Mercury Bioaccumulation in Female Breast Cancer Is Associated to CXCR4 Expression
Abstract
1. Introduction
2. Results
2.1. Histopathological Assessment
2.2. Mercury Bioaccumulation in Breast Cancer
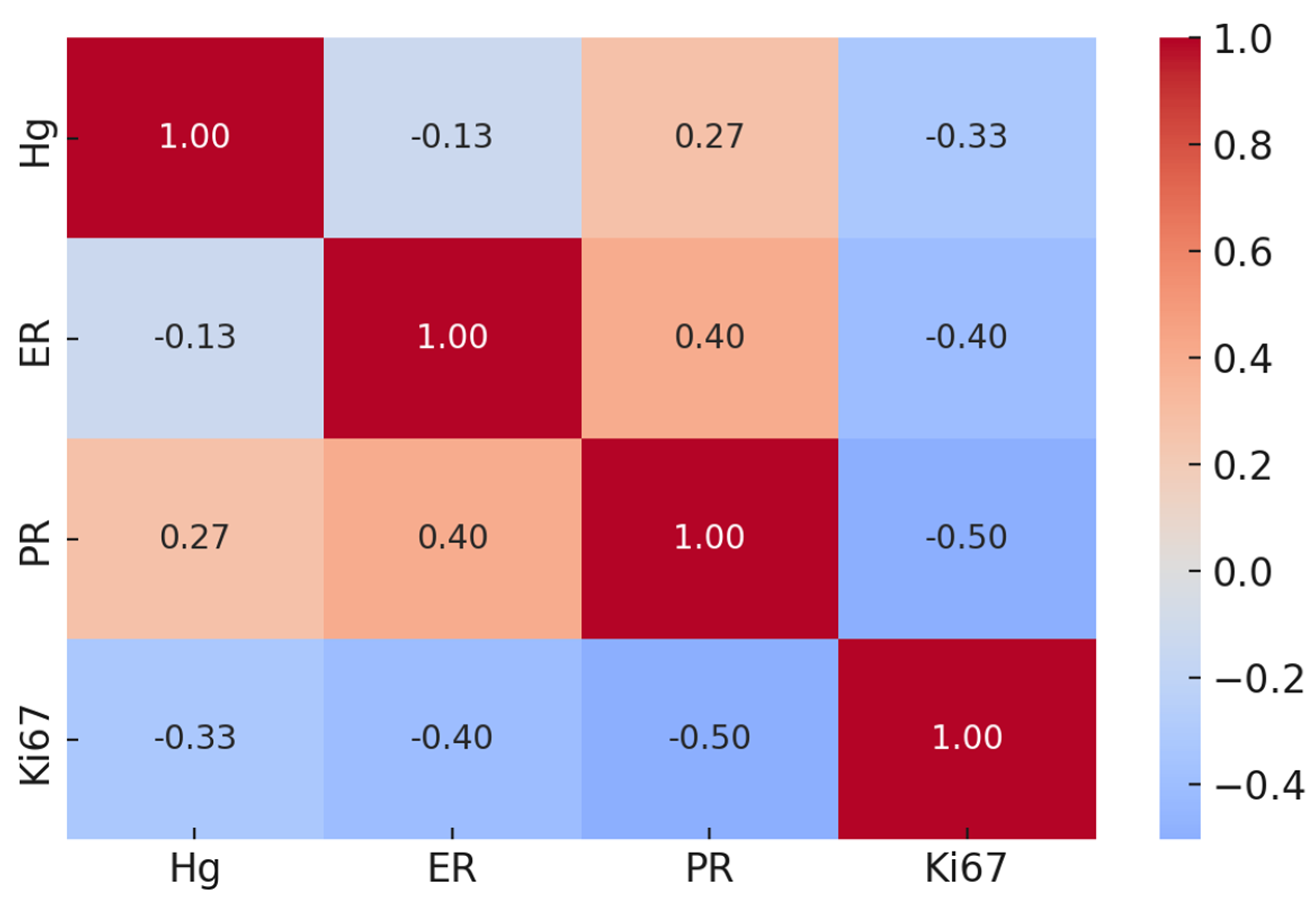
2.3. Mercury Bioaccumulation and Traditional Prognostic Markers in Breast Cancer
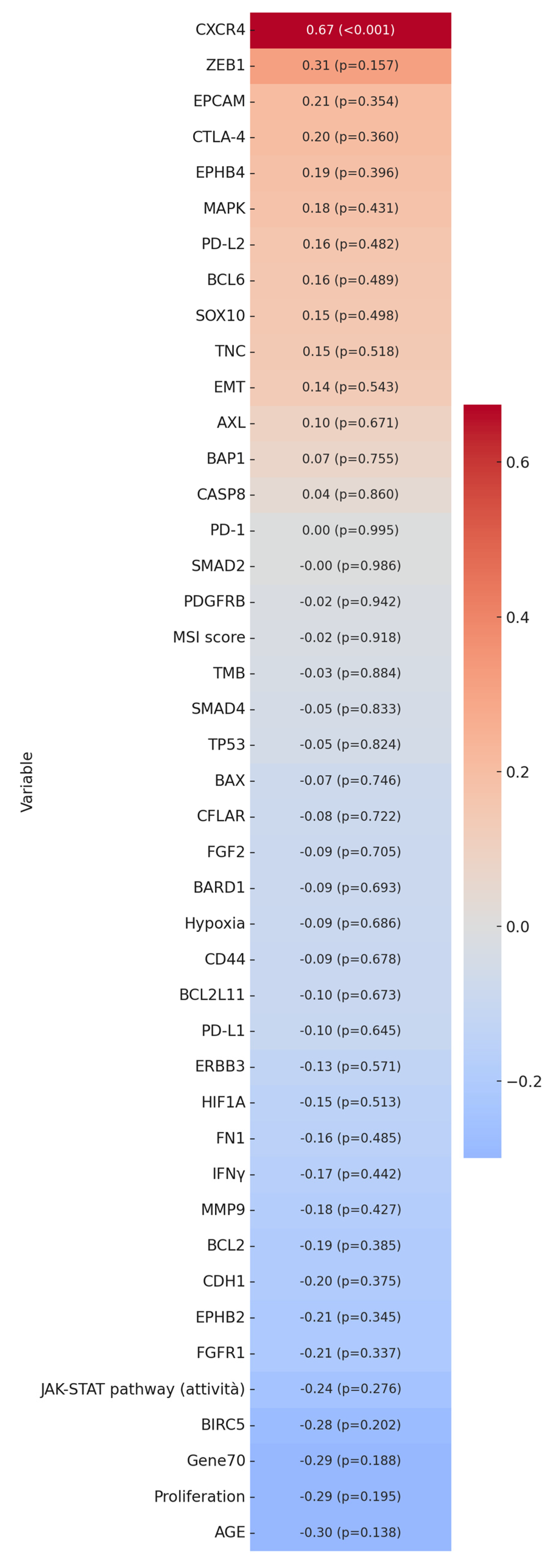
2.4. Mercury Bioaccumulation and Emerging Prognostic Factors in Breast Cancer
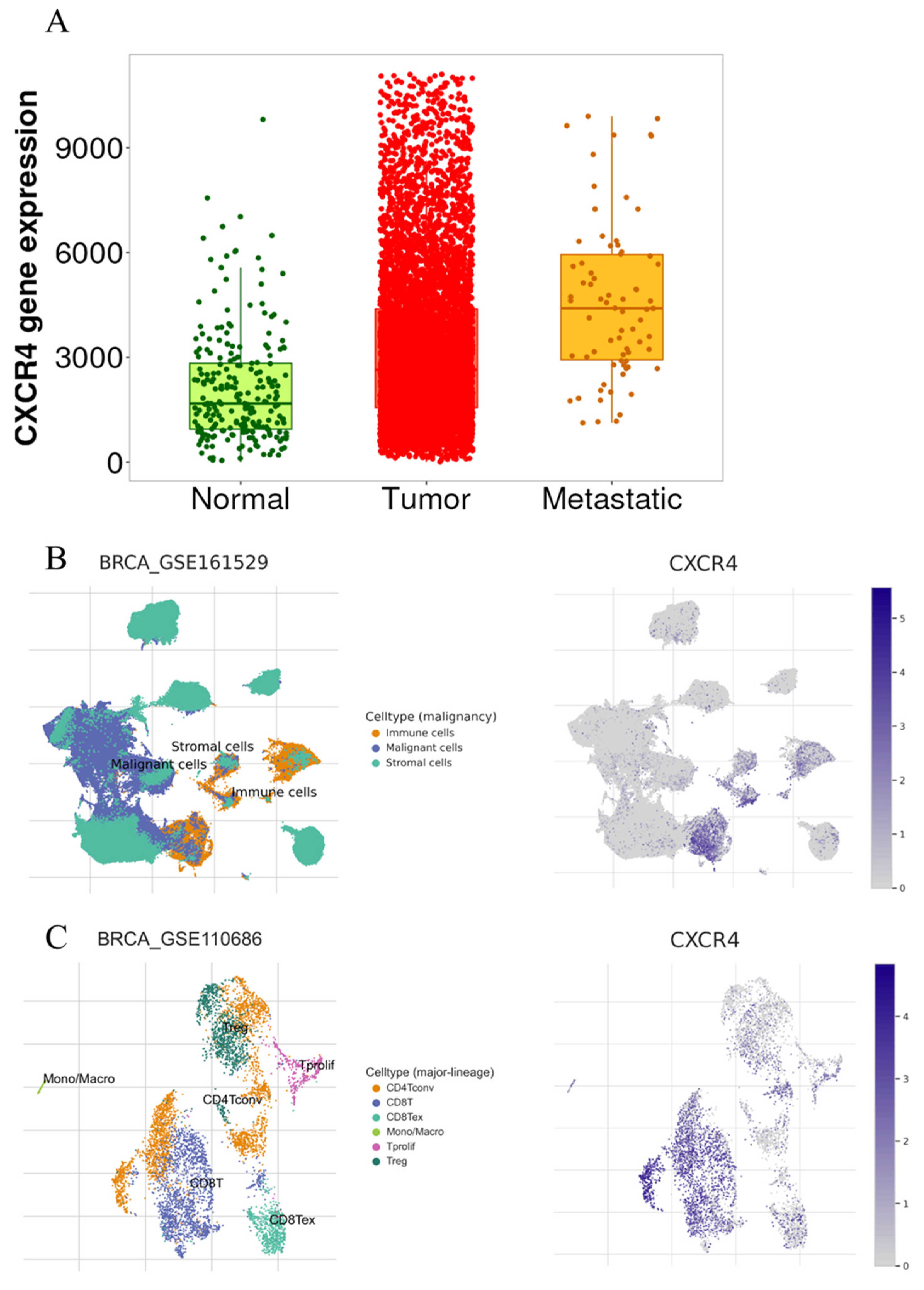
2.5. Bioinformatic Analysis
3. Discussion
4. Materials and Methods
4.1. Samples Collection and Study Design
