P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development
Abstract
1. Introduction
2. Results
2.1. Phenotypic Features of ys Mutants
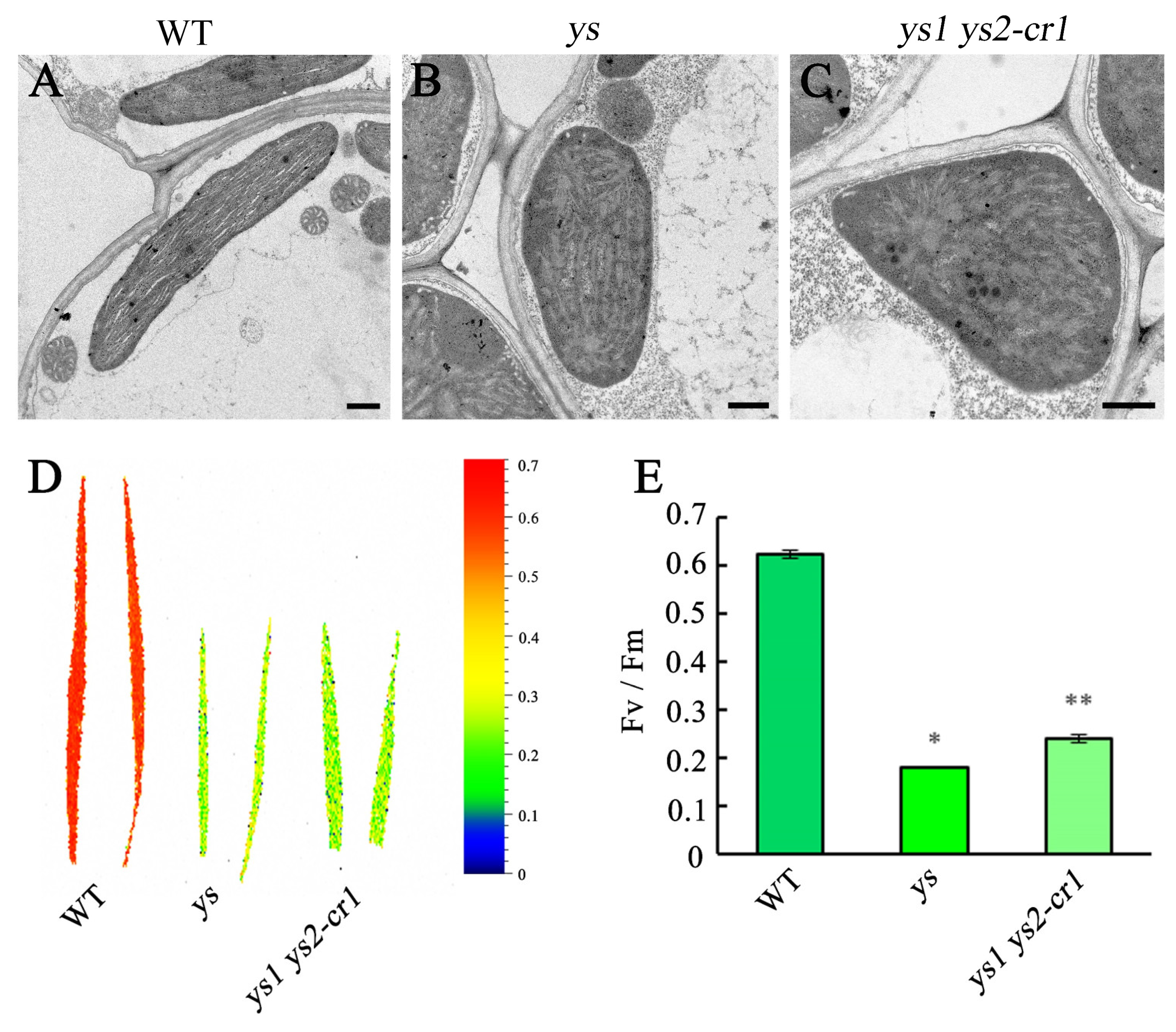
2.2. The Chloroplast Thylakoid Structure Was Collapsed in ys
2.3. Photosynthetic Activity Was Reduced in ys
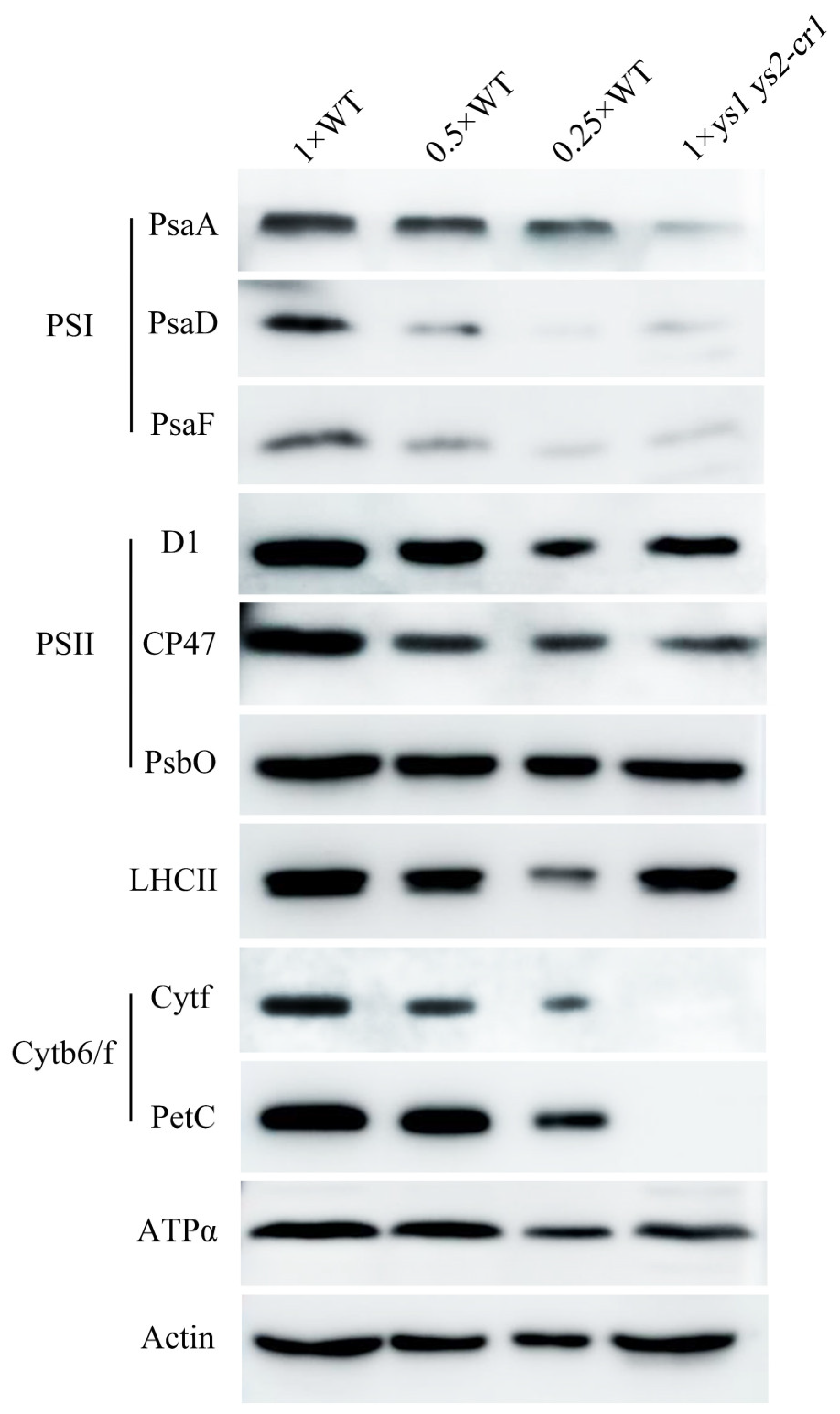
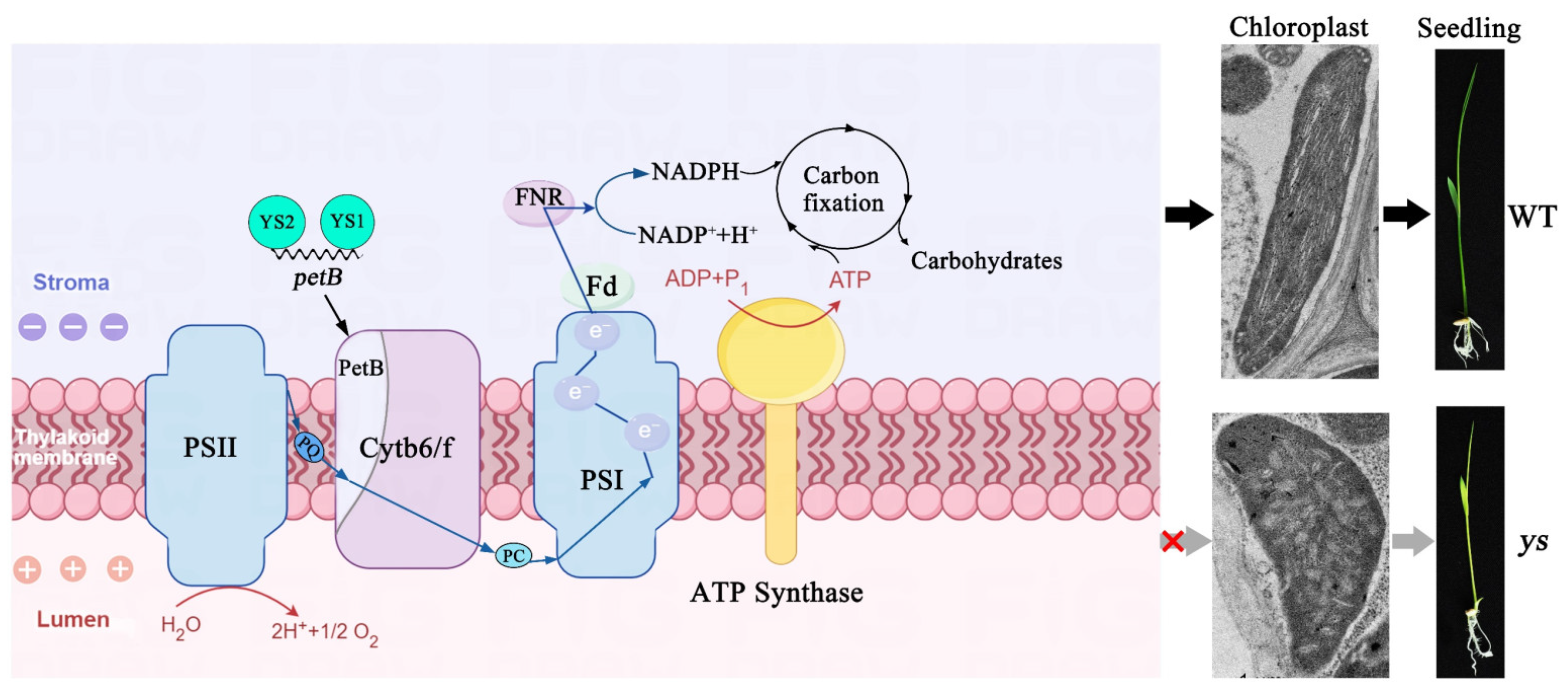
2.4. PSI and Cytochrome b6/f Complex Were Impaired in ys
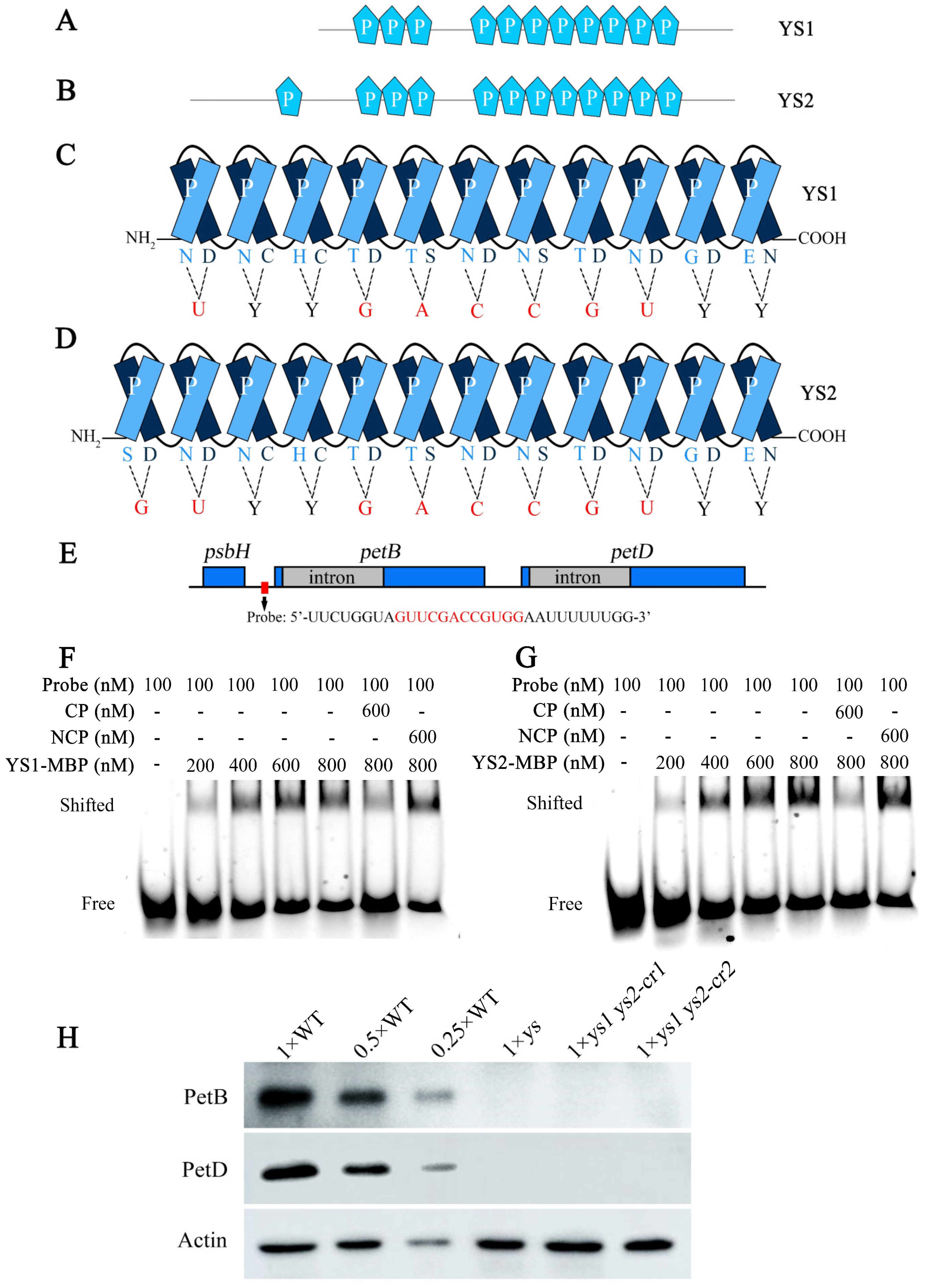
2.5. YS1 and YS2 Encode Two Highly Conserved P-Type PPR Proteins
2.6. YS1 and YS2 Were Predominantly Expressed in Leaves
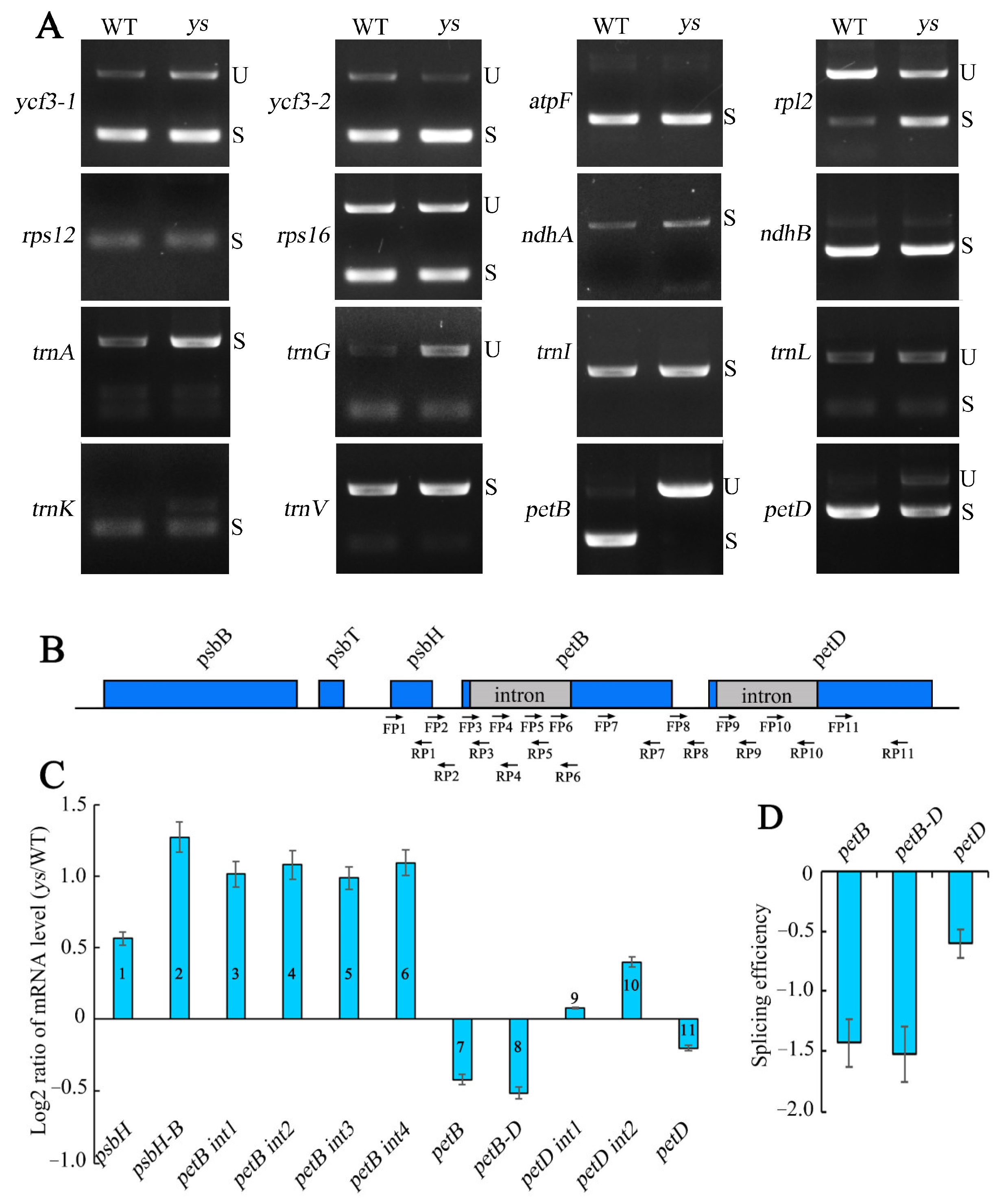
2.7. petB and petD Intron Splicing Were Impeded in ys Mutants
2.8. YS1 and YS2 Directly Bound to the psbH-petB Intergenic Region Transcript
2.9. Photosynthetic Electron Transfer Was Damaged in ys Mutants
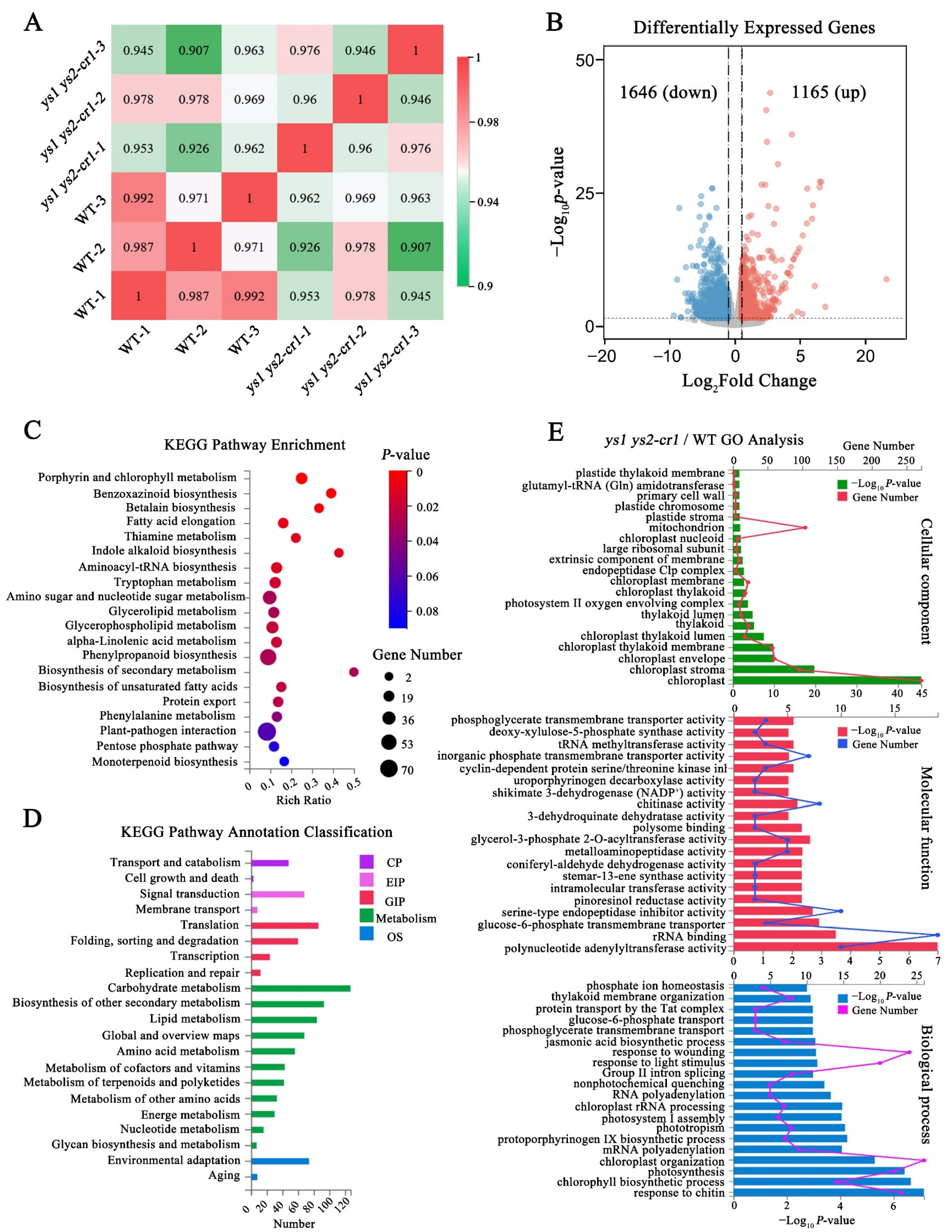
2.10. Altered Expression Profile in ys Mutants
2.11. Carbohydrate Contents in Photosynthesis Were Decreased in ys
3. Discussion
3.1. YS1 and YS2 Participated in the Splicing of the petB Intron
3.2. YS1 and YS2 Were Essential for Functional PSI and Cytochrome b6/f Complex
3.3. YS1 and YS2 Were Duplicated Genes and Essential for Rice Chloroplast Development
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Measurement of Chlorophyll and Carotenoid Contents
4.3. Transmission Electron Microscopy (TEM)
4.4. Detection of Photosynthetic Efficiency
4.5. Protein Isolation and Immunoblot Analysis
4.6. Phylogenetic Analysis
4.7. PPR Motif Alignment Among Homologous Proteins
4.8. Total RNA Extraction and RT-PCR
4.9. Prokaryotic Protein Expression, Purification, and RNA Electrophoretic Mobility Shift Assays (REMSA)
4.10. Detection of NADPH Concentration
4.11. Transcriptome Sequencing and Analysis
4.12. Determination of Sugars and Starch Contents
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stadnichuk, I.N.; Kusnetsov, V.V. Endosymbiotic origin of chloroplasts in plant cells’ evolution. Russ. J. Plant Physl. 2021, 68, 1–6. [Google Scholar] [CrossRef]
