Metabolomics Combined with Photosynthetic Analysis Reveals Potential Mechanisms of Phenolic Compound Accumulation in Lonicera japonica Induced by Nitrate Nitrogen Supply
Abstract
1. Introduction
2. Results
2.1. Plant Growth and Physiology
2.2. Metabolomic Profile Analyses
2.3. Key Differential Metabolites and Pathways
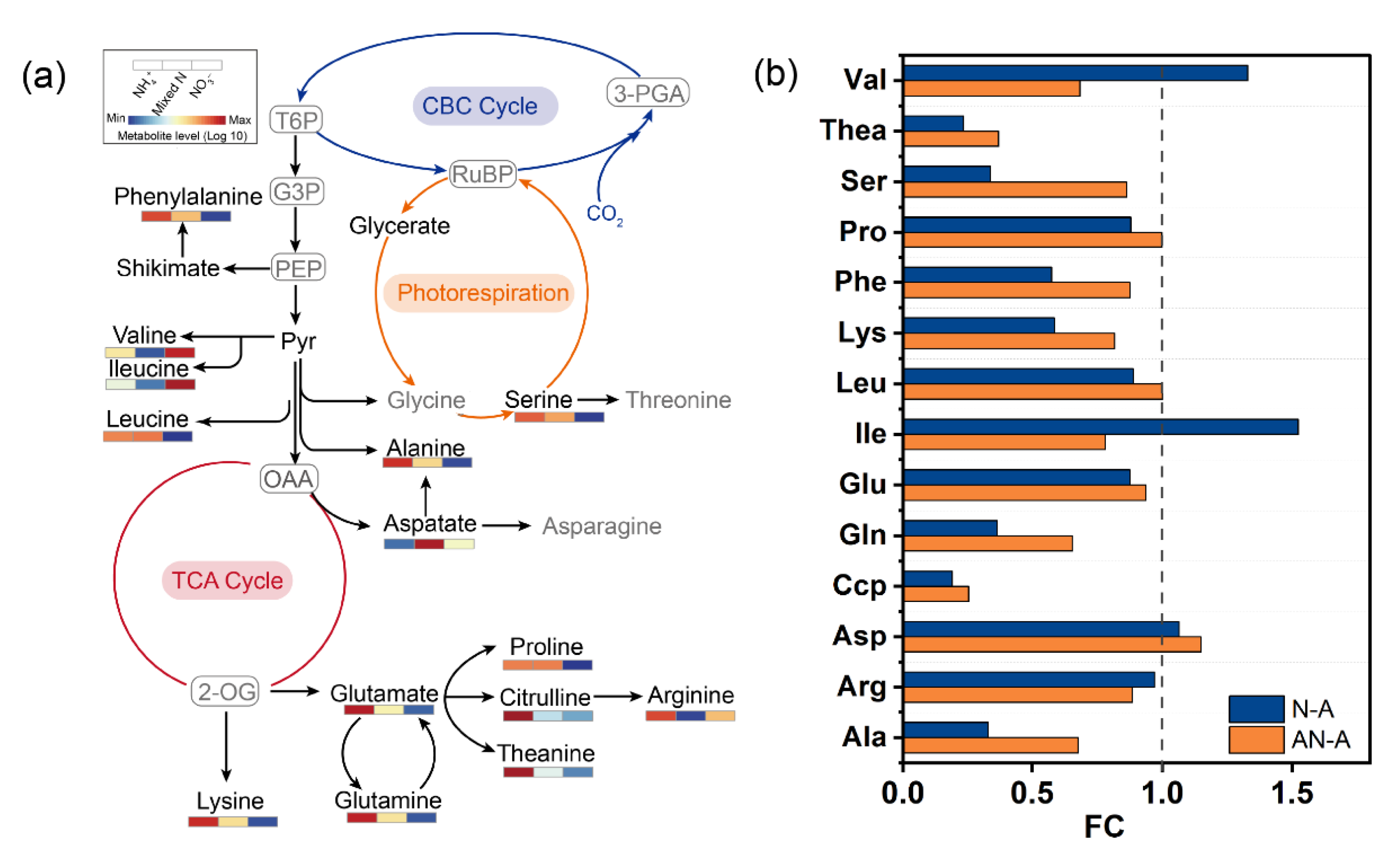
2.4. Amino Acid Metabolism
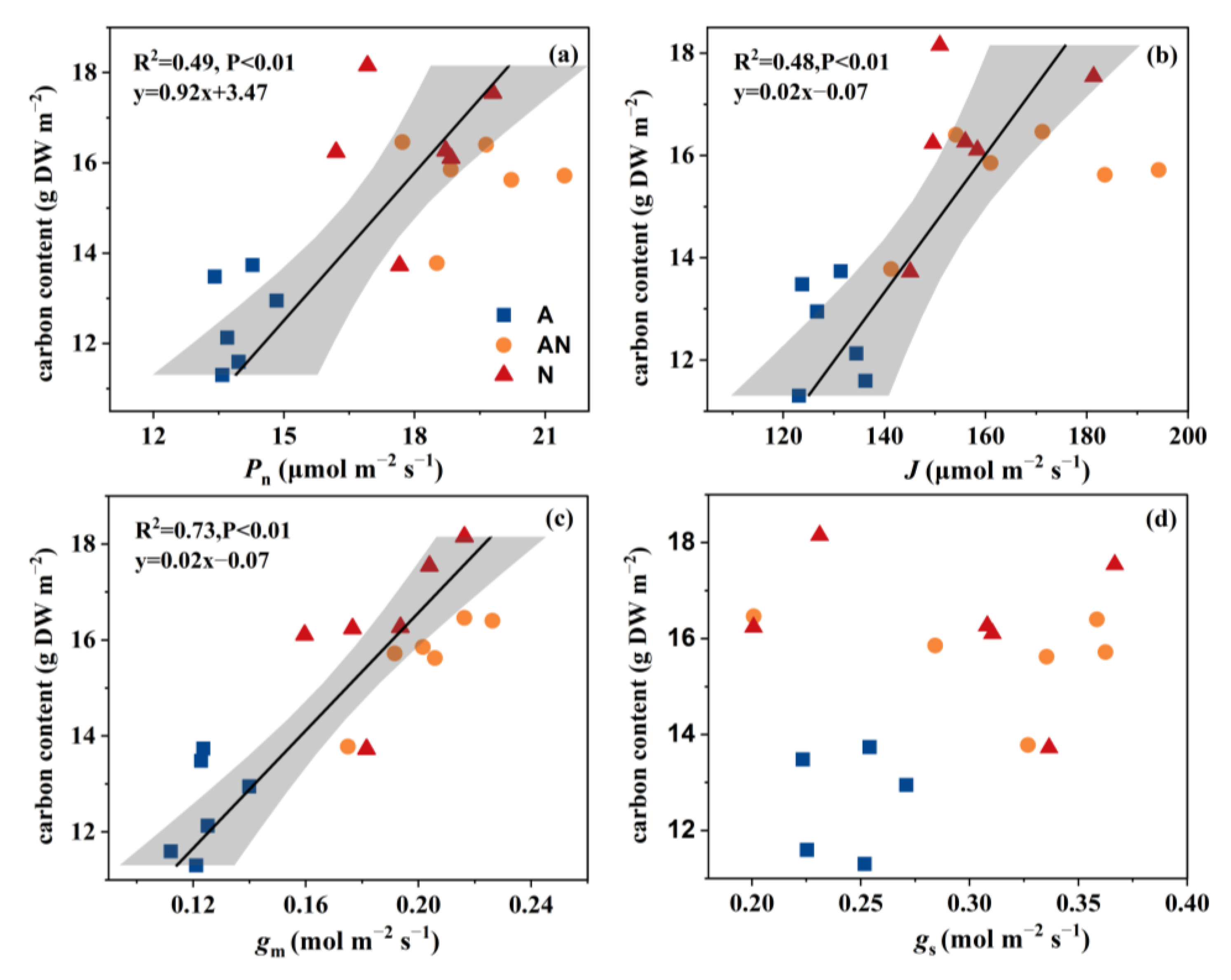
2.5. C Metabolism
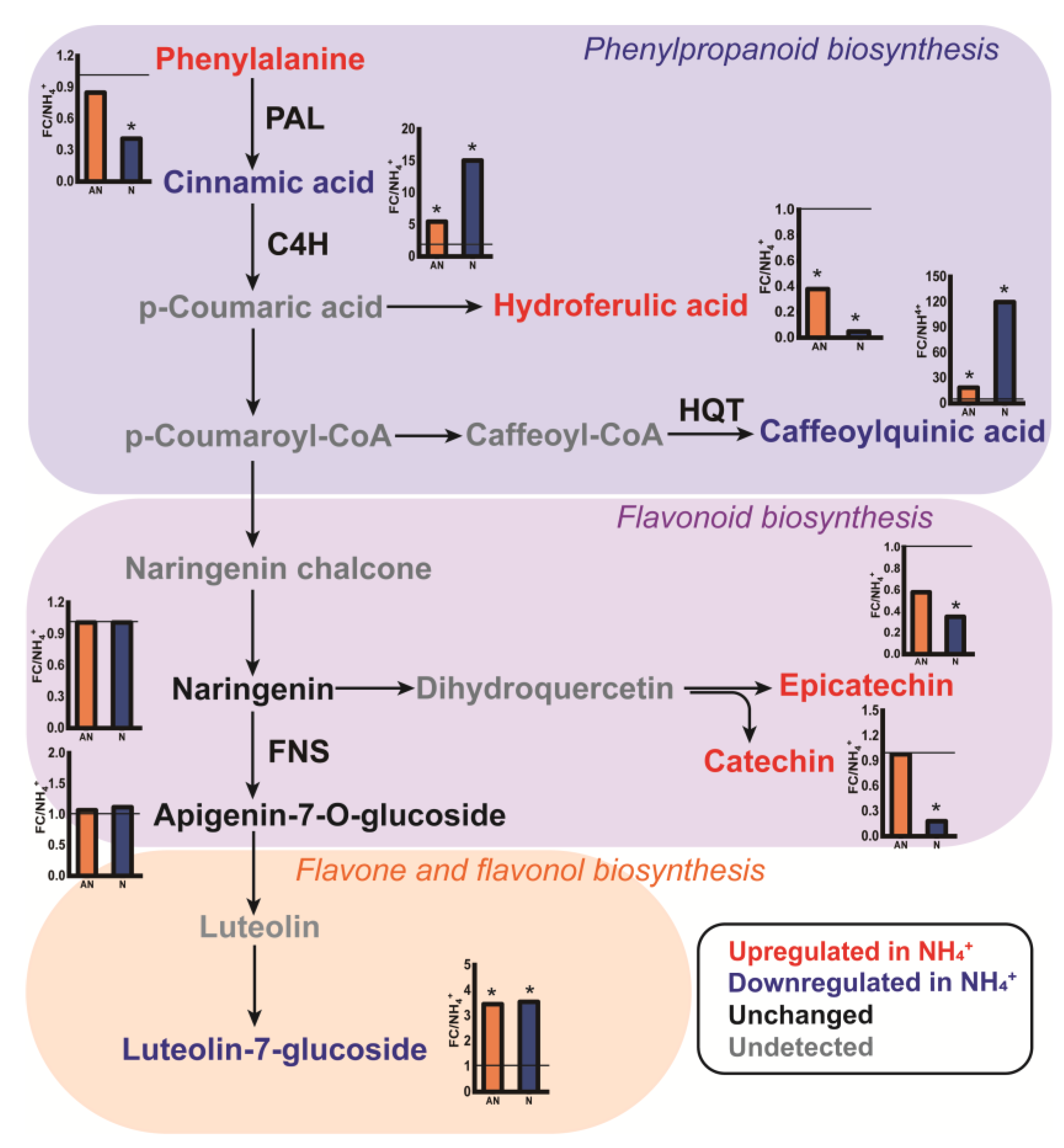
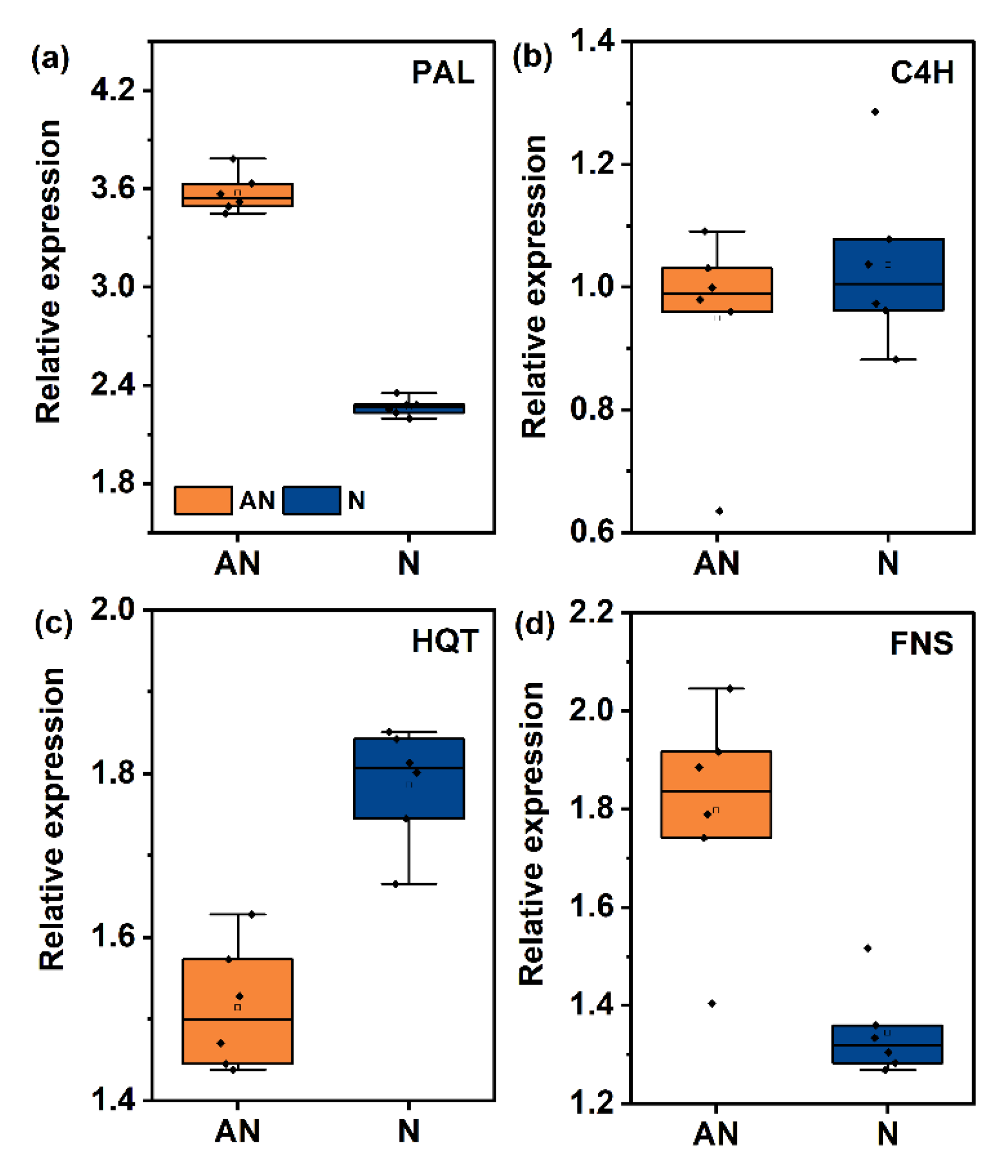
2.6. Biosynthesis and Accumulation of Phenolic Metabolites
3. Discussion
3.1. Photosynthesis and C/N Metabolism in L. japonica in Response to N Source
3.2. Phenolic Metabolism Patterns in L. japonica in Response to the N Source
4. Materials and Methods
4.1. Plant Material and Growth Condition
4.2. Gas Exchange and Fluorescence Measurements
4.3. Measurement of the CO2 Compensation Point in the Absence of Mitochondrial Respiration (Γ*)
4.4. Measurement of Leaf Area and Biomass
4.5. Physiology Measurement
4.6. Metabolomic Analyses
4.7. Total RNA Extraction and qPCR Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| C | carbon |
| CNB | carbon–nutrition balance |
| C4H | cinnamate 4−hydroxylase |
| CGA | chlorogenic acids |
| DEMs | differential expressed metabolites |
| ETR | electron transfer rate |
| FC | fold change |
| FNS | flavone synthase |
| gm | mesophyll conductance |
| GS | glutamine synthetase |
| GOGAT | glutamate synthetase |
| HQT | hydroxycinnamoyl–CoA quinate transferase |
| N | nitrogen |
| NR | nitrate reductase |
| NiR | nitrite reductase |
| OPLS−DA | orthogonal partial least squares discriminant analyses |
| PAL | phenylalanine ammonia−lyase |
| PCA | principal component analyses |
| PEPC | phosphoenolpyruvate carboxylase |
| Pn | net photosynthetic rates |
| PPFD | photosynthetic photon flux density |
| SPS | sucrose phosphate synthase |
| VIP | variable importance in the project |
| Γ* | CO2 compensation point in the absence of mitochondrial respiration |
References
- Pu, X.; Li, Z.; Tian, Y.; Gao, R.; Hao, L.; Hu, Y.; Song, J. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol. 2020, 227, 930–943. [Google Scholar] [CrossRef]