4.2. Histology, Immunohistochemistry (IHC), and Fluorescent In Situ Hybridization (FISH)
4.3. Inductively Coupled Plasma Mass Spectrometry
4.4. Nucleic Acid Extraction and Quality Assessment
4.5. Library Preparation and Next-Generation Sequencing (NGS)
4.6. Molecular Investigation
4.7. Bioinformatic Analysis
4.8. Statistical Analysis
5. Conclusions
Limits of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Romaniuk, A.; Lyndin, M.; Sikora, V.; Lyndina, Y.; Romaniuk, S.; Sikora, K. Heavy metals effect on breast cancer progression. J. Occup. Med. Toxicol. 2017, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Koual, M.; Tomkiewicz, C.; Cano-Sancho, G.; Antignac, J.P.; Bats, A.S.; Coumoul, X. Environmental chemicals, breast cancer progression and drug resistance. Environ. Health 2020, 19, 117. [Google Scholar] [CrossRef]
- ECHA. Available online: https://echa.europa.eu/it/substance-information/-/substanceinfo/100.028.278 (accessed on 7 April 2025).
- EPA. Available online: https://www.epa.gov/mercury (accessed on 7 April 2025).
- ECHA. Available online: https://echa.europa.eu/it/home (accessed on 7 April 2025).