- Sato, N. Endosymbiotic Theories of Organelles Revisited: Retrospects and Prospects; Springer Nature: Berlin, Germany, 2020. [Google Scholar]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.-Y. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef]
- Mukhopadhyay, J.; Hausner, G. Organellar introns in fungi, algae, and plants. Cells 2021, 10, 2001. [Google Scholar] [CrossRef]
- De Longevialle, A.F.; Small, I.D.; Lurin, C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 2010, 3, 691–705. [Google Scholar] [CrossRef]
- Zeng, C.; Jiao, Q.; Jia, T.; Hu, X. Updated progress on group II intron splicing factors in plant chloroplasts. Curr. Issues Mol. Biol. 2022, 44, 4229–4239. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Liu, S.; Teng, Q.; Li, S.; Jiang, Y. Functions of PPR proteins in plant growth and development. Int. J. Mol. Sci. 2021, 22, 11274. [Google Scholar] [CrossRef]
- Lurin, C.; Andreés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveal their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Williams-Carrier, R.E.; Williams-Voelker, P.M.; Kroeger, T.S.; Vichas, A.; Barkan, A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 2006, 18, 2650–2663. [Google Scholar] [CrossRef] [PubMed]
- De Longevialle, A.F.; Hendrickson, L.; Taylor, N.L.; Delannoy, E.; Lurin, C.; Badger, M.; Millar, A.H.; Small, L. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 2008, 56, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Tadini, L.; Ferrari, R.; Lehniger, M.-K.; Mizzotti, C.; Moratti, F.; Resentini, F.; Colombo, M.; Costa, A.; Masiero, S.; Pesaresi, P. Trans-splicing of plastid rps12 transcripts, mediated by At PPR4, is essential for embryo patterning in Arabidopsis thaliana. Planta 2018, 248, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, S.J.; des Francs-Small, C.C.; Whitby, M.; Small, I.; Kang, H. The coordinated action of PPR4 and EMB2654 on each intron half mediates trans-splicing of rps12 transcripts in plant chloroplasts. Plant J. 2019, 100, 1193–1207. [Google Scholar] [CrossRef]