- Schierenbeck, K. Japanese honeysuckle (Lonicera japonica) as an invasive species; history, ecology, and context. Crit. Rev. Plant Sci. 2004, 23, 391–400. [Google Scholar] [CrossRef]
- Lee, Y.; Chang, M.; Yeh, Y.; Huang, C.; Lin, F.; Huang, J. Honeysuckle Aqueous Extracts Induced Let-7a Suppress EV71 Replication and Pathogenesis In Vitro and In Vivo and Is Predicted to Inhibit SARS-CoV-2. Viruses 2021, 13, 308. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Kuang, X.; Wang, W.; Wan, C.; Li, W. Comparison of chemical constitution and bioactivity among different parts of Lonicera japonica Thunb. J. Sci. Food Agric. 2020, 100, 614–622. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, G.; Zhu, P.; Zhu, W.; Li, Y.; Fan, X. Metabolite profiles and antibacterial and antioxidant activities of leaf extracts of five Lonicera species: A comparative study. Chem. Biol. Tech. Agri. 2023, 10, 91. [Google Scholar] [CrossRef]
- Albuquerque, B.; Heleno, S.; Oliveira, M.; Barros, L.; Ferreira, I. Phenolic compounds: Current industrial applications, limitations and future challenges. Food Func. 2021, 12, 14–29. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant. 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Cheynier, V. Phenolic compounds: From plants to foods. Phytochem. Rev. 2012, 11, 153–177. [Google Scholar] [CrossRef]
- Bistgani, Z.; Hashemi, M.; DaCosta, M.; Craker, L.; Maggi, F.; Morshedloo, M. Effect of salinity stress on the physiological characteristics, phenolic compounds and antioxidant activity of Thymus vulgaris L. and Thymus daenensis Celak. Ind. Crops Prod. 2019, 135, 311–320. [Google Scholar] [CrossRef]
- Nichols, S.; Hofmann, R.; Williams, W. Physiological drought resistance and accumulation of leaf phenolics in white clover interspecific hybrids. Environ. Exp. Bot. 2015, 119, 40–47. [Google Scholar] [CrossRef]
- Chandler, S.; Dodds, J. The effect of phosphate, nitrogen and sucrose on the production of phenolics and solasodine in callus cultures of Solanum laciniatum. Plant Cell Rep. 1983, 2, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Sobuj, N.; Virjamo, V.; Zhang, Y.; Nybakken, L.; Julkunen, R. Impacts of elevated temperature and CO2 concentration on growth and phenolics in the sexually dimorphic Populus tremula (L.). Environ. Exp. Bot. 2018, 146, 34–44. [Google Scholar] [CrossRef]
