Towards Precision in Sarcopenia Assessment: The Challenges of Multimodal Data Analysis in the Era of AI
Abstract
1. Introduction
2. Methodology for Selection of Studies
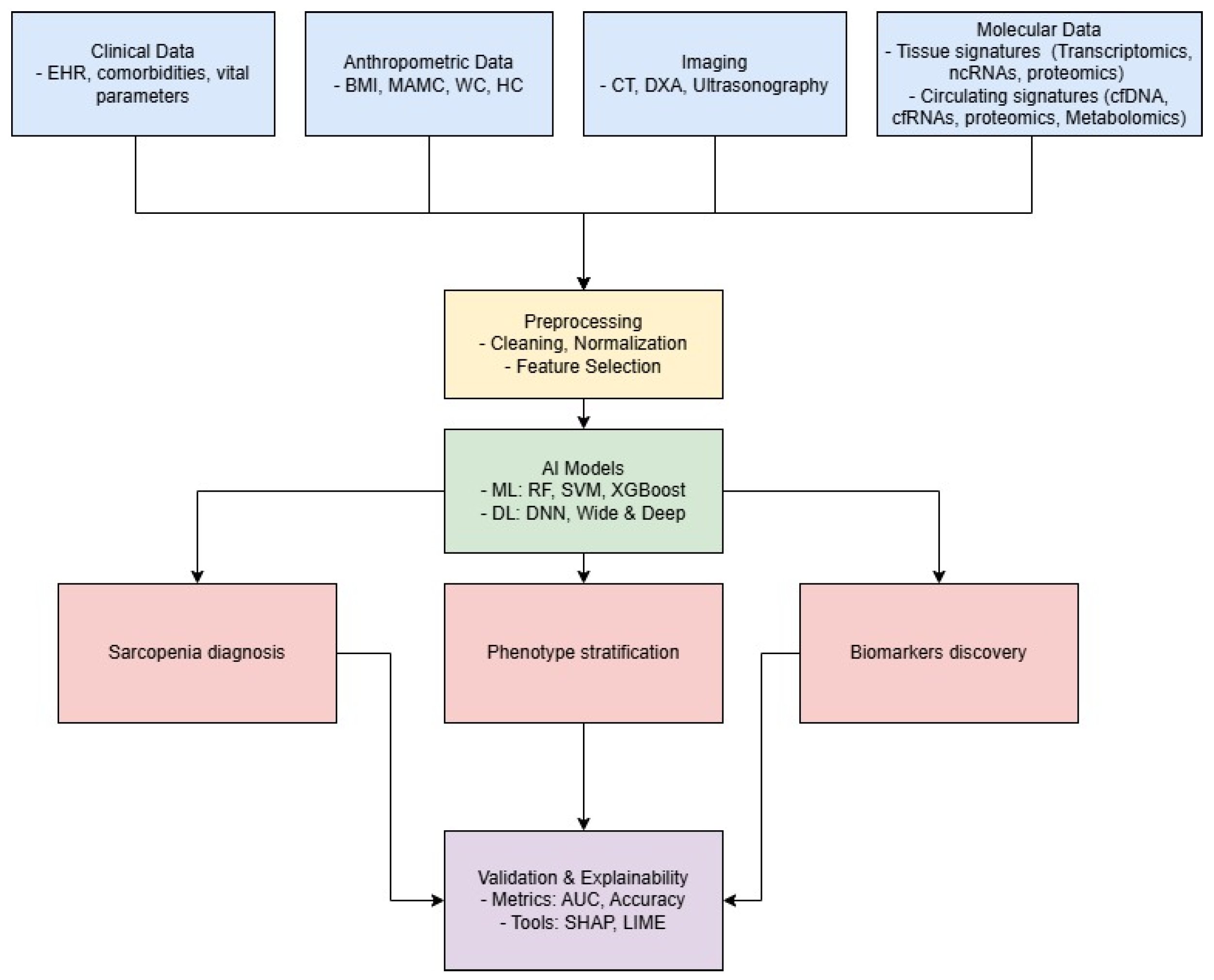
3. AI for Comprehensive Data Integration in Sarcopenia
4. Application of AI for Identifying Molecular Biomarkers
5. Circulating Biomarkers: Update and New Frontiers for AI in Sarcopenia Research
| Circulating Signature | Biological Source | Expression Profile | Ref. |
|---|---|---|---|
| D3-creatinine | urine | downregulation | [57] |
| ALDOA | serum | upregulation | [58] |
| CTSD | serum | upregulation | [58] |
| P3NP | serum | upregulation | [59] |
| IL6 | serum | upregulation | [60,61] |
| TNF | serum | upregulation | [61] |
| CAF | serum | upregulation | [62] |
| VCAM1 | serum | upregulation | [63] |
| GDF15 | serum | upregulation | [64] |
| CETP | serum | upregulation | [65] |
| APOA2 | serum | downregulation | [65] |
| IGF1 | serum | downregulation | [66] |
| GH | serum | downregulation | [66] |
| Cf-mtDNA | plasma | high levels | [53] |
| miR-28-5p | plasma | upregulation | [45] |
| miR-1-3p | plasma | upregulation | [54,67] |
| miR-133a | plasma | downregulation | [44] |
| miR-133a-3p | serum | downregulation | [68] |
| miR-200a-3p | serum | downregulation | [68] |
| miR-434-3p | plasma | downregulation | [44] |
| miR-455-3p | plasma | downregulation | [44] |
| miR-486 | plasma | downregulation | [55] |
| miR-146a | plasma | downregulation | [55] |
| miR-21 | serum | upregulation | [69] |
| traumatic acid | plasma | high levels | [70] |
| ceramides | plasma | high levels | [71] |
| sphyngomielins | plasma | high levels | [71] |
| sphyngomielins | plasma | high and low levels depending on lipid | [72] |
| sterol ST(d14:0/25:5) | plasma | high levels | [72] |
| phosphatidylcholines | plasma | high and low levels depending on lipid | [72] |
| phosphatidylserines | plasma | high and low levels depending on lipid | [72] |
| PI 32:1 | plasma | high levels | [73] |
| isoleucine | plasma | low levels | [73] |
| 1-methylhistamine/3-methylhistamine | plasma | high levels | [73] |
| carnosine | plasma | low levels | [73] |
| creatinine | plasma | low levels | [73] |
| arginine | serum | low levels | [43] |
| cystin | serum | low levels | [43] |
| taurin | serum | high levels | [43] |
| hypoxanthine | plasma | high levels | [56] |
| hypoxanthine | serum | high levels | [74] |
| L-2-amino-3-oxobutanoic acid | plasma | low levels | [56,74] |
| PC(14:0/20:2(11Z,14Z)) | plasma | low levels | [56] |
| LysoPC(17:0) | plasma | low levels | [56] |
| palmitic acid | plasma | low levels | [56] |
| mannose | serum | high levels | [74] |
| galactose | serum | high levels | [74] |
| triethanolamine | serum | low levels | [74] |
| homogentisic acid | serum | low levels | [74] |
| oleoyl ethanolamide | plasma | high levels | [42] |
| stearoyl ethanolamide | plasma | low levels | [42] |
| docosahexaenoylethanolamide | plasma | low levels | [42] |
