Genome-Wide Association Study to Identify Soybean Lodging Resistance Loci and Candidate Genes
Abstract
1. Introduction
2. Results
2.1. Phenotype Variations of Lodging Traits in Soybean
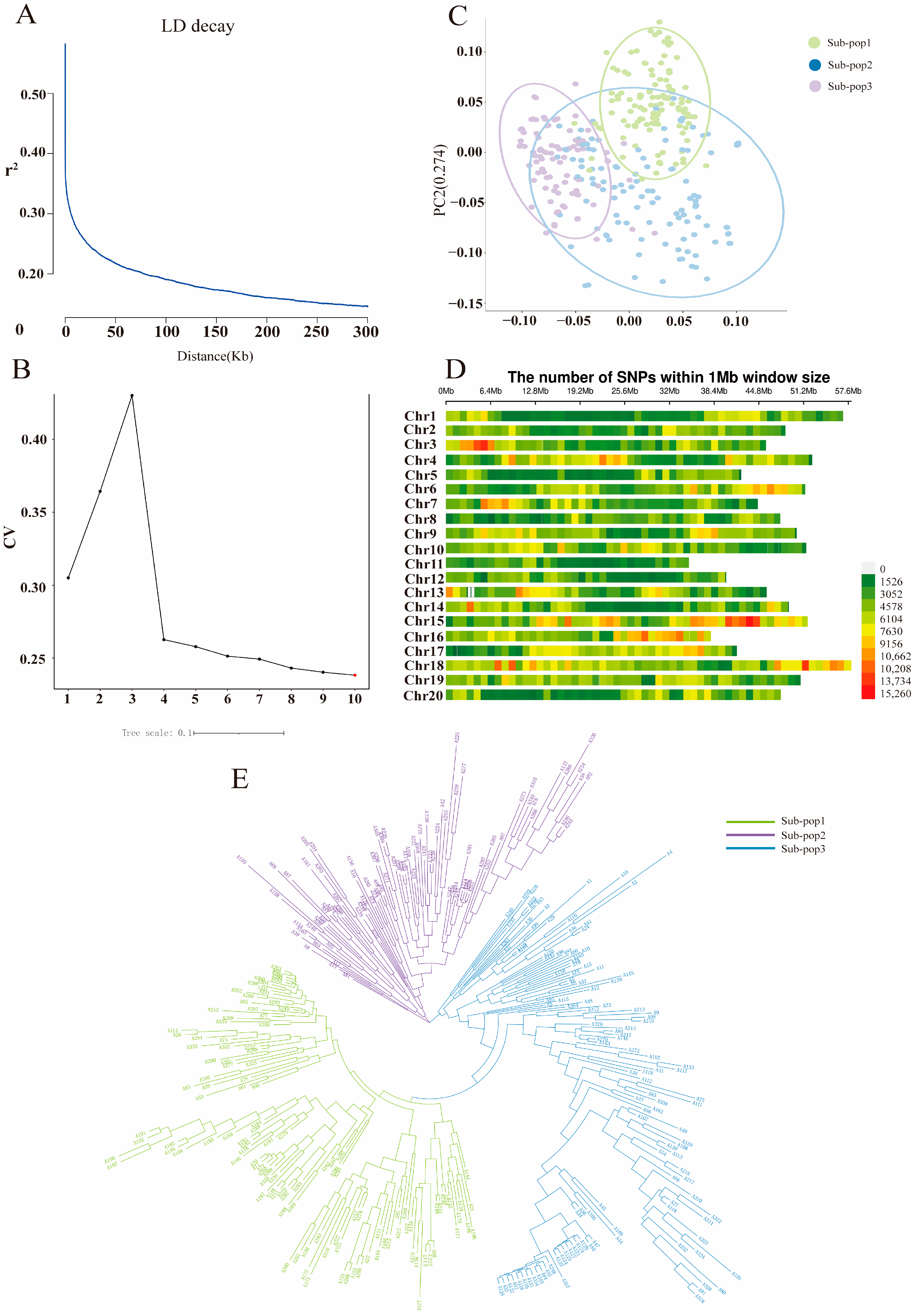
2.2. Linkage Disequilibrium, Population Genetic Structure, and SNP Distribution
2.3. Genome-Wide Association Analysis
2.4. Screening and Identification of Candidate Genes
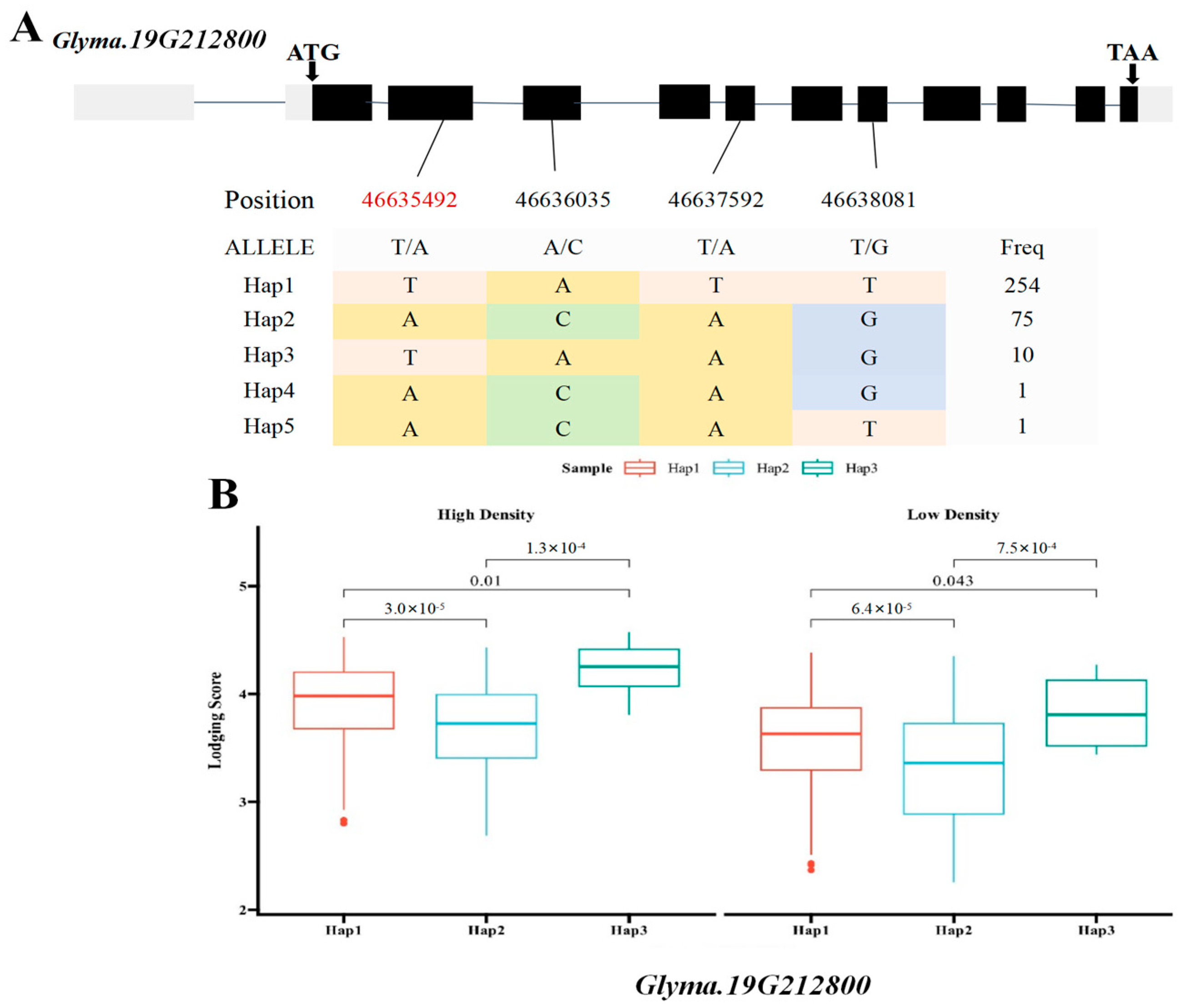
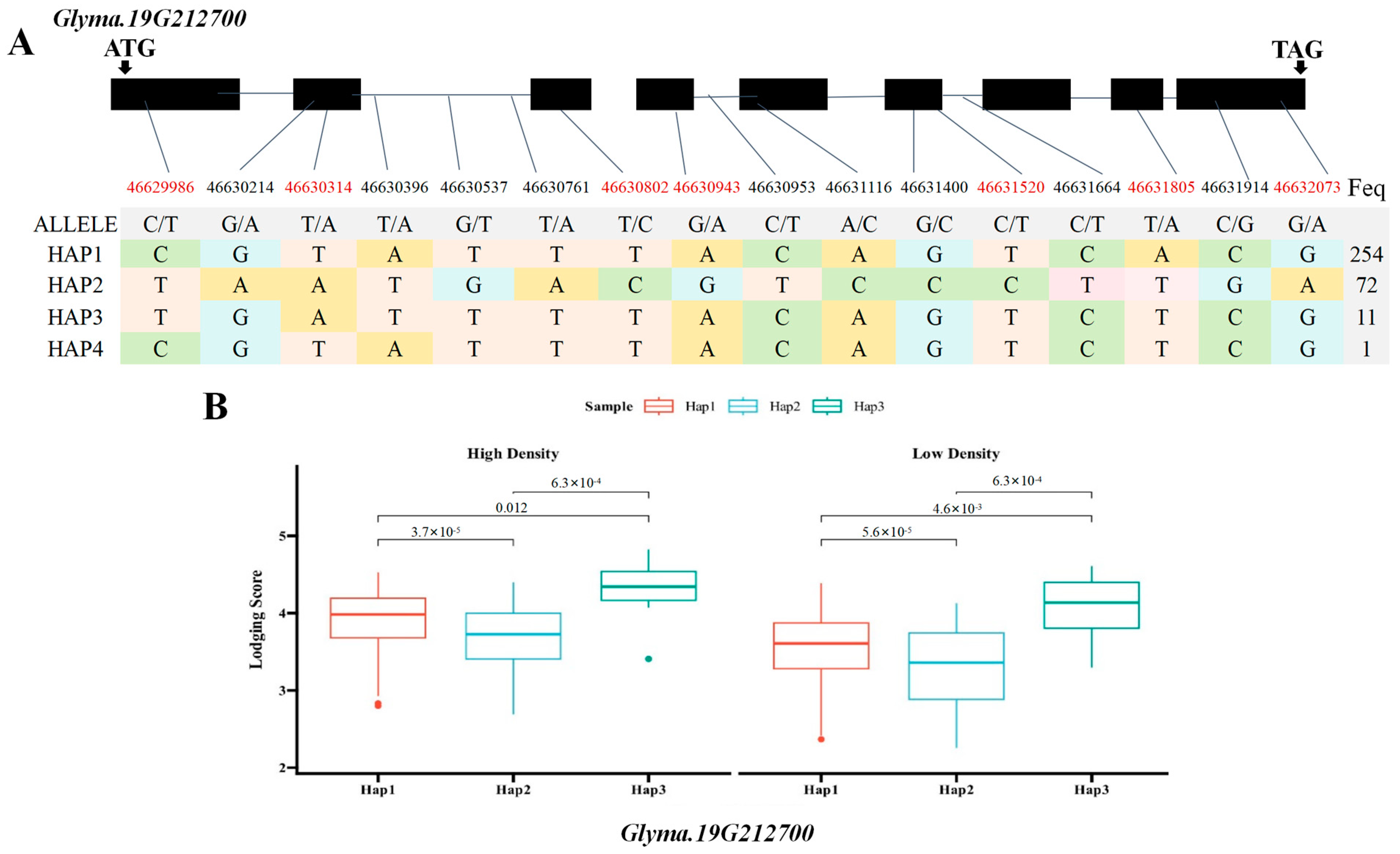
2.5. Haplotype Analysis of Candidate Genes Glyma.19G212800 and Glyma.19G212700
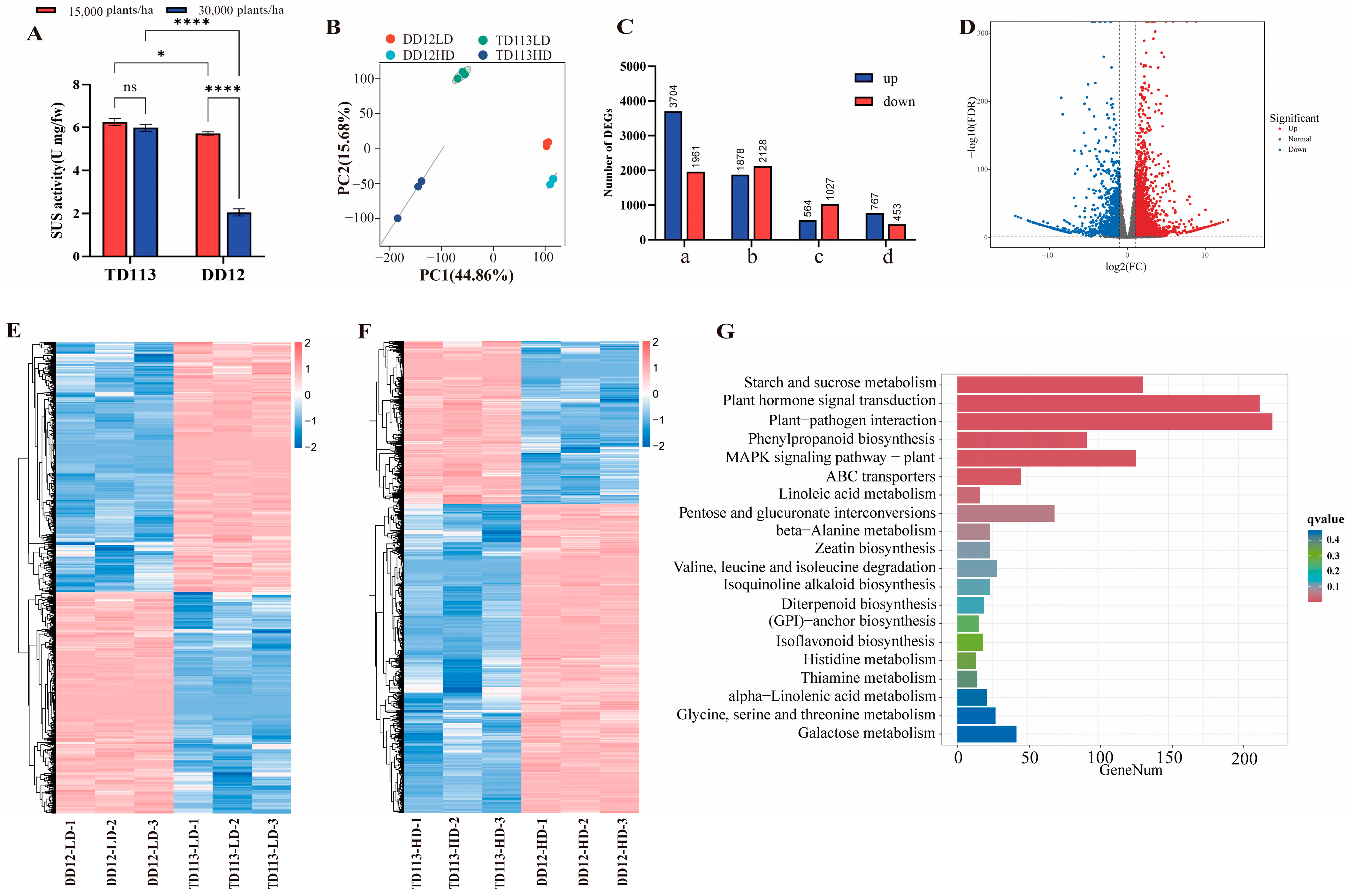
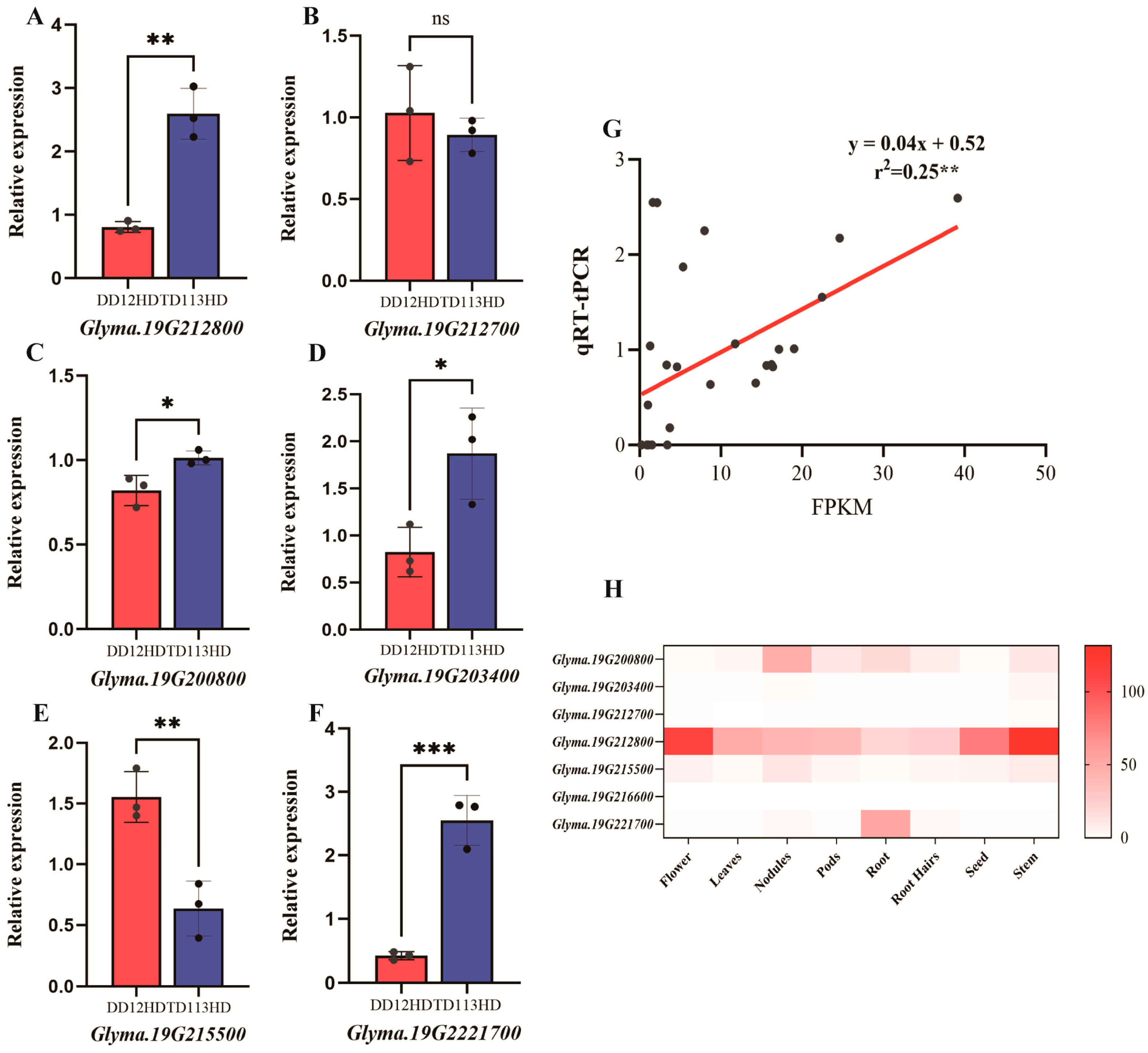
2.6. Transcriptome Analysis and qRT-PCR Validation
3. Discussion
3.1. Analysis of Genetic Variation for Lodging Score Traits
3.2. Comparative Analysis of Loci Associated with Lodging Resistance
3.3. Analysis of Candidate Genes for Lodging Resistance
3.4. Haplotype Analysis, Transcriptome Profiling, and qRT-PCR Validation of Candidate Genes
4. Materials and Methods
4.1. Plant Materials, Field Experiments and the Measurement of Trait
4.2. Genotyping and SNP Calling
4.3. Linkage Disequilibrium, Population Genetic Structure, and SNP Distribution
4.4. Genome-Wide Association Study
4.5. Candidate Gene Identification and Haplotype Analysis
4.6. RNA Sequencing and Quantitative Real-Time PCR Analysis
4.7. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bandillo, N.B.; Anderson, J.E.; Kantar, M.B.; Stupar, R.M.; Specht, J.E.; Graef, G.L.; Lorenz, A.J. Dissecting the Genetic Basis of Local Adaptation in Soybean. Sci. Rep. 2017, 7, 17195. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cheng, B.; Zhou, T.; Jing, S.; Liu, R.; Gao, Y.; Deng, C.; Ye, W.; Luo, Z.; Raza, A.; et al. Quantifying the effects of plant density on soybean lodging resistance and growth dynamics in maize-soybean strip intercropping. Front. Plant Sci. 2023, 14, 1264378. [Google Scholar] [CrossRef]
- Zhao, T.; Gai, J.; Li, H.; Xing, H.; Qiu, J. Advances in breeding for super high-yielding soybean cultivars. Sci. Agric. Sin. 2006, 39, 29–37. [Google Scholar]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, C.; Xia, J.; Wu, L.; Xu, G.; Wu, W.; Li, D.; Qin, W.; Han, X.; Chen, Q.; et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2019, 365, 658–664. [Google Scholar] [CrossRef]
- Gao, J.; Yang, S.; Cheng, W.; Fu, Y.; Leng, J.; Yuan, X.; Jiang, N.; Ma, J.; Feng, X. GmILPA1, encoding an APC8-like protein, controls leaf petiole angle in soybean. Plant Physiol. 2017, 174, 1167–1176. [Google Scholar] [PubMed]