- Tan, J.; Tan, Z.; Wu, F.; Sheng, P.; Heng, Y.; Wang, X.; Ren, Y.; Wang, J.; Guo, X.; Zhang, X.; et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. Plant 2014, 7, 1329–1349. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, Y.; Zhou, K.; Liu, L.; Wang, J.; Xu, Y.; Zhang, H.; Zhang, L.; Feng, Z.; Wang, L.; et al. WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front. Plant Sci. 2017, 8, 1116. [Google Scholar] [CrossRef]
- Liu, X.; Lan, J.; Huang, Y.; Cao, P.; Zhou, C.; Ren, Y.; He, N.; Liu, S.; Tian, Y.; Nguyen, T.; et al. WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J. Exp. Bot. 2018, 69, 3949–3961. [Google Scholar] [CrossRef]
- Huang, W.; Zhu, Y.; Wu, W.; Li, X.; Zhang, D.; Yin, P.; Huang, J. The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant Physiol. 2018, 177, 684–697. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, W.; Wen, K.; Chen, G.; Sun, J.; Tian, Y.; Tang, W.; Yu, J.; An, H.; Wu, T.; et al. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 2017, 95, 345–357. [Google Scholar] [CrossRef]
- Wang, D.; Liu, H.; Zhai, G.; Wang, L.; Shao, J.; Tao, Y. OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J. Genet. Genom. 2016, 43, 601–608. [Google Scholar] [CrossRef]
- Barkan, A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988, 7, 2637–2644. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Ke, X.; Li, L.; Liu, Y.; Yuan, S.; Wang, Q.; Hou, X.; Zhao, J. ALBINO EMBRYO AND SEEDLING is required for RNA splicing and chloroplast homeostasis in Arabidopsis. Plant Physiol. 2023, 193, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Felder, S.; Meierhoff, K.; Sane, A.P.; Meurer, J.; Driemel, C.; Plucken, H.; Klaff, P.; Stein, B.; Bechtold, N.; Westhoff, P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell 2001, 13, 2127–2141. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Walker, M.; Nolasco, M.; Johnson, D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994, 13, 3170–3181. [Google Scholar] [CrossRef]