- Ghasemzadeh, A.; Jaafar, H.; Rahmat, A.; Wahab, P.; Halim, M. Effect of different light intensities on total phenolics and flavonoids synthesis and anti-oxidant activities in young ginger varieties (Zingiber officinale Roscoe). Int. J. Mol. Sci. 2010, 11, 3885–3897. [Google Scholar] [CrossRef]
- Tcherkez, G.; Carroll, A.; Abadie, C.; Mainguet, S.; Davanture, M.; Zivy, M. Protein synthesis increases with photosynthesis via the stimulation of translation initiation. Plant Sci. 2020, 291, 110352. [Google Scholar] [CrossRef]
- Guo, S.; Zhou, Y.; Gao, Y.; Li, Y.; Shen, Q. New insights into the nitrogen form effect on photosynthesis and photorespiration. Pedosphere 2007, 17, 601–610. [Google Scholar] [CrossRef]
- Wang, D.; Liu, H.; Li, S.; Zhai, G.; Shao, J.; Tao, Y. Characterization and molecular cloning of a serine hydroxymethyltransferase 1 (OsSHM1) in rice. J. Integr. Plant Biol. 2015, 57, 745–756. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B. Induction of phenolic metabolites and physiological changes in chamomile plants in relation to nitrogen nutrition. Food Chem. 2014, 142, 334–341. [Google Scholar] [CrossRef]
- Qin, J.; Yue, X.; Ling, Y.; Zhou, Y.; Li, N.; Shang, X.; Fang, S. Nitrogen form and ratio impact phenolic accumulation and relative gene expression in Cyclocarya paliurus. Trees 2021, 35, 685–696. [Google Scholar] [CrossRef]
- Lang, C.; Merkt, N.; Klaiber, I.; Pfannstiel, J.; Zörb, C. Different forms of nitrogen application affect metabolite patterns in grapevine leaves and the sensory of wine. Plant Physiol. Biochem. 2019, 143, 308–319. [Google Scholar] [CrossRef]
- Zhu, Z.; Yu, M.; Chen, Y.; Guo, Q.; Zhang, L.; Shi, H.; Liu, L. Effects of ammonium to nitrate ratio on growth, nitrogen metabolism, photosynthetic efficiency and bioactive phytochemical production of Prunella vulgaris. Pharm. Biol. 2014, 52, 1518–1525. [Google Scholar] [CrossRef]