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Daly, R.M.; Iuliano, S.; Fyfe, J.; Scott, D.; Kirk, B.; Thompson, M.; Dent, E.; Fetterplace, K.; Wright, O.; Lynch, G.; et al. Screening, Diagnosis and Management of Sarcopenia and Frailty in Hospitalized Older Adults: Recommendations from the Australian and New Zealand Society for Sarcopenia and Frailty Research (ANZSSFR) Expert Working Group. J. Nutr. Health Aging 2022, 26, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Sayer, A.A.; Cooper, R.; Arai, H.; Cawthon, P.M.; Ntsama Essomba, M.J.; Fielding, R.A.; Grounds, M.D.; Witham, M.D.; Cruz-Jentoft, A.J. Sarcopenia. Nat. Rev. Dis. Prim. 2024, 10, 68. [Google Scholar] [CrossRef]
- Tarantino, G.; Sinatti, G.; Citro, V.; Santini, S.; Balsano, C. Sarcopenia, a condition shared by various diseases: Can we alleviate or delay the progression? Intern. Emerg. Med. 2023, 18, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Sabatino, A.; Cuppari, L.; Stenvinkel, P.; Lindholm, B.; Avesani, C.M. Sarcopenia in chronic kidney disease: What have we learned so far? J. Nephrol. 2020, 34, 1347–1372. [Google Scholar] [CrossRef]
- Nagano, A.; Wakabayashi, H.; Maeda, K.; Kokura, Y.; Miyazaki, S.; Mori, T.; Fujiwara, D. Respiratory Sarcopenia and Sarcopenic Respiratory Disability: Concepts, Diagnosis, and Treatment. J. Nutr. Health Aging 2021, 25, 507–515. [Google Scholar] [CrossRef] [PubMed]
- An, H.J.; Tizaoui, K.; Terrazzino, S.; Cargnin, S.; Lee, K.H.; Nam, S.W.; Kim, J.S.; Yang, J.W.; Lee, J.Y.; Smith, L.; et al. Sarcopenia in Autoimmune and Rheumatic Diseases: A Comprehensive Review. Int. J. Mol. Sci. 2020, 21, 5678. [Google Scholar] [CrossRef]
- Wiedmer, P.; Jung, T.; Castro, J.P.; Pomatto, L.C.; Sun, P.Y.; Davies, K.J.; Grune, T. Sarcopenia—Molecular mechanisms and open questions. Ageing Res. Rev. 2021, 65, 101200. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2018, 48, 16–31. [Google Scholar] [CrossRef]
- Chen, L.K.; Woo, J.; Assantachai, P.; Auyeung, T.W.; Chou, M.Y.; Iijima, K.; Jang, H.C.; Kang, L.; Kim, M.; Kim, S.; et al. Asian Working Group for Sarcopenia: 2019 Consensus Update on Sarcopenia Diagnosis and Treatment. J. Am. Med. Dir. Assoc. 2020, 21, 300–307.e2. [Google Scholar] [CrossRef]
- Bhasin, S.; Travison, T.G.; Manini, T.M.; Patel, S.; Pencina, K.M.; Fielding, R.A.; Magaziner, J.M.; Newman, A.B.; Kiel, D.P.; Cooper, C.; et al. Sarcopenia Definition: The Position Statements of the Sarcopenia Definition and Outcomes Consortium. J. Am. Geriatr. Soc. 2020, 68, 1410–1418. [Google Scholar] [CrossRef]
- Kufel, J.; Bargieł-Łączek, K.; Kocot, S.; Koźlik, M.; Bartnikowska, W.; Janik, M.; Czogalik, L.; Dudek, P.; Magiera, M.; Lis, A.; et al. What Is Machine Learning, Artificial Neural Networks and Deep Learning?—Examples of Practical Applications in Medicine. Diagnostics 2023, 13, 2582. [Google Scholar] [CrossRef] [PubMed]
- Tolles, J.; Meurer, W.J. Logistic Regression. JAMA 2016, 316, 533–534. [Google Scholar] [CrossRef]
- Kweon, S.; Kim, Y.; Jang, M.j.; Kim, Y.; Kim, K.; Choi, S.; Chun, C.; Khang, Y.H.; Oh, K. Data Resource Profile: The Korea National Health and Nutrition Examination Survey (KNHANES). Int. J. Epidemiol. 2014, 43, 69–77. [Google Scholar] [CrossRef]