- Asano, T.; Miyao, A.; Hirochika, H.; Kikuchi, S.; Kadowaki, K.-I. A pentatricopeptide repeat gene of rice is required for splicing of chloroplast transcripts and RNA editing of ndhA. Plant Biotechnol. 2013, 30, 57–64. [Google Scholar] [CrossRef]
- Wang, Z.-W.; Lv, J.; Xie, S.-Z.; Zhang, Y.; Qiu, Z.-N.; Chen, P.; Cui, Y.-T.; Niu, Y.-F.; Hu, S.-K.; Jiang, H.-Z.; et al. OsSLA4 encodes a pentatricopeptide repeat protein essential for early chloroplast development and seedling growth in rice. Plant Growth Regul. 2018, 84, 249–260. [Google Scholar] [CrossRef]
- Yu, L.; Nie, Y.; Jiao, J.; Jian, L.; Zhao, J. The sequencing-based mapping method for effectively cloning plant mutated genes. Int. J. Mol. Sci. 2021, 22, 6224. [Google Scholar] [CrossRef]
- Fromme, P.; Grotjohann, I. Overview of photosynthesis. J. Am. Chem. Soc. 2008, 20, 1–22. [Google Scholar]
- Yan, J.; Yao, Y.; Hong, S.; Yang, Y.; Shen, C.; Zhang, Q.; Zhang, D.; Zou, T.; Yin, P. Delineation of pentatricopeptide repeat codes for target RNA prediction. Nucleic Acids Res. 2019, 47, 3728–3738. [Google Scholar] [CrossRef]
- Mulo, P. Chloroplast-targeted ferredoxin-NADP+ oxidoreductase (FNR): Structure, function and location. Biochim. Biophys. Acta (BBA)-Bioenergetics 2011, 1807, 927–934. [Google Scholar] [CrossRef]
- Rodriguez-Heredia, M.; Saccon, F.; Wilson, S.; Finazzi, G.; Ruban, A.V.; Hanke, G.T. Protection of photosystem I during sudden light stress depends on ferredoxin: NADP (H) reductase abundance and interactions. Plant Physiol. 2022, 188, 1028–1042. [Google Scholar] [CrossRef] [PubMed]
