| BrJAZ9b | BraA07g036560.3.5C | A07 | 2677 | 7 | 804 | 267 | 28,813 | 9.76 |
| BrJAZ10a | BraA02g004800.3.5C | A02 | 1532 | 4 | 555 | 187 | 21,005 | 10.07 |
| BrJAZ10b | BraA03g005750.3.5C | A03 | 1783 | 4 | 552 | 183 | 20,569 | 9.95 |
| BrJAZ10c | BraA10g025610.3.5C | A10 | 1557 | 5 | 591 | 196 | 21,819 | 9.91 |
| BrJAZ12a | BraA02g009340.3.5C | A02 | 1810 | 5 | 666 | 221 | 23,083 | 4.97 |
| BrJAZ12b | BraA10g019850.3.5C | A10 | 1431 | 4 | 528 | 195 | 20,395 | 7.93 |
| BrJAZ13 | BraA05g025740.3.5C | A05 | 549 | 3 | 372 | 123 | 14,005 | 9.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Liang, C.; Feng, Q.; Wang, X.; Li, K.; Li, Z.; Zhang, Y.; Zhang, Y.; Liu, Y. Genome-Wide Analysis of the JAZ Family in Brassica rapa and the Roles of BrJAZ1a and 6b in Response to Stresses. Int. J. Mol. Sci. 2026, 27, 289. https://doi.org/10.3390/ijms27010289
Liang C, Feng Q, Wang X, Li K, Li Z, Zhang Y, Zhang Y, Liu Y. Genome-Wide Analysis of the JAZ Family in Brassica rapa and the Roles of BrJAZ1a and 6b in Response to Stresses. International Journal of Molecular Sciences. 2026; 27(1):289. https://doi.org/10.3390/ijms27010289
Chicago/Turabian StyleLiang, Chuang, Qingchang Feng, Xingliang Wang, Kaixin Li, Zhixu Li, Yan Zhang, Yaowei Zhang, and Yan Liu. 2026. "Genome-Wide Analysis of the JAZ Family in Brassica rapa and the Roles of BrJAZ1a and 6b in Response to Stresses" International Journal of Molecular Sciences 27, no. 1: 289. https://doi.org/10.3390/ijms27010289
APA StyleLiang, C., Feng, Q., Wang, X., Li, K., Li, Z., Zhang, Y., Zhang, Y., & Liu, Y. (2026). Genome-Wide Analysis of the JAZ Family in Brassica rapa and the Roles of BrJAZ1a and 6b in Response to Stresses. International Journal of Molecular Sciences, 27(1), 289. https://doi.org/10.3390/ijms27010289