- Coradduzza, D.; Congiargiu, A.; Azara, E.; Mammani, I.M.A.; De Miglio, M.R.; Zinellu, A.; Carru, C.; Medici, S. Heavy metals in biological samples of cancer patients: A systematic literature review. Biometals 2024, 37, 803–817. [Google Scholar] [CrossRef]
- Pamphlett, R.; Satgunaseelan, L.; Kum Jew, S.; Doble, P.A.; Bishop, D.P. Elemental bioimaging shows mercury and other toxic metals in normal breast tissue and in breast cancers. PLoS ONE 2020, 15, e0228226. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, J.G.; Novotny, J.; Stejskal, V.; Lätsch, A.; Blaurock-Busch, E.; Eisenmann-Klein, M. Increased levels of transition metals in breast cancer tissue. Neuro Endocrinol. Lett. 2006, 27 (Suppl. S1), 36–39, Erratum in Neuro Endocrinol Lett. 2007, 28, iii. [Google Scholar]
- Mohammadi, M.; Riyahi Bakhtiari, A.; Khodabandeh, S. Concentration of cd, pb, hg, and se in different parts of human breast cancer tissues. J. Toxicol. 2014, 2014, 413870. [Google Scholar] [CrossRef]
- Pamphlett, R.; Bishop, D.P. Elemental biomapping of human tissues suggests toxic metals such as mercury play a role in the pathogenesis of cancer. Front. Oncol. 2024, 14, 1420451. [Google Scholar] [CrossRef]
- Tang, P.; Tse, G.M. Immunohistochemical Surrogates for Molecular Classification of Breast Carcinoma: A 2015 Update. Arch. Pathol. Lab. Med. 2016, 140, 806–814. [Google Scholar] [CrossRef]
- Mak, M.P.; Tong, P.; Diao, L.; Cardnell, R.J.; Gibbons, D.L.; William, W.N.; Skoulidis, F.; Parra, E.R.; Rodriguez-Canales, J.; Wistuba, I.I.; et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2016, 22, 609–620. [Google Scholar] [CrossRef]
- Buffa, F.M.; Harris, A.L.; West, C.M.; Miller, C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br. J. Cancer 2010, 102, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- van‘t Veer, L.J.; Dai, H.; van de Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Coyte, R.M.; Darrah, T.H.; Barrett, E.; O’Connor, T.G.; Olesik, J.W.; Salafia, C.M.; Shah, R.; Love, T.; Miller, R.K. Comparison of trace element concentrations in paired formalin-fixed paraffin-embedded and frozen human placentae. Placenta 2023, 131, 98–103. [Google Scholar] [CrossRef]
- Bonfiglio, R.; Giacobbi, E.; Palumbo, V.; Casciardi, S.; Sisto, R.; Servadei, F.; Scioli, M.P.; Schiaroli, S.; Cornella, E.; Cervelli, G.; et al. Aluminum Concentration Is Associated with Tumor Mutational Burden and the Expression of Immune Response Biomarkers in Colorectal Cancers. Int. J. Mol. Sci. 2024, 25, 13388. [Google Scholar] [CrossRef]
- Bonfiglio, R.; Sisto, R.; Casciardi, S.; Palumbo, V.; Scioli, M.P.; Giacobbi, E.; Servadei, F.; Melino, G.; Mauriello, A.; Scimeca, M. Aluminium bioaccumulation in colon cancer, impinging on epithelial-mesenchymal-transition and cell death. Sci. Total Environ. 2024, 908, 168335. [Google Scholar] [CrossRef]
- Bonfiglio, R.; Scimeca, M.; Mauriello, A. The impact of environmental pollution on cancer: Risk mitigation strategies to consider. Sci. Total Environ. 2023, 902, 166219. [Google Scholar] [CrossRef]
- Balkwill, F.R. The chemokine system and cancer. J. Pathol. 2012, 226, 148–157. [Google Scholar] [CrossRef]
- Sun, Y.; Mao, X.; Fan, C.; Liu, C.; Guo, A.; Guan, S.; Jin, Q.; Li, B.; Yao, F.; Jin, F. CXCL12-CXCR4 axis promotes the natural selection of breast cancer cell metastasis. Tumour Biol. 2014, 35, 7765–7773. [Google Scholar] [CrossRef]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Zhao, J. The Role of chemokine receptor CXCR4 in breast cancer metastasis. Am. J. Cancer Res. 2013, 3, 46–57. [Google Scholar] [PubMed]
- Suva, L.J.; Griffin, R.J.; Makhoul, I. Mechanisms of bone metastases of breast cancer. Endocr. Relat. Cancer 2009, 16, 703–713. [Google Scholar] [CrossRef]
- Shi, Q.; Yang, W.; Ouyang, Y.; Liu, Y.; Cai, Z. CXCR4 promotes tumor stemness maintenance and CDK4/6 inhibitors resistance in ER-positive breast cancer. Breast Cancer Res. 2025, 27, 15. [Google Scholar] [CrossRef]