- Chung, H.; Jo, Y.; Ryu, D.; Jeong, C.; Choe, S.; Lee, J. Artificial-intelligence-driven discovery of prognostic biomarker for sarcopenia. J. Cachexia Sarcopenia Muscle 2021, 12, 2220–2230. [Google Scholar] [CrossRef]
- Luo, X.; Ding, H.; Broyles, A.; Warden, S.J.; Moorthi, R.N.; Imel, E.A. Using machine learning to detect sarcopenia from electronic health records. Digit. Health 2023, 9, 20552076231197098. [Google Scholar] [CrossRef] [PubMed]
- Onishi, S.; Kuwahara, T.; Tajika, M.; Tanaka, T.; Yamada, K.; Shimizu, M.; Niwa, Y.; Yamaguchi, R. Artificial intelligence for body composition assessment focusing on sarcopenia. Sci. Rep. 2025, 15, 1324. [Google Scholar] [CrossRef]
- Wu, L.W.; OuYoung, T.; Chiu, Y.C.; Hsieh, H.F.; Hsiu, H. Discrimination between possible sarcopenia and metabolic syndrome using the arterial pulse spectrum and machine-learning analysis. Sci. Rep. 2022, 12, 21452. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Olea, C.; García-Zapirain Soto, B.; Carballo Lozano, C.; Zuñiga, C. Automatic Classification of Sarcopenia Level in Older Adults: A Case Study at Tijuana General Hospital. Int. J. Environ. Res. Public Health 2019, 16, 3275. [Google Scholar] [CrossRef]
- Ding, X.; Liu, J.; Yang, F.; Cao, J. Random radial basis function kernel-based support vector machine. J. Frankl. Inst. 2021, 358, 10121–10140. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Y.; Zhu, J.; Fang, Y. Machine learning-based prediction of sarcopenia in community-dwelling middle-aged and older adults: Findings from the CHARLS. Psychogeriatrics 2025, 25, e13205. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, Y.; Smith, J.P.; Strauss, J.; Yang, G. Cohort Profile: The China Health and Retirement Longitudinal Study (CHARLS). Int. J. Epidemiol. 2014, 43, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Zupo, R.; Moroni, A.; Castellana, F.; Gasparri, C.; Catino, F.; Lampignano, L.; Perna, S.; Clodoveo, M.L.; Sardone, R.; Rondanelli, M. A Machine-Learning Approach to Target Clinical and Biological Features Associated with Sarcopenia: Findings from Northern and Southern Italian Aging Populations. Metabolites 2023, 13, 565. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Moroni, A.; Perna, S.; Azzolino, D.; Gasparri, C.; Zupo, R.; Micheletti Cremasco, M.; Rondanelli, M. Discovering the Individualized Factors Associated with Sarcopenia and Sarcopenic Obesity Phenotypes—A Machine Learning Approach. Nutrients 2023, 15, 4536. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yoo, J.I.; Ha, Y.C. Sarcopenia feature selection and risk prediction using machine learning. Medicine 2019, 98, e17699. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support Vector Machines for Classification. In Efficient Learning Machines: Theories, Concepts, and Applications for Engineers and System Designers; Apress: Berkeley, CA, USA, 2015; pp. 39–66. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 4765–4774. [Google Scholar]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16, New York, NY, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, M.; Liu, Q.; Ding, F.; Hou, L.; Deng, Y.; Cui, T.; Han, Y.; Pang, W.; Ye, W.; et al. Machine and deep learning-based clinical characteristics and laboratory markers for the prediction of sarcopenia. Chin. Med. J. 2023, 136, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.T.; Koc, L.; Harmsen, J.; Shaked, T.; Chandra, T.; Aradhye, H.; Anderson, G.; Corrado, G.; Chai, W.; Ispir, M.; et al. Wide & Deep Learning for Recommender Systems. In Proceedings of the 1st Workshop on Deep Learning for Recommender Systems, DLRS 2016, New York, NY, USA, 15 September 2016; pp. 7–10. [Google Scholar] [CrossRef]
- Hou, L.; Liu, X.; Zhang, Y.; Zhao, W.; Xia, X.; Chen, X.; Lin, X.; Yue, J.; Ge, N.; Dong, B. Cohort Profile: West China Health and Aging Trend (WCHAT). J. Nutr. Health Aging 2021, 25, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Porciello, G.; Di Lauro, T.; Luongo, A.; Coluccia, S.; Prete, M.; Abbadessa, L.; Coppola, E.; Di Martino, A.; Mozzillo, A.L.; Racca, E.; et al. Optimizing Nutritional Care with Machine Learning: Identifying Sarcopenia Risk Through Body Composition Parameters in Cancer Patients-Insights from the NUTritional and Sarcopenia RIsk SCREENing Project (NUTRISCREEN). Nutrients 2025, 17, 1376. [Google Scholar] [CrossRef]
- Gu, Y.; Su, S.; Wang, X.; Mao, J.; Ni, X.; Li, A.; Liang, Y.; Zeng, X. Comparative study of XGBoost and logistic regression for predicting sarcopenia in postsurgical gastric cancer patients. Sci. Rep. 2025, 15, 12808. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Z.; Hu, X.; Zhang, Y. Epigenetic characterization of sarcopenia-associated genes based on machine learning and network screening. Eur. J. Med. Res. 2024, 29, 54. [Google Scholar] [CrossRef]
- Lin, S.; Chen, C.; Cai, X.; Yang, F.; Fan, Y. Development and Verification of a Combined Diagnostic Model for Sarcopenia with Random Forest and Artificial Neural Network. Comput. Math. Methods Med. 2022, 2022, 2957731. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD ’16, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. Game theory, on-line prediction and boosting. In Proceedings of the Ninth Annual Conference on Computational Learning Theory, COLT ’96, New York, NY, USA, 28 June–1 July 1996; pp. 325–332. [Google Scholar] [CrossRef]
- Ahn, S.; Sung, Y.; Song, W. Machine Learning-Based Identification of Diagnostic Biomarkers for Korean Male Sarcopenia Through Integrative DNA Methylation and Methylation Risk Score: From the Korean Genomic Epidemiology Study (KoGES). J. Korean Med. Sci. 2024, 39, e200. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Granic, A.; Suetterlin, K.; Shavlakadze, T.; Grounds, M.D.; Sayer, A.A. Hallmarks of ageing in human skeletal muscle and implications for understanding the pathophysiology of sarcopenia in women and men. Clin. Sci. 2023, 137, 1721–1751. [Google Scholar] [CrossRef]