- Li, S.; Sun, Z.; Sang, Q.; Qin, C.; Kong, L.; Huang, X.; Liu, H.; Su, T.; Li, H.; He, M.; et al. Soybean reduced internode 1 determines internode length and improves grain yield at dense planting. Nat. Commun. 2023, 14, 7939. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Akhter, M.; Sneller, C.H. Yield and yield components of early maturing soybean genotypes in the mid-south. Crop Sci. 1996, 36, 877–882. [Google Scholar] [CrossRef]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef]
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Xu, Z.; Liu, B.; Kong, F.; Tsubokura, Y.; Watanabe, S.; Xia, Z.; Harada, K.; Kanazawa, A.; Yamada, T.; et al. Genetic variation in four maturity genes affects photoperiod insensitivity and PHYA-regulated post-flowering responses of soybean. BMC Plant Biol. 2013, 13, 91. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tang, X.; Zhang, X.; Wang, H.; Ji, W.; Xu, C.; Yang, Z.; Li, P. Genome-wide association study identifies novel candidate loci or genes affecting stalk strength in maize. Crop J. 2023, 11, 220–227. [Google Scholar] [CrossRef]
- Kato, S.; Samanfar, B.; Morrison, M.J.; Bekele, W.A.; Torkamaneh, D.; Rajcan, I.; O’donoughue, L.; Belzile, F.; Cober, E.R. Genome-wide association study to identify soybean stem pushing resistance and lodging resistance loci. Can. J. Plant Sci. 2021, 101, 663–670. [Google Scholar] [CrossRef]
- Kim, S.-H.; Tayade, R.; Kang, B.-H.; Hahn, B.-S.; Ha, B.-K.; Kim, Y.-H. Genome-Wide Association Studies of Seven Root Traits in Soybean (Glycine max L.) Landraces. Int. J. Mol. Sci. 2023, 24, 873. [Google Scholar] [CrossRef]
- Ban, Y.-W.; Roy, N.S.; Yang, H.; Choi, H.-K.; Kim, J.-H.; Babu, P.; Ha, K.-S.; Ham, J.-K.; Park, K.C.; Choi, I.-Y. Comparative transcriptome analysis reveals higher expression of stress and defense responsive genes in dwarf soybeans obtained from the crossing of G. max and G. soja. Genes Genom. 2019, 41, 1315–1327. [Google Scholar] [CrossRef]
- Konno, T.; Homma, K. Impact assessment of main stem elongation and wind speed on lodging of soybean cultivar ‘Miyagishirome’. Plant Prod. Sci. 2024, 27, 185–196. [Google Scholar] [CrossRef]
- Specht, J.; Chase, K.; Macrander, M.; Graef, G.; Chung, J.; Markwell, J.; Germann, M.; Orf, J.; Lark, K. Soybean Response to Water: A QTL Analysis of Drought Tolerance. Crop Sci. 2001, 41, 493–509. [Google Scholar] [CrossRef]
- Lee, S.; Jun, T.H.; Michel, A.P.; Mian, M.A.R. SNP markers linked to QTL conditioning plant height, lodging, and maturity in soybean. Euphytica 2014, 203, 521–532. [Google Scholar] [CrossRef]