- Patrick, J.W.; Botha, F.C.; Birch, R.G. Metabolic engineering of sugars and simple sugar derivatives in plants. Plant Biotechnol. J. 2013, 11, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Stoppel, R.; Lezhneva, L.; Schwenkert, S.; Torabi, S.; Felder, S.; Meierhoff, K.; Westhoff, P.; Meurer, J. Recruitment of a ribosomal release factor for light-and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 2011, 23, 2680–2695. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. PrfB3: A nuclear-encoded protein recruited for regulation of transcript stability in Arabidopsis chloroplasts. Plant Cell 2011, 7, 2474. [Google Scholar] [CrossRef]
- Jiang, J.; Chai, X.; Manavski, N.; Williams-Carrier, R.; He, B.; Brachmann, A.; Ji, D.; Ouyang, M.; Liu, Y.; Barkan, A.; et al. An RNA chaperone-like protein plays critical roles in chloroplast mRNA stability and translation in Arabidopsis and maize. Plant Cell 2019, 31, 1308–1327. [Google Scholar] [CrossRef]
- Zito, F.; Kuras, R.; Choquet, Y.; Kössel, H.; Wollman, F.-A. Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 1997, 33, 79–86. [Google Scholar] [CrossRef]
- Brzezowski, P.; Ksas, B.; Havaux, M.; Grimm, B.; Chazaux, M.; Peltier, G.; Johnson, X.; Alric, J. The function of PROTOPORPHYRINOGEN IX OXIDASE in chlorophyll biosynthesis requires oxidised plastoquinone in Chlamydomonas reinhardtii. Commun. Biol. 2019, 2, 159. [Google Scholar] [CrossRef]
- Meierhoff, K.; Felder, S.; Nakamura, T.; Bechtold, N.; Schuster, G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 2003, 15, 1480–1495. [Google Scholar] [CrossRef]
- Nakamura, T.; Meierhoff, K.; Westhoff, P.; Schuster, G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003, 270, 4070–4081. [Google Scholar] [CrossRef]
- Ostheimer, G.J.; Williams-Carrier, R.; Belcher, S.; Osborne, E.; Gierke, J.; Barkan, A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003, 22, 3919–3929. [Google Scholar] [CrossRef]