- Dalle, S.; Rossmeislova, L.; Koppo, K. The Role of Inflammation in Age-Related Sarcopenia. Front. Physiol. 2017, 8, 1045. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.A.; Lee, S.H.; Koh, J.M.; Kwon, S.H.; Lee, Y.; Cho, H.J.; Kim, H.; Kim, S.J.; Lee, J.H.; Yoo, H.J.; et al. Fatty acid amides as potential circulating biomarkers for sarcopenia. J. Cachexia Sarcopenia Muscle 2023, 14, 1558–1568. [Google Scholar] [CrossRef]
- Hua, C.; Chen, Y.; Sun, Z.; Shi, Z.; Song, Q.; Shen, L.; Lu, W.; Wang, Z.; Zang, J. Associations of serum arginine acid with sarcopenia in Chinese eldely women. Nutr. Metab. 2024, 21, 63. [Google Scholar] [CrossRef]
- Qaisar, R.; Karim, A.; Muhammad, T.; Shah, I.; Khan, J. Circulating MicroRNAs as Biomarkers of Accelerated Sarcopenia in Chronic Heart Failure. Glob. Heart 2021, 16, 56. [Google Scholar] [CrossRef]
- Seco-Cervera, M.; Ibáñez-Cabellos, J.S.; Pallardo, F.V.; García-Giménez, J.L.; Aulinas, A.; Martel-Duguech, L.; Webb, S.M.; Valassi, E. Circulating miR-28-5p is overexpressed in patients with sarcopenia despite long-term remission of Cushing’s syndrome: A pilot study. Front. Endocrinol. 2024, 15, 1410080. [Google Scholar] [CrossRef]
- Murgia, M.; Brocca, L.; Monti, E.; Franchi, M.V.; Zwiebel, M.; Steigerwald, S.; Giacomello, E.; Sartori, R.; Zampieri, S.; Capovilla, G.; et al. Plasma proteome profiling of healthy subjects undergoing bed rest reveals unloading-dependent changes linked to muscle atrophy. J. Cachexia Sarcopenia Muscle 2023, 14, 439–451. [Google Scholar] [CrossRef]
- Petermann-Rocha, F.; Balntzi, V.; Gray, S.R.; Lara, J.; Ho, F.K.; Pell, J.P.; Celis-Morales, C. Global prevalence of sarcopenia and severe sarcopenia: A systematic review and meta-analysis. J. Cachexia Sarcopenia Muscle 2022, 13, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Smith, L.; Hamer, M. Gender-specific risk factors for incident sarcopenia: 8-year follow-up of the English longitudinal study of ageing. J. Epidemiol. Community Health 2019, 73, 86–88. [Google Scholar] [CrossRef]
- Salamanna, F.; Contartese, D.; Ruffilli, A.; Barile, F.; Bellavia, D.; Marchese, L.; Manzetti, M.; Viroli, G.; Faldini, C.; Giavaresi, G. Sharing Circulating Micro-RNAs between Osteoporosis and Sarcopenia: A Systematic Review. Life 2023, 13, 602. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.L.; Esa, M.S.; Li, K.H.C.; Krishnan, S.R.G.; Elgallab, G.M.; Pearce, M.S.; Young, D.A.; Birrell, F.N. Osteoporosis, fracture, osteoarthritis & sarcopenia: A systematic review of circulating microRNA association. Bone 2021, 152, 116068. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Boehm, I.; Fernandes, M.; Rutter, A.; Skipworth, R.J.E.; Husi, H. Gene Ontology (GO)-Driven Inference of Candidate Proteomic Markers Associated with Muscle Atrophy Conditions. Molecules 2022, 27, 5514. [Google Scholar] [CrossRef]
- Huang, H.; Chen, Y.; Xu, W.; Cao, L.; Qian, K.; Bischof, E.; Kennedy, B.K.; Pu, J. Decoding aging clocks: New insights from metabolomics. Cell Metab. 2025, 37, 34–58. [Google Scholar] [CrossRef]
- Fan, Z.; Yang, J.Y.; Guo, Y.; Liu, Y.X.; Zhong, X.Y. Altered levels of circulating mitochondrial DNA in elderly people with sarcopenia: Association with mitochondrial impairment. Exp. Gerontol. 2022, 163, 111802. [Google Scholar] [CrossRef]