- Bazakos, C.; Hanemian, M.; Trontin, C.; Jiménez-Gómez, J.M.; Loudet, O. New Strategies and Tools in Quantitative Genetics: How to Go from the Phenotype to the Genotype. Annu. Rev. Plant Biol. 2017, 68, 435–455. [Google Scholar] [CrossRef]
- Wu, J.; Yu, R.; Wang, H.; Zhou, C.; Huang, S.; Jiao, H.; Yu, S.; Nie, X.; Wang, Q.; Liu, S.; et al. A large-scale genomic association analysis identifies the candidate causal genes conferring stripe rust resistance under multiple field environments. Plant Biotechnol. J. 2021, 19, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, H.; Fan, X.; Huang, W.; Yang, J.; Zheng, Y.; Wen, Z.; Li, Y.; Wang, D.; Wang, S.; et al. Selection of soybean elite cultivars based on phenotypic and genomic characters related to lodging tolerance. Plant Breed. 2017, 136, 526–538. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cui, Z.; Lin, J.; Liu, M.; Jin, Y.; Zhang, A.; Gao, Y.; Cao, H.; Ruan, Y. Genome-wide association study reveals the genetic basis of brace root angle and diameter in maize. Front. Genet. 2022, 13, 963852. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Dong, Z.; Wang, C.; Wei, X.; Long, Y.; Wan, X. Genetic structure and molecular mechanism underlying the stalk lodging traits in maize (Zea mays L.). Comput. Struct. Biotechnol. J. 2023, 21, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Jian, H.; Lu, K.; Yin, N.; Wang, J.; Duan, X.; Li, W.; Liu, L.; Xu, X.; Wang, R.; et al. Genetic and transcriptomic analyses of lignin- and lodging-related traits in Brassica napus. Theor. Appl. Genet. 2017, 130, 1961–1973. [Google Scholar] [CrossRef]
- Rashid, M.A.R.; Zhao, Y.; Azeem, F.; Zhao, Y.; Ahmed, H.G.M.-D.; Atif, R.M.; Pan, Y.; Zhu, X.; Liang, Y.; Zhang, H.; et al. Unveiling the genetic architecture for lodging resistance in rice (Oryza sativa L.) by genome-wide association analyses. Front. Genet. 2022, 13, 960007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, P.; Zhao, L.; Chen, Q.; Nong, S.; Li, Q.; Wang, L. Integrated VIS/NIR Spectrum and Genome-Wide Association Study for Genetic Dissection of Cellulose Crystallinity in Wheat Stems. Int. J. Mol. Sci. 2024, 25, 3028. [Google Scholar] [CrossRef]
- Singh, D.; Wang, X.; Kumar, U.; Gao, L.; Noor, M.; Imtiaz, M.; Singh, R.P.; Poland, J. High-Throughput Phenotyping Enabled Genetic Dissection of Crop Lodging in Wheat. Front. Plant Sci. 2019, 10, 394. [Google Scholar] [CrossRef]
- Cheng, B.; Raza, A.; Wang, L.; Xu, M.; Lu, J.; Gao, Y.; Qin, S.; Zhang, Y.; Ahmad, I.; Zhou, T.; et al. Effects of Multiple Planting Densities on Lignin Metabolism and Lodging Resistance of the Strip Intercropped Soybean Stem. Agronomy 2020, 10, 1177. [Google Scholar] [CrossRef]