- Prinsi, B.; Morgutti, S.; Negrini, N.; Faoro, F.; Espen, L. Insight into composition of bioactive phenolic compounds in leaves and flowers of green and purple basil. Plants 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, F.; Wan, Q.; Ruan, J. Transcriptome analysis using RNA-Seq revealed the effects of nitrogen form on major secondary metabolite biosynthesis in tea (Camellia sinensis) plants. Acta Physiol. Plant. 2018, 40, 1–17. [Google Scholar] [CrossRef]
- Domínguez, M.; Aparicio, P.; Lamsfus, C.; Cruz, C.; Martins Loução, M.; Moran, J. Nitrogen nutrition and antioxidant metabolism in ammonium-tolerant and -sensitive plants. Physiol. Plant. 2008, 132, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, K.; Ishihara, A. Metabolic engineering of the tryptophan and phenylalanine biosynthetic pathways in rice. Plant Biotech. 2009, 26, 523–533. [Google Scholar] [CrossRef]
- Li, D.; Zhang, X.; Qu, H. Delaying the biosynthesis of aromatic secondary metabolites in postharvest strawberry fruit exposed to elevated CO2 atmosphere. Food Chem. 2020, 306, 125611. [Google Scholar] [CrossRef]
- Palumbo, M.; Putz, F.; Talcott, S. Nitrogen fertilizer and gender effects on the secondary metabolism of yaupon, a caffeine-containing North American holly. Oecologia 2007, 151, 1–9. [Google Scholar] [CrossRef]
- Zhong, C.; Jian, S.; Chen, D.; Huang, X.; Miao, H. Organic nitrogen sources promote andrographolide biosynthesis by reducing nitrogen metabolism and increasing carbon accumulation in Andrographis paniculata. Plant Physiol. Bioch. 2021, 164, 82–91. [Google Scholar] [CrossRef]
- Cao, Y.; Pan, Y.; Wang, M.; Liu, T.; Meng, X.; Guo, S. The effects of different nitrogen forms on chlorophyll fluorescence and photosystem II in Lonicera japonica. J. Plant Growth Regul. 2023, 42, 4106–4117. [Google Scholar] [CrossRef]
- Lu, Z.; Hu, W.; Ren, T.; Zhu, C.; Li, X.; Cong, R.; Lu, J. Impact of K deficiency on leaves and siliques photosynthesis via metabolomics in Brassica napus. Environ. Exp. Bot. 2019, 158, 89–98. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.; Pan, Q.; Sun, X.; Chen, H.; Chen, F.; Mi, G. Increased biomass accumulation in maize grown in mixed nitrogen supply is mediated by auxin synthesis. J. Exp. Bot. 2019, 70, 1859–1873. [Google Scholar] [CrossRef]