- Daras, G.; Rigas, S.; Alatzas, A.; Samiotaki, M.; Chatzopoulos, D.; Tsitsekian, D.; Papadaki, V.; Templalexis, D.; Banilas, G.; Athanasiadou, A.-M.; et al. LEFKOTHEA regulates nuclear and chloroplast mRNA splicing in plants. Dev. Cell 2019, 50, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Watkins, K.P.; Kroeger, T.S.; Cooke, A.M.; Williams-Carrier, R.E.; Friso, G.; Belcher, S.E.; van Wijk, K.J.; Barkan, A. A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 2007, 19, 2606–2623. [Google Scholar] [CrossRef] [PubMed]
- Kroeger, T.S.; Watkins, K.P.; Friso, G.; van Wijk, K.J.; Barkan, A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 4537–4542. [Google Scholar] [CrossRef] [PubMed]
- Tikhonov, A.N. Induction events and short-term regulation of electron transport in chloroplasts: An overview. Photosynth. Res. 2015, 125, 65–94. [Google Scholar] [CrossRef]
- Goldschmidt-Clermont, M.; Bassi, R. Sharing light between two photosystems: Mechanism of state transitions. Curr. Opin. Plant Biol. 2015, 25, 71–78. [Google Scholar] [CrossRef]
- Rochaix, J.D. Regulation of photosynthetic electron transport. Biochim. Biophys. Acta (BBA)-Bioenergetics 2011, 1807, 375–383. [Google Scholar] [CrossRef]
- Kozuleva, M.A.; Ivanov, B.N. The mechanisms of oxygen reduction in the terminal reducing segment of the chloroplast photosynthetic electron transport chain. Plant Cell Physiol. 2016, 57, 1397–1404. [Google Scholar] [CrossRef]
- Yang, X.; Che, Y.; García, V.J.; Shen, J.; Zheng, Y.; Su, Z.; Zhu, L.; Luan, S.; Hou, X. Cyclophilin 37 maintains electron transport via the cytochrome b6/f complex under high light in Arabidopsis. Plant Physiol. 2023, 192, 2803–2821. [Google Scholar] [CrossRef]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef]
- Nandha, B.; Finazzi, G.; Joliot, P.; Hald, S.; Johnson, G.N. The role of PGR5 in the redox poising of photosynthetic electron transport. Biochim. Biophys. Acta (BBA)-Bioenergetics 2007, 1767, 1252–1259. [Google Scholar] [CrossRef]
- DalCorso, G.; Pesaresi, P.; Masiero, S.; Aseeva, E.; Schünemann, D.; Finazzi, G.; Joliot, P.; Barbato, R.; Leister, D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 2008, 132, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Fisk, D.G.; Walker, M.B.; Barkan, A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999, 18, 2621–2630. [Google Scholar] [CrossRef]