- Shan, F.; Sun, K.; Gong, S.; Wang, C.; Ma, C.; Zhang, R.; Yan, C. Effects of Shading on the Internode Critical for Soybean (Glycine max) Lodging. Agronomy 2022, 12, 492. [Google Scholar] [CrossRef]
- Saitoh, K.; Nishimura, K.; Kitahara, T. Effect of Lodging on Seed Yield of Field-Grown Soybean—Artificial Lodging and Lodging Preventing Treatments. Jpn. J. Crop Sci. 2012, 81, 27–32. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “green revolution” for soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Vuong, T.D.; Sonah, H.; Meinhardt, C.G.; Deshmukh, R.; Kadam, S.; Nelson, R.L.; Shannon, J.G.; Nguyen, H.T. Genetic architecture of cyst nematode resistance revealed by genome-wide association study in soybean. BMC Genom. 2015, 16, 593. [Google Scholar] [CrossRef]
- Fang, C.; Ma, Y.; Wu, S.; Liu, Z.; Wang, Z.; Yang, R.; Hu, G.; Zhou, Z.; Yu, H.; Zhang, M.; et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Bailey, M.A.; Mian, M.A.R.; Carter, T.E., Jr.; Ashley, D.A.; Hussey, R.S.; Parrott, W.A.; Boerma, H.R. Molecular markers associated with soybean plant height, lodging, and maturity across locations. Crop Sci. 1996, 36, 728–735. [Google Scholar] [CrossRef]
- Mansur, L.M.; Lark, K.G.; Kross, H.; Oliveira, A. Interval mapping of quantitative trait loci for reproductive, morphological, and seed traits of soybean (Glycine max L.). Theor. Appl. Genet. 1993, 86, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-S.; Diers, B.W.; Hyten, D.L.; Mian, M.A.R.; Shannon, J.G.; Nelson, R.L. Identification of positive yield QTL alleles from exotic soybean germplasm in two backcross populations. Theor. Appl. Genet. 2012, 125, 1353–1369. [Google Scholar] [CrossRef]
- Orf, J.H.; Chase, K.; Jarvik, T.; Mansur, L.M.; Cregan, P.B.; Adler, F.R.; Lark, K.G. Genetics of Soybean Agronomic Traits: I. Comparison of Three Related Recombinant Inbred Populations. Crop Sci. 1999, 39, 1642–1651. [Google Scholar] [CrossRef]
- Li, D.; Pfeiffer, T.W.; Cornelius, P.L. Soybean QTL for Yield and Yield Components Associated with Glycine soja Alleles. Crop Sci. 2008, 48, 571–581. [Google Scholar] [CrossRef]
- Mansur, L.M.; Orf, J.H.; Chase, K.; Jarvik, T.; Cregan, P.B.; Lark, K.G. Genetic Mapping of Agronomic Traits Using Recombinant Inbred Lines of Soybean. Crop Sci. 1996, 36, 1327–1336. [Google Scholar] [CrossRef]