- Turner, M.; Heuberger, A.; Kirkwood, J.; Collins, C.; Wolfrum, E.; Broeckling, C.; Jahn, C. Non-targeted metabolomics in diverse sorghum breeding lines indicates primary and secondary metabolite profiles are associated with plant biomass accumulation and photosynthesis. Front. Plant Sci. 2016, 7, 953. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, H.; Wu, J.; Xu, C. Paclobutrazol improves leaf carbon-use efficiency by increasing mesophyll conductance rate, while abscisic acid antagonizes this increased rate. Photosynthetica 2020, 58, 762–768. [Google Scholar] [CrossRef]
- Hanson, A.; Gage, D.; Shachar-Hill, Y. Plant one-carbon metabolism and its engineering. Trends Plant Sci. 2000, 5, 206–213. [Google Scholar] [CrossRef]
- Rubio-Wilhelmi, M.; Sanchez-Rodriguez, E.; Leyva, R.; Blasco, B.; Romero, L.; Ruiz, M. Response of carbon and nitrogen-rich metabolites to nitrogen deficiency in PSARK::IPT tobacco plants. Plant Physiol. Biochem. 2012, 57, 231–237. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef]
- Li, X.; Yan, X.; Liu, J. Effects of nitrogen forms on nitrogen metabolism-related enzymes and growth of Phellodendron amurense seedlings. Chin. Bull. Bot. 2006, 23, 255. [Google Scholar]
- Foyer, C.; Parry, M.; Noctor, G. Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 2003, 54, 585–593. [Google Scholar] [CrossRef]
- Poucet, T.; González-Moro, M.; Cabasson, C.; Beauvoit, B.; Gibon, Y.; Dieuaide-Noubhani, M.; Marino, D. Ammonium supply induces differential metabolic adaptive responses in tomato according to leaf phenological stage. J. Exp. Bot. 2021, 72, 3185–3199. [Google Scholar] [CrossRef]
- Izui, K.; Matsumura, H.; Furumoto, T. Phosphoenolpyruvate carboxylase: A new era of structural biology. Annu. Rev. Plant Biol. 2004, 55, 69–84. [Google Scholar] [CrossRef]
- Masumoto, C.; Gantt, E. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc. Natl. Acad. Sci. USA 2010, 107, 5226–5231. [Google Scholar] [CrossRef]