- Xu, R.; Cui, S.; Chen, L.; Chen, X.C.; Ma, L.L.; Yang, H.N.; Wen, F.M. Circulating miRNA-1-3p as Biomarker of Accelerated Sarcopenia in Patients Diagnosed with Chronic Heart Failure. Rev. Investig. Clin. Organo Del Hosp. Enfermedades Nutr. 2022, 74, 276–283. [Google Scholar] [CrossRef]
- Liu, H.C.; Han, D.S.; Hsu, C.C.; Wang, J.S. Circulating MicroRNA-486 and MicroRNA-146a serve as potential biomarkers of sarcopenia in the older adults. BMC Geriatr. 2021, 21, 86. [Google Scholar] [CrossRef]
- Han, P.; Chen, X.; Liang, Z.; Liu, Y.; Yu, X.; Song, P.; Zhao, Y.; Zhang, H.; Zhu, S.; Shi, X.; et al. Metabolic signatures and risk of sarcopenia in suburb-dwelling older individuals by LC-MS-based untargeted metabonomics. Front. Endocrinol. 2024, 15, 1308841. [Google Scholar] [CrossRef]
- Clark, R.V.; Walker, A.C.; O’Connor-Semmes, R.L.; Leonard, M.S.; Miller, R.R.; Stimpson, S.A.; Turner, S.M.; Ravussin, E.; Cefalu, W.T.; Hellerstein, M.K.; et al. Total body skeletal muscle mass: Estimation by creatine (methyl-d3) dilution in humans. J. Appl. Physiol. 2014, 116, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- L’hôte, C.; Cordier, B.; Labasse, A.; Boileau, C.; Costes, B.; Henrotin, Y. Identification of new biomarkers for sarcopenia and characterization of cathepsin D biomarker. Jcsm Rapid Commun. 2021, 4, 122–132. [Google Scholar] [CrossRef]
- Pellegrino, R.; Paganelli, R.; Di Iorio, A.; Bandinelli, S.; Moretti, A.; Iolascon, G.; Sparvieri, E.; Tarantino, D.; Ferrucci, L. Muscle quality, physical performance, and comorbidity are predicted by circulating procollagen type III N-terminal peptide (P3NP): The InCHIANTI follow-up study. GeroScience 2024, 46, 1259–1269. [Google Scholar] [CrossRef]
- Ding, J.; Yang, G.; Sun, W.; Li, Y.; Wang, N.; Wang, J.; Zhao, Y. Association of interleukin-6 with sarcopenia and its components in older adults: A systematic review and meta-analysis of cross-sectional studies. Ann. Med. 2024, 56, 2384664. [Google Scholar] [CrossRef] [PubMed]
- Bellanti, F.; Lo Buglio, A.; Quiete, S.; Dobrakowski, M.; Kasperczyk, A.; Kasperczyk, S.; Vendemiale, G. Sarcopenia Is Associated with Changes in Circulating Markers of Antioxidant/Oxidant Balance and Innate Immune Response. Antioxidants 2023, 12, 1992. [Google Scholar] [CrossRef] [PubMed]
- Landi, F.; Calvani, R.; Lorenzi, M.; Martone, A.M.; Tosato, M.; Drey, M.; D’Angelo, E.; Capoluongo, E.; Russo, A.; Bernabei, R.; et al. Serum levels of C-terminal agrin fragment (CAF) are associated with sarcopenia in older multimorbid community-dwellers: Results from the ilSIRENTE study. Exp. Gerontol. 2016, 79, 31–36. [Google Scholar] [CrossRef]
- Hsu, B.G.; Wang, C.H.; Lai, Y.H.; Kuo, C.H.; Lin, Y.L. Association of endothelial dysfunction and peripheral arterial disease with sarcopenia in chronic kidney disease. J. Cachexia Sarcopenia Muscle 2024, 15, 1199–1208. [Google Scholar] [CrossRef]
- Deng, M.; Bian, Y.; Zhang, Q.; Zhou, X.; Hou, G. Growth Differentiation Factor-15 as a Biomarker for Sarcopenia in Patients with Chronic Obstructive Pulmonary Disease. Front. Nutr. 2022, 9, 897097. [Google Scholar] [CrossRef]