- He, W.; Chai, Q.; Zhao, C.; Yu, A.; Fan, Z.; Yin, W.; Hu, F.; Fan, H.; Sun, Y.; Wang, F. Blue light regulated lignin and cellulose content of soybean petioles and stems under low light intensity. Funct. Plant Biol. 2024, 51, FP23091. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Bailey, M.A.; Mian, M.A.R.; Shipe, E.R.; Ashley, D.A.; Parrott, W.A.; Hussey, R.S.; Boerma, H.R. Identification of quan-titative trait loci for plant height, lodging, and maturity in a soybean population segregating for growth habit. Theor. Appl. Genet. 1996, 92, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhao, K.; Wang, J.; Mu, W.; Zhan, Y.; Li, W.; Teng, W.; Zhao, X.; Han, Y. Identification of quantitative trait loci underlying lodging of soybean across multiple environments. Crop. Pasture Sci. 2022, 73, 652–662. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.-C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Jiao, X.; Lyu, Y.; Wu, X.; Li, H.; Cheng, L.; Zhang, C.; Yuan, L.; Jiang, R.; Jiang, B.; Rengel, Z.; et al. Grain production versus resource and environmental costs: Towards increasing sustainability of nutrient use in China. J. Exp. Bot. 2016, 67, 4935–4949. [Google Scholar] [CrossRef]
- Panthee, D.R.; Pantalone, V.R.; Saxton, A.M.; West, D.R.; Sams, C.E. Quantitative trait loci for agronomic traits in soybean. Plant Breed. 2007, 126, 51–57. [Google Scholar] [CrossRef]
- Fang, T.; Bai, Y.; Huang, W.; Wu, Y.; Yuan, Z.; Luan, X.; Liu, X.; Sun, L. Identification of Potential Gene Regulatory Pathways Affecting the Ratio of Four-Seed Pod in Soybean. Front. Genet. 2021, 12, 717770. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Chen, K.; Tian, F.; Yang, X.; Cao, L.; Ruan, N.; Dang, Z.; Yin, X.; Huang, Y.; et al. The DEP1 Mutation Improves Stem Lodging Resistance and Biomass Saccharification by Affecting Cell Wall Biosynthesis in Rice. Rice 2024, 17, 35. [Google Scholar] [CrossRef]
- Chai, S.; Yao, Q.; Zhang, X.; Xiao, X.; Fan, X.; Zeng, J.; Sha, L.; Kang, H.; Zhang, H.; Li, J.; et al. The semi-dwarfing gene Rht-dp from dwarf polish wheat (Triticum polonicum L.) is the "Green Revolution” gene Rht-B1b. BMC Genom. 2021, 22, 63. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Chin-Atkins, A.N.; Wilson, I.W.; Chapple, R.; Dennis, E.S.; Chaudhury, A. The Arabidopsis AMP1 Gene Encodes a Putative Glutamate Carboxypeptidase. Plant Cell 2001, 13, 2115–2125. [Google Scholar] [CrossRef]
- Wang, P.; Yang, Y.; Li, D.; Xu, J.; Gu, R.; Zheng, J.; Fu, J.; Wang, J.; Zhang, H. Cloning of a new allele of ZmAMP1 and evaluation of its breeding value in hybrid maize. Crop J. 2023, 11, 157–165. [Google Scholar] [CrossRef]