- Ruan, J.; Haerdter, R.; Gerendás, J. Impact of nitrogen supply on carbon/nitrogen allocation: A case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Biol. 2010, 12, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 2015, 25, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhou, G.; Qin, H.; Guan, Y.; Wang, T.; Ni, W.; Ge, S. Metabolomics combined with physiology and transcriptomics reveal key metabolic pathway responses in apple plants exposure to different selenium concentrations. J. Hazard. Mater. 2024, 464, 132953. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Giglione, C.; Lee, D.; An, S.; Jeong, D.H.; Meinnel, T.; An, G. Rice peptide deformylase PDF1B is crucial for development of chloroplasts. Plant Cell Physiol. 2008, 49, 1536–1546. [Google Scholar] [CrossRef]
- Balasundram, N.; Sundram, K.; Samman, S. Phenolic compounds in plants and agri-industrial by-products: Antioxidant activity, occurrence, and potential uses. Food Chem. 2006, 99, 191–203. [Google Scholar] [CrossRef]
- Kong, D.; Li, Y.; Wang, M.; Bai, M.; Zou, R.; Tang, H.; Wu, H. Effects of light intensity on leaf photosynthetic characteristics, chloroplast structure, and alkaloid content of Mahonia bodinieri (Gagnep.) Laferr. Acta Physiol. Plant. 2016, 38, 120. [Google Scholar] [CrossRef]
- Matt, P.; Krapp, A.; Haake, V.; Mock, H.; Stitt, M. Decreased Rubisco activity leads to dramatic changes of nitrate metabolism, amino acid metabolism and the levels of phenylpropanoids and nicotine in tobacco antisense RBCS transformants. Plant J. 2002, 30, 663–677. [Google Scholar] [CrossRef]
- Montanari, M.; Degl Innocenti, E.; Maggini, R.; Pacifici, S.; Pardossi, A.; Guidi, L. Effect of nitrate fertilization and saline stress on the contents of active constituents of Echinacea angustifolia DC. Food Chem. 2008, 107, 1461–1466. [Google Scholar] [CrossRef]
- Fritz, C.; Palacios-Rojas, N.; Feil, R.; Stitt, M. Regulation of secondary metabolism by the carbon-nitrogen status in tobacco: Nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 2006, 46, 533–548. [Google Scholar] [CrossRef]
- Moura, J.; Bonine, C.; Fernandes Viana, J.; Dornelas, M.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Liu, Q.; Zheng, L.; He, F.; Zhao, F.; Shen, Z.; Zheng, L. Transcriptional and physiological analyses identify a regulatory role for hydrogen peroxide in the lignin biosynthesis of copper-stressed rice roots. Plant Soil 2015, 387, 323–336. [Google Scholar] [CrossRef]