- Wu, J.; Cao, L.; Wang, J.; Wang, Y.; Hao, H.; Huang, L. Characterization of serum protein expression profiles in the early sarcopenia older adults with low grip strength: A cross-sectional study. BMC Musculoskelet. Disord. 2022, 23, 894. [Google Scholar] [CrossRef]
- Bian, A.; Ma, Y.; Zhou, X.; Guo, Y.; Wang, W.; Zhang, Y.; Wang, X. Association between sarcopenia and levels of growth hormone and insulin-like growth factor-1 in the elderly. BMC Musculoskelet. Disord. 2020, 21, 214. [Google Scholar] [CrossRef]
- Siracusa, J.; Koulmann, N.; Banzet, S. Circulating myomiRs: A new class of biomarkers to monitor skeletal muscle in physiology and medicine. J. Cachexia Sarcopenia Muscle 2018, 9, 20–27. [Google Scholar] [CrossRef]
- Millet, M.; Auroux, M.; Beaudart, C.; Demonceau, C.; Ladang, A.; Cavalier, E.; Reginster, J.Y.; Bruyère, O.; Chapurlat, R.; Rousseau, J.C. Association of circulating hsa-miRNAs with sarcopenia: The SarcoPhAge study. Aging Clin. Exp. Res. 2024, 36, 70. [Google Scholar] [CrossRef]
- Okugawa, Y.; Yao, L.; Toiyama, Y.; Yamamoto, A.; Shigemori, T.; Yin, C.; Omura, Y.; Ide, S.; Kitajima, T.; Shimura, T.; et al. Prognostic impact of sarcopenia and its correlation with circulating miR-21 in colorectal cancer patients. Oncol. Rep. 2018, 39, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.S.; Wang, S.Y.; Chang, C.H.; Chen, C.Y.; Wen, C.J.; Chen, G.Y.; Kuo, C.H.; Tseng, Y.J.; Chen, C.Y. Identification of traumatic acid as a potential plasma biomarker for sarcopenia using a metabolomics-based approach. J. Cachexia Sarcopenia Muscle 2022, 13, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Koh, J.M.; Cho, H.J.; Kim, H.; Lee, Y.S.; Kim, S.J.; Yoon, P.W.; Kim, W.; Bae, S.J.; Kim, H.K.; et al. Sphingolipid metabolites as potential circulating biomarkers for sarcopenia in men. J. Cachexia Sarcopenia Muscle 2024, 15, 2476–2486. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, Z.; He, P.; Mao, X.; Jing, X.; Hu, Y.; Jing, L. LC/MS-Based Untargeted Lipidomics Reveals Lipid Signatures of Sarcopenia. Int. J. Mol. Sci. 2024, 25, 8793. [Google Scholar] [CrossRef]
- Hsu, W.H.; Wang, S.Y.; Chao, Y.M.; Chang, K.V.; Han, D.S.; Lin, Y.L. Novel metabolic and lipidomic biomarkers of sarcopenia. J. Cachexia Sarcopenia Muscle 2024, 15, 2175–2186. [Google Scholar] [CrossRef]
- Shida, T.; Yoshida, Y.; Ohta, T.; Kojima, N.; Osuka, Y.; Takekoshi, K.; Sasai, H. Identification of a novel biomarker for sarcopenia diagnosis using serum metabolomic analysis: A pilot study. Eur. Geriatr. Med. 2024, 15, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef]
- Sayer, A.A.; Cruz-Jentoft, A. Sarcopenia definition, diagnosis and treatment: Consensus is growing. Age Ageing 2022, 51, afac220. [Google Scholar] [CrossRef]
- Rashidi, H.H.; Pantanowitz, J.; Chamanzar, A.; Fennell, B.; Wang, Y.; Gullapalli, R.R.; Tafti, A.; Deebajah, M.; Albahra, S.; Glassy, E.; et al. Generative Artificial Intellegence (AI) in Pathology and Medicine: A Deeper Dive. Mod. Pathol. 2025, 38, 100687. [Google Scholar] [CrossRef] [PubMed]