- Zemzoumi, K.; Frontini, M.; Bellorini, M.; Mantovani, R. NF-Y histone fold α1 helices help impart CCAAT specificity. J. Mol. Biol. 1999, 286, 327–337. [Google Scholar] [CrossRef]
- Wang, W.; Xu, B.; Wang, H.; Li, J.; Huang, H.; Xu, L. YUCCA Genes Are Expressed in Response to Leaf Adaxial-Abaxial Juxtaposition and Are Required for Leaf Margin Development. Plant Physiol. 2011, 157, 1805–1819. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, X.; Zhu, M.; Xu, M.; Wang, L. Transcription factors NF-YA2 and NF-YA10 regulate leaf growth via auxin signaling in Arabidopsis. Sci. Rep. 2017, 7, 1395. [Google Scholar] [CrossRef]
- Nowak, K.; Wójcik, A.M.; Konopka, K.; Jarosz, A.; Dombert, K.; Gaj, M.D. miR156-SPL and miR169-NF-YA Modules Regulate the Induction of Somatic Embryogenesis in Arabidopsis via LEC- and Auxin-Related Pathways. Int. J. Mol. Sci. 2024, 25, 9217. [Google Scholar] [CrossRef] [PubMed]
- Knob, A.; Terrasan, C.R.F.; Carmona, E.C. β-Xylosidases from filamentous fungi: An overview. World J. Microbiol. Biotechnol. 2010, 26, 389–407. [Google Scholar] [CrossRef]
- Chen, J.; Qu, C.; Chang, R.; Suo, J.; Yu, J.; Sun, X.; Liu, G.; Xu, Z. Genome-wide identification of BXL genes in Populus trichocarpa and their expression under different nitrogen treatments. 3 Biotech 2020, 10, 57. [Google Scholar] [CrossRef]
- Zhou, J.; Zhong, R.; Ye, Z.-H. Arabidopsis NAC Domain Proteins, VND1 to VND5, Are Transcriptional Regulators of Secondary Wall Biosynthesis in Vessels. PLoS ONE 2014, 9, e105726. [Google Scholar] [CrossRef]
- Sakamoto, S.; Takata, N.; Oshima, Y.; Yoshida, K.; Taniguchi, T.; Mitsuda, N. Wood reinforcement of poplar by rice NAC transcription factor. Sci. Rep. 2016, 6, 19925. [Google Scholar] [CrossRef]
- Sakamoto, S.; Nomura, T.; Kato, Y.; Ogita, S.; Mitsuda, N. High-transcriptional activation ability of bamboo SECONDARY WALL NAC transcription factors is derived from C-terminal domain. Plant Biotechnol. 2022, 39, 229–240. [Google Scholar] [CrossRef]
- Zeng, X.; Sheng, J.; Zhu, F.; Wei, T.; Zhao, L.; Hu, X.; Zheng, X.; Zhou, F.; Hu, Z.; Diao, Y.; et al. Genetic, transcriptional, and regulatory landscape of monolignol biosynthesis pathway in Miscanthus × giganteus. Biotechnol. Biofuels 2020, 13, 179. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Singh, V.K.; Saxena, R.K.; Khan, A.W.; Abbai, R.; Chitikineni, A.; Desai, A.; Molla, J.; Upadhyaya, H.D.; Kumar, A.; et al. Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan L.). Plant Biotechnol. J. 2020, 18, 2482–2490. [Google Scholar] [CrossRef]