- Zhang, J.Z. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Mackie Ogilvie, C.; Scriven, P.N. Meiotic outcomes in reciprocal translocation carriers ascertained in 3-day human embryos. Eur. J. Hum. Genet. 2002, 10, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xue, C.; Liu, G.; Liu, X.; Zhang, M.; Wang, X.; Zhang, T.; Gong, Z. Segmental duplication of chromosome 11 and its implications for cell division and genome-wide expression in rice. Sci. Rep. 2017, 7, 2689. [Google Scholar] [CrossRef]
- Sheltzer, J.M.; Blank, H.M.; Pfau, S.J.; Tange, Y.; George, B.M.; Humpton, T.J.; Brito, I.L.; Hiraoka, Y.; Niwa, O.; Amon, A. Aneuploidy drives genomic instability in yeast. Science 2011, 333, 1026–1030. [Google Scholar] [CrossRef]
- Guo, M.; Birchler, J.A. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 1994, 266, 1999–2002. [Google Scholar] [CrossRef]
- Birchler, J.A.; Hiebert, J.C.; Paigen, K. Analysis of autosomal dosage compensation involving the alcohol dehydrogenase locus in Drosophila melanogaster. Genetics 1990, 124, 677–686. [Google Scholar] [CrossRef]
- Chen, G.; Zou, Y.; Hu, J.; Ding, Y. Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genom. 2018, 19, 720. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.; Hao, B.; Ge, S.; Luo, J. Duplication and DNA segmental loss in the rice genome: Implications for diploidization. New Phytol. 2005, 165, 937–946. [Google Scholar] [CrossRef]
- Mao, D.; Yu, H.; Liu, T.; Yang, G.; Xing, Y. Two complementary recessive genes in duplicated segments control etiolation in rice. Theor. Appl. Genet. 2011, 122, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Yu, L.; Mao, L.; Zou, W.; Zhang, X.; Zhao, J. Vitamin B1 THIAMIN REQUIRING1 synthase mediates the maintenance of chloroplast function by regulating sugar and fatty acid metabolism in rice. J. Integr. Plant Biol. 2022, 64, 1575–1595. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, R.K.; Tuteja, N. Development of Agrobacterium-mediated transformation technology for mature seed-derived callus tissues of indica rice cultivar IR64. GM Crops Food 2012, 3, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Analysis 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Wang, Z.; Lin, X.; Li, Z.; Ren, Y.; Zhao, J. The translocase of the inner mitochondrial membrane 22-2 is required for mitochondrial membrane function during Arabidopsis seed development. J. Exp. Bot. 2023, 74, 4427–4448. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhu, L.; Wang, Q.; Hou, X. Autophagy-Related 2 regulates chlorophyll degradation under abiotic stress conditions in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4515. [Google Scholar] [CrossRef]
- Hou, X.; Fu, A.; Garcia, V.J.; Buchanan, B.B.; Luan, S. PSB27: A thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc. Natl. Acad. Sci. USA 2015, 112, 1613–1618. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Nie, Y.; Yu, L.; Yue, X.; Hou, X.; Zhao, J. P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development. Int. J. Mol. Sci. 2025, 26, 4459. https://doi.org/10.3390/ijms26094459
Sun H, Nie Y, Yu L, Yue X, Hou X, Zhao J. P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development. International Journal of Molecular Sciences. 2025; 26(9):4459. https://doi.org/10.3390/ijms26094459
Chicago/Turabian StyleSun, Hui, Yanshen Nie, Li Yu, Xiaohong Yue, Xin Hou, and Jie Zhao. 2025. "P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development" International Journal of Molecular Sciences 26, no. 9: 4459. https://doi.org/10.3390/ijms26094459
APA StyleSun, H., Nie, Y., Yu, L., Yue, X., Hou, X., & Zhao, J. (2025). P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development. International Journal of Molecular Sciences, 26(9), 4459. https://doi.org/10.3390/ijms26094459