- Panche, A.; Diwan, A.; Chandra, S. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Yang, H.; Wei, Z.; Yang, H.; Fan, S.; Li, W. Effects of Different Nitrogen Forms on Blackberry Fruit Quality. Foods 2023, 12, 2318. [Google Scholar] [CrossRef] [PubMed]
- Fallovo, C.; Schreiner, M.; Schwarz, D.; Colla, G.; Krumbein, A. Phytochemical changes induced by different nitrogen supply forms and radiation levels in two leafy Brassica species. J. Agric. Food Chem. 2011, 59, 4198–4207. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, Y.; Zhang, Z.; Zhang, Y.; Jiang, Y.; Sun, Z. Transcriptomics and metabolomics reveal the primary and secondary metabolism changes in Glycyrrhiza uralensis with different forms of nitrogen utilization. Front. Plant Sci. 2023, 14, 1229253. [Google Scholar] [CrossRef]
- Tang, D.; Liu, M.; Zhang, Q.; Ma, L.; Shi, Y.; Ruan, J. Preferential assimilation of NH4+ over NO3− in tea plant associated with genes involved in nitrogen transportation, utilization and catechins biosynthesis. Plant Sci. 2020, 291, 110369. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Lu, Y.; Qiu, Q.; Fan, D.; Wang, X.; Zheng, X. Influence of different nitrogen sources on carbon and nitrogen metabolism and gene expression in tea plants (Camellia sinensis L.). Plant Physiol. Biochem. 2021, 167, 561–566. [Google Scholar] [CrossRef]
- Han, J.; Gu, L.; Warren, J.M.; Guha, A.; Mclennan, D.; Zhang, W.; Zhang, Y. The roles of photochemical and non-photochemical quenching in regulating photosynthesis depend on the phases of fluctuating light conditions. Tree Physiol. 2022, 42, 848–861. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Wang, B.; Li, Y.; Ding, L.; Wang, M.; Guo, S. Leaf nitrate accumulation influences the photorespiration of rice (Oryza sativa L.) seedlings. Plant Soil. 2020, 456, 323–338. [Google Scholar] [CrossRef]



| Treatment | Leaf Biomass (g Leaf−1 FW) | Leaf Area (cm2) | C Content (g m−2 DW) | N Content (g m−2 DW) | NH4+ (μmol g−1 FW) | NO3− (μmol g−1 FW) | C/N Ratio |
|---|---|---|---|---|---|---|---|
| A | 0.16 ± 0.02 b | 16.6 ± 1.9 a | 12.5 ± 1.0 b | 1.7 ± 0.1 a | 4.9 ± 0.4 a | 17.0 ± 1.4 b | 7.2 ± 0.2 c |