- Hayashi, T.; Yoshida, K.; Woopark, Y.; Konishi, T.; Baba, K. Cellulose Metabolism in Plants. Int. Rev. Cytol. 2005, 247, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Guo, W.; Zhu, H.; Ruan, Y.; Zhang, T. Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 2012, 10, 301–312. [Google Scholar] [CrossRef]
- Fan, C.; Feng, S.; Huang, J.; Wang, Y.; Wu, L.; Li, X.; Wang, L.; Tu, Y.; Xia, T.; Li, J.; et al. AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice. Biotechnol. Biofuels 2017, 10, 221. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.; Zhou, R.; Lu, A.; Huang, B.; Liu, L.; Chen, L.; Luo, B.; Huang, J.; Tian, Z. SEQdata-BEACON: A comprehensive database of sequencing performance and statistical tools for performance evaluation and yield simulation in BGISEQ-500. BioData Min. 2019, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Zhang, R.; Jia, G.; Diao, X. geneHapR: An R package for gene haplotypic statistics and visualization. BMC Bioinform. 2023, 24, 199. [Google Scholar] [CrossRef]
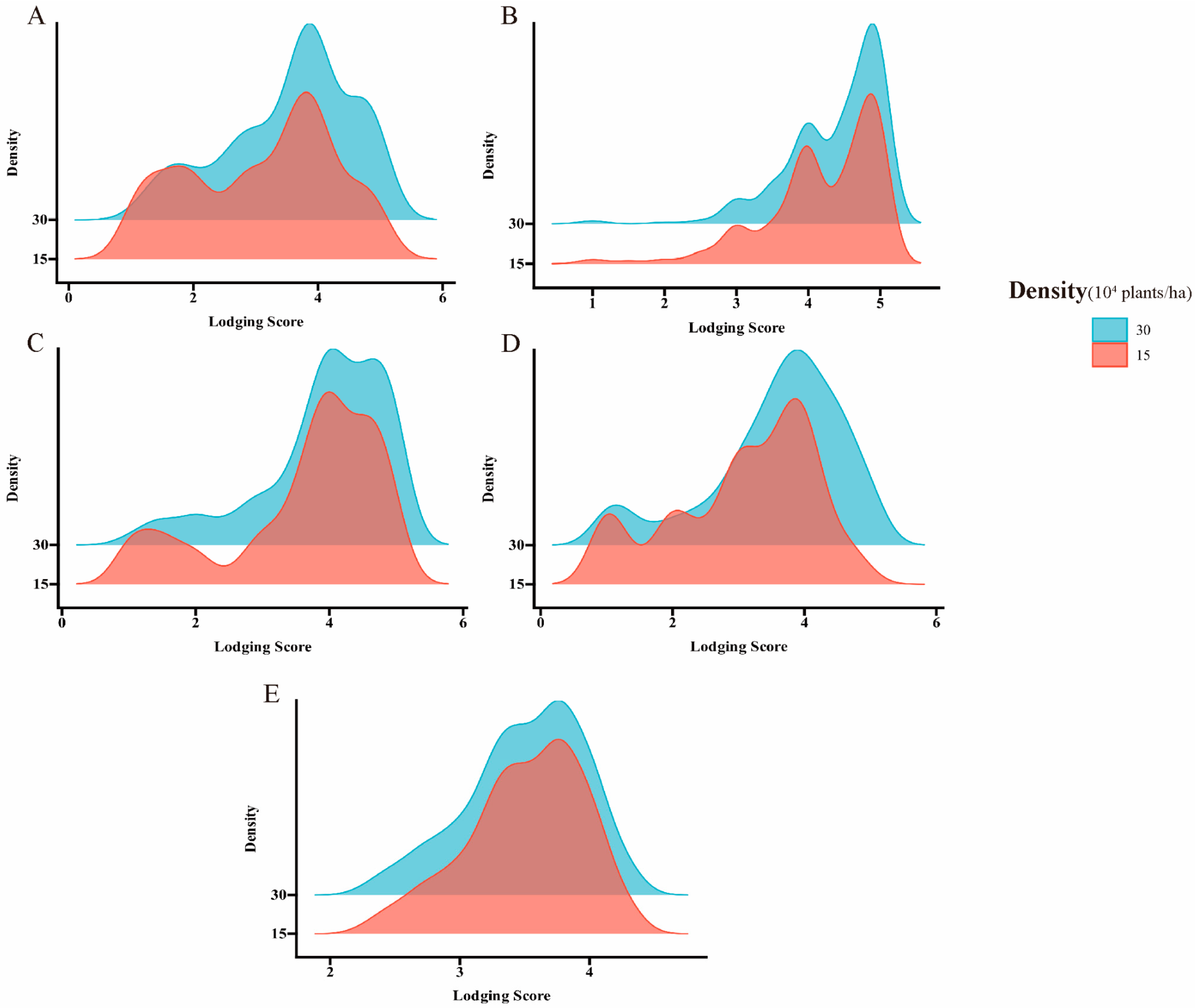
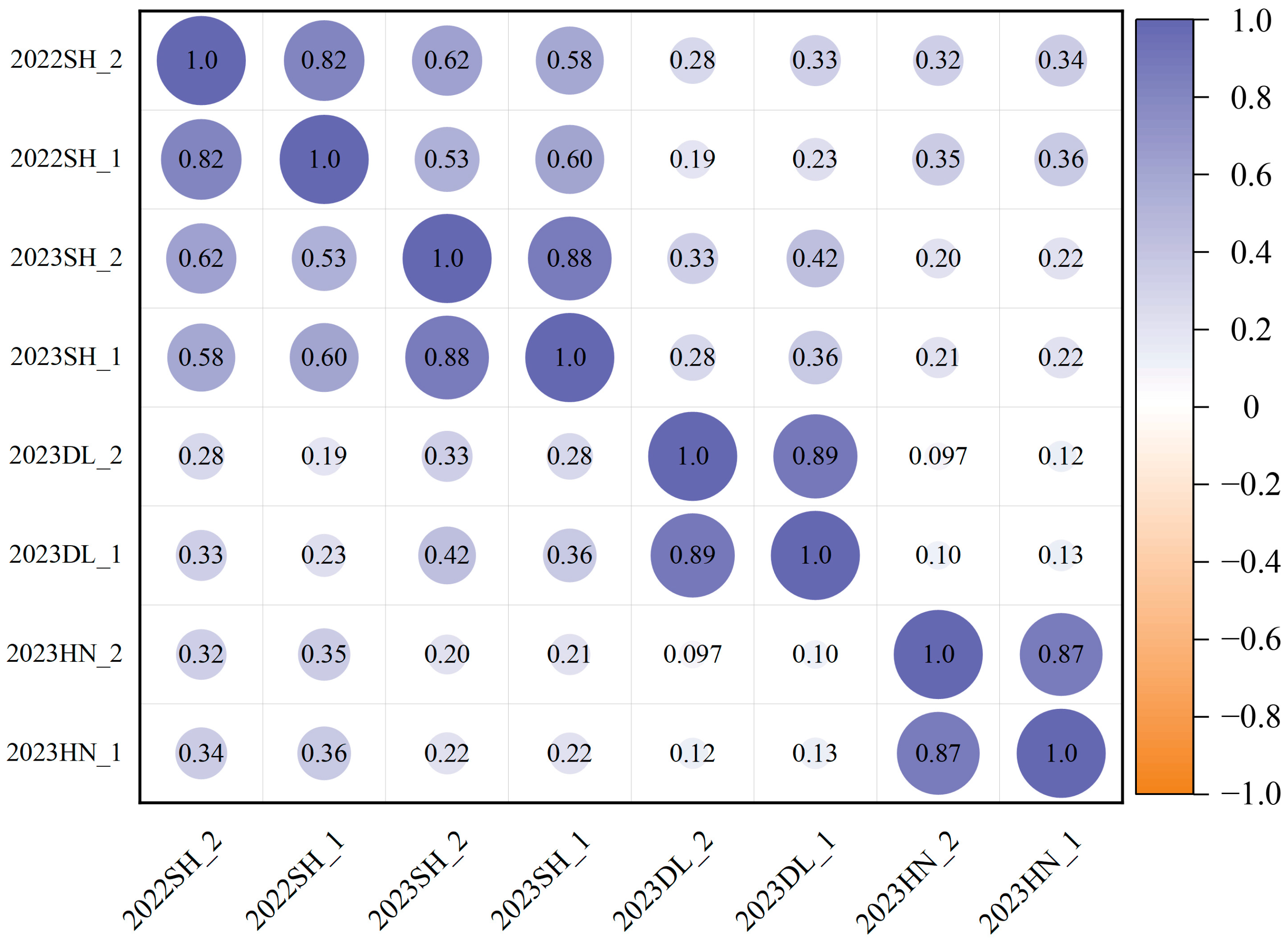


| Trait | Environments | Density (×104 Plants/ha) | Range | Mean | SD | CV% | H2 |
|---|---|---|---|---|---|---|---|
| Lodging Score | DL2023 | 30 | 1.00–5.00 | 3.919 | 0.944 | 24.13 | 0.55 |
| SH2023 | 30 | 1.00–5.00 | 4.398 | 0.642 | 14.61 | ||
| HN2023 | 30 | 1.00–5.00 | 3.588 | 1.014 | 28.22 | ||
| SH2022 | 30 | 1.10–5.00 | 3.502 | 1.037 | 29.64 | ||
| Mean | 30 | 1.05–5.00 | 3.852 | 0.909 | 24.10 | ||
| DL2023 | 15 | 1.00–5.00 | 3.612 | 1.158 | 32.15 | 0.52 | |
| SH2023 | 15 | 1.00–5.00 | 4.219 | 0.781 | 18.51 | ||
| HN2023 | 15 | 1.00–5.00 | 3.131 | 1.058 | 33.86 | ||
| SH2022 | 15 | 1.00–5.00 | 3.073 | 1.159 | 37.70 | ||
| Mean | 15 | 1.00–5.00 | 3.509 | 1.039 | 30.59 |
| Chr. | Peak Loci | PEAK Position | −log 10 (p) | PVE (%) | MAF |
|---|---|---|---|---|---|
| 1 | Chr01:39325847 | 39,325,847 | 5.31–6.03 | 6.03–6.91 | 0.022 |
| 1 | Chr01:44922215 | 44,922,215 | 5.76–5.21 | 5.90–6.60 | 0.034 |
| 2 | Chr02:2604732 | 2,604,732 | 5.53–5.54 | 6.31–6.32 | 0.019 |
| 2 | Chr02:15458372 | 15,458,372 | 6.48–6.81 | 6.66–7.47 | 0.028 |
| 3 | Chr03.:35831309 | 35,831,309 | 5.31–5.62 | 6.01–6.42 | 0.092 |
| 3 | Chr03:36451125 | 36,451,125 | 6.30–7.19 | 7.04–8.35 | 0.158 |
| 4 | Chr04:47442227 | 47,442,227 | 5.31–5.69 | 6.04–6.50 | 0.021 |
| 7 | Chr07:20616713 | 20,616,713 | 5.32–5.37 | 6.03–6.11 | 0.015 |
| 8 | Chr08:30754555 | 3,075,4555 | 5.99–6.51 | 6.86–7.57 | 0.024 |
| 8 | Chr08:32270717 | 3,2270,717 | 5.14–6.12 | 5.81–7.04 | 0.013 |
| 9 | Chr09:35877303 | 35,877,303 | 5.52–6.26 | 6.29–7.19 | 0.021 |