| Dataset | AI Model | Performance | Key Predictors | Ref. |
|---|---|---|---|---|
| EHR from 1304 patients | RF, SVM | AUC > 90% | Diagnoses, medications, lab tests | [15] |
| 166 patients, 99 variables | RBF SVM | Accuracy: 82.5%, F1: 90.2%, precision: 82.8% | Age, hypertension, MNA, sodium | [18] |
| 133 subjects, BPW signals | LDA, Scoring System | AUC: 0.77 (LDA), 0.83 (scoring) | APS | [17] |
| CHARLS | XGBoost | AUROC: 0.759 | MMSE, drinking habits, BUN | [20] |
| WCHAT cohort, XMAT validation | Wide and Deep | AUC: 0.97, ACC: 91.1% | MAMC, CC, TSF, AST/ALT ratio | [29] |
| Italian ageing populations | RF (3 models) | ACC 89.89%, sensitivity 14.50%, specificity 99.37% | Albumin, CRP, vitamin D, folates | [22] |
| 1510 patients | LR | S: 0.33, SO: 0.19 OS: 0.267 | BMI | [24] |
| KNHANES (4020 patients) | LR, RF, SVM, GBM | AUC [men–women] RF: 0.82–0.78 SVM: 0.8–0.81 GB: 0.81–0.81 LR: 0.82–0.80 | BMI, RBC, nutrient intake, water intake | [25] |
| 3096 Japanese patients | CNN | ACC: 0.88 | MAMC, CC, TSF, AST/ALT ratio | [16] |
| 879 oncological patients | PCA + K-means | ACC: PC1 (59%), PC2 (24%), PC3 (15%) | Advanced age, lung, gynecological, gastroint. cancer, diabetes, malnutrition | [32] |
| 231 post-surgical patients | XGBoost vs. LASSO | AUROC: 0.98 | Serum albumin, diabetes, type surgery, nutritional score, ECOG status | [33] |
| Dataset | AI Model | Key Biomarkers | Performance | Ref. |
|---|---|---|---|---|
|
-GSE1428 -GSE136344 | LASSO, SVM-RFE | MYH8, HOXB2, CDKN1A | AUC > 0.7 | [34] |
| -GSE111017 | DNN (DSnet-v1) | 27 AI-featured genes (e.g., H4C3, PSMA6, CENPC, VPS35L) | Acc. 0.96 Sens. 1.000 Spec. 0.94 AUC: 0.99 | [14] |
|
-GSE8479 -GSE9103 -GSE38718 -GSE1428 | RF, ANN | MT1X, CISD1, WISP2 | AUC: 0.999 (train), 0.85 (test) | [35] |
|
509 Korean males | RFECV, Ensemble ML | 8 CpG sites | AUC: 0.94 | [38] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, V.; Letteri, I.; Santini, S.J.; Sinatti, G.; Balsano, C. Towards Precision in Sarcopenia Assessment: The Challenges of Multimodal Data Analysis in the Era of AI. Int. J. Mol. Sci. 2025, 26, 4428. https://doi.org/10.3390/ijms26094428
Caputo V, Letteri I, Santini SJ, Sinatti G, Balsano C. Towards Precision in Sarcopenia Assessment: The Challenges of Multimodal Data Analysis in the Era of AI. International Journal of Molecular Sciences. 2025; 26(9):4428. https://doi.org/10.3390/ijms26094428
Chicago/Turabian StyleCaputo, Valerio, Ivan Letteri, Silvano Junior Santini, Gaia Sinatti, and Clara Balsano. 2025. "Towards Precision in Sarcopenia Assessment: The Challenges of Multimodal Data Analysis in the Era of AI" International Journal of Molecular Sciences 26, no. 9: 4428. https://doi.org/10.3390/ijms26094428
APA StyleCaputo, V., Letteri, I., Santini, S. J., Sinatti, G., & Balsano, C. (2025). Towards Precision in Sarcopenia Assessment: The Challenges of Multimodal Data Analysis in the Era of AI. International Journal of Molecular Sciences, 26(9), 4428. https://doi.org/10.3390/ijms26094428