| AN | 0.22 ± 0.05 a | 17.8 ± 2.5 a | 15.6 ± 1.0 a | 1.8 ± 0.1 a | 4.6 ± 0.5 a | 18.2 ± 1.5 b | 8.7 ± 0.7 b |
| N | 0.22 ± 0.03 a | 16.3 ± 1.5 a | 16.3 ± 1.5 a | 1.5 ± 0.1 b | 5.0 ± 0.3 a | 25.1 ± 1.1 a | 10.7 ± 0.5 a |
| Treatment | Pn (μmol m−2 s−1) | gs( mol m−2 s−1) | gm (mol m−2 s−1) | ETR (μmol m−2 s−1) | Γ* (μmol mol−1) |
|---|---|---|---|---|---|
| A | 14.0 ± 0.5 b | 0.23 ± 0.04 b | 0.12 ± 0.01 c | 129 ± 6 b | 43.8 ± 0.7 b |
| AN | 19.3 ± 1.5 a | 0.35 ± 0.12 a | 0.21 ± 0.01 a | 168 ± 19 a | 43.1 ± 1.0 b |
| N | 18.2 ± 1.3 a | 0.29 ± 0.06 ab | 0.19 ± 0.01 b | 157 ± 13 a | 46.2 ± 0.8 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Yang, Y.; Tan, Z.; Feng, X.; Tian, Z.; Liu, T.; Pan, Y.; Wang, M.; Su, X.; Liang, H.; et al. Metabolomics Combined with Photosynthetic Analysis Reveals Potential Mechanisms of Phenolic Compound Accumulation in Lonicera japonica Induced by Nitrate Nitrogen Supply. Int. J. Mol. Sci. 2025, 26, 4464. https://doi.org/10.3390/ijms26094464
Cao Y, Yang Y, Tan Z, Feng X, Tian Z, Liu T, Pan Y, Wang M, Su X, Liang H, et al. Metabolomics Combined with Photosynthetic Analysis Reveals Potential Mechanisms of Phenolic Compound Accumulation in Lonicera japonica Induced by Nitrate Nitrogen Supply. International Journal of Molecular Sciences. 2025; 26(9):4464. https://doi.org/10.3390/ijms26094464
Chicago/Turabian StyleCao, Yiwen, Yating Yang, Zhengwei Tan, Xihan Feng, Zhiyao Tian, Tianheng Liu, Yonghui Pan, Min Wang, Xiaoyu Su, Huizhen Liang, and et al. 2025. "Metabolomics Combined with Photosynthetic Analysis Reveals Potential Mechanisms of Phenolic Compound Accumulation in Lonicera japonica Induced by Nitrate Nitrogen Supply" International Journal of Molecular Sciences 26, no. 9: 4464. https://doi.org/10.3390/ijms26094464
APA StyleCao, Y., Yang, Y., Tan, Z., Feng, X., Tian, Z., Liu, T., Pan, Y., Wang, M., Su, X., Liang, H., & Guo, S. (2025). Metabolomics Combined with Photosynthetic Analysis Reveals Potential Mechanisms of Phenolic Compound Accumulation in Lonicera japonica Induced by Nitrate Nitrogen Supply. International Journal of Molecular Sciences, 26(9), 4464. https://doi.org/10.3390/ijms26094464