| 10 | Chr10:5376997 | 5,376,997 | 5.45–6.27 | 6.19–7.22 | 0.075 |
| 11 | Chr11:3581424 | 3,581,424 | 5.61–6.85 | 6.39–7.94 | 0.050 |
| 14 | Chr1.:326408 | 326,408 | 6.21–6.22 | 7.13–7.14 | 0.411 |
| 14 | Chr14:617529 | 617,529 | 5.21–5.55 | 5.90–6.33 | 0.450 |
| 15 | Chr15:49852803 | 49,852,803 | 5.59–5.75 | 6.38–6.61 | 0.058 |
| 18 | Chr18:15099487 | 15,099,487 | 5.46–5.60 | 5.73–6.22 | 0.040 |
| 19 | Chr19:44477717 | 44,477,717 | 5.84–7.09 | 6.69–8.20 | 0.432 |
| 19 | Chr19:46692661 | 46,692,661 | 7.85–9.45 | 9.16–10.99 | 0.327 |
| 20 | Chr20:7716540 | 7,716,540 | 5.28–6.45 | 5.98–7.44 | 0.120 |
| SNP Location | Chr. | Location | Gene ID | Gene Annotation |
|---|---|---|---|---|
| Chr19.:44477717 | Chr19 | 45,769,759 | Glyma.19G200800 | Nuclear factor Y, subunit A10 |
| Chr19.:46692661 | Chr19 | 45,999,764 | Glyma.19G203400 | Encodes glutamate carboxypeptidase |
| Chr19.:46692661 | Chr19 | 46,629,986 | Glyma.19G212700 | Glycosyl hydrolase 9B13 |
| Chr19.:46692661 | Chr19 | 46,635,492 | Glyma.19G212800 | Sucrose synthase 3 |
| Chr19.:46692661 | Chr19 | 46,856,657 | Glyma.19G215500 | Beta-xylosidase 2 |
| Chr19.:46692661 | Chr19 | 46,960,586 | Glyma.19G216600 | SET domain-containing protein |
| Chr19.:46692661 | Chr19 | 47,352,760 | Glyma.19G221700 | WRKY family transcription factor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Z.; Qi, N.; Li, R.; Gao, R.; Huang, J.; Zhao, W.; Zhang, H.; Wang, H.; Ao, X.; Yao, X.; et al. Genome-Wide Association Study to Identify Soybean Lodging Resistance Loci and Candidate Genes. Int. J. Mol. Sci. 2025, 26, 4446. https://doi.org/10.3390/ijms26094446
Liang Z, Qi N, Li R, Gao R, Huang J, Zhao W, Zhang H, Wang H, Ao X, Yao X, et al. Genome-Wide Association Study to Identify Soybean Lodging Resistance Loci and Candidate Genes. International Journal of Molecular Sciences. 2025; 26(9):4446. https://doi.org/10.3390/ijms26094446
Chicago/Turabian StyleLiang, Zicong, Nianhua Qi, Ruoning Li, Ruijia Gao, Junxia Huang, Wei Zhao, Huijun Zhang, Haiying Wang, Xue Ao, Xingdong Yao, and et al. 2025. "Genome-Wide Association Study to Identify Soybean Lodging Resistance Loci and Candidate Genes" International Journal of Molecular Sciences 26, no. 9: 4446. https://doi.org/10.3390/ijms26094446
APA StyleLiang, Z., Qi, N., Li, R., Gao, R., Huang, J., Zhao, W., Zhang, H., Wang, H., Ao, X., Yao, X., & Xie, F. (2025). Genome-Wide Association Study to Identify Soybean Lodging Resistance Loci and Candidate Genes. International Journal of Molecular Sciences, 26(9), 4446. https://doi.org/10.3390/ijms26094446