- Queiroz, M.I.C.; Sales, M.V.S.; Barros, E.D.S.S.; D’Amato, F.O.S.; Gonçalves, C.M.; Ursulino, J.S.; Bueno, N.B.; Marinho, C.; Rocha, U.; Aquino, T.M.; et al. Exposure to a contaminated environment and its relationship with human health: Mercury effect on loss of functionality and increased oxidative stress of blood cells. J. Hazard Mater. 2025, 492, 138088. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Zhang, L.; Liu, S.; Wang, F.; Liu, H.; Zhou, F.; Wu, F.; Zhang, H.; Fan, D.; Nie, Y.; et al. Platinum drugs upregulate CXCR4 and PD-L1 expression via ROS-dependent pathways, with implications for novel combined treatment in gastric cancer. J. Pathol. Clin. Res. 2025, 11, e70015. [Google Scholar] [CrossRef]
- Skalny, A.V.; Aschner, M.; Sekacheva, M.I.; Santamaria, A.; Barbosa, F.; Ferrer, B.; Aaseth, J.; Paoliello, M.M.B.; Rocha, J.B.T.; Tinkov, A.A. Mercury and cancer: Where are we now after two decades of research? Food Chem. Toxicol. 2022, 164, 113001. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Cancer Epigenetics, Tumor Immunity, and Immunotherapy. Trends Cancer 2020, 6, 580–592. [Google Scholar] [CrossRef]
- Lee, J.Y.; Tokumoto, M.; Hwang, G.W.; Kim, M.S.; Takahashi, T.; Naganuma, A.; Yoshida, M.; Satoh, M. Effect of Metallothionein-III on Mercury-Induced Chemokine Gene Expression. Toxics 2018, 6, 48. [Google Scholar] [CrossRef]
- von der Heyde, S.; Raman, N.; Gabelia, N.; Matias-Guiu, X.; Yoshino, T.; Tsukada, Y.; Melino, G.; Marshall, J.L.; Wellstein, A.; Juhl, H.; et al. Tumor specimen cold ischemia time impacts molecular cancer drug target discovery. Cell Death Dis. 2024, 15, 691. [Google Scholar] [CrossRef] [PubMed]
- Servadei, F.; Anemona, L.; Cardellini, M.; Scimeca, M.; Montanaro, M.; Rovella, V.; Di Daniele, F.; Giacobbi, E.; Legramante, I.M.; Noce, A.; et al. The risk of carotid plaque instability in patients with metabolic syndrome is higher in women with hypertriglyceridemia. Cardiovasc. Diabetol. 2021, 20, 98. [Google Scholar] [CrossRef]
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology 2020, 77, 181–185. [Google Scholar] [CrossRef]
- Vasconcelos, I.; Hussainzada, A.; Berger, S.; Fietze, E.; Linke, J.; Siedentopf, F.; Schoenegg, W. The St. Gallen surrogate classification for breast cancer subtypes successfully predicts tumor presenting features, nodal involvement, recurrence patterns and disease free survival. Breast 2016, 29, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Wolff, A.C.; Somerfield, M.R.; Dowsett, M.; Hammond, M.E.H.; Hayes, D.F.; McShane, L.M.; Saphner, T.J.; Spears, P.A.; Allison, K.H. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: ASCO-College of American Pathologists Guideline Update. J. Clin. Oncol. 2023, 41, 3867–3872. [Google Scholar] [CrossRef]
- Scimeca, M.; Bischof, J.; Bonfiglio, R.; Nale, E.; Iacovelli, V.; Carilli, M.; Vittori, M.; Agostini, M.; Rovella, V.; Servadei, F.; et al. Molecular profiling of a bladder cancer with very high tumour mutational burden. Cell Death Discov. 2024, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Rovella, V.; Smirnov, A.; Buonomo, O.C.; Mauriello, A.; Perretta, T.; Shi, Y.; Woodmsith, J.; Bischof, J.; TORCENTRE; et al. A BRCA2 germline mutation and high expression of immune checkpoints in a TNBC patient. Cell Death Discov. 2023, 9, 370. [Google Scholar] [CrossRef]
- Giovannini, S.; Smirnov, A.; Concetti, L.; Scimeca, M.; Mauriello, A.; Bischof, J.; Rovella, V.; Melino, G.; Buonomo, C.O.; Candi, E.; et al. A comprehensive molecular characterization of a claudin-low luminal B breast tumor. Biol. Direct. 2024, 19, 66. [Google Scholar] [CrossRef]
- Manders, F.; Brandsma, A.M.; de Kanter, J.; Verheul, M.; Oka, R.; van Roosmalen, M.J.; van der Roest, B.; van Hoeck, A.; Cuppen, E.; van Boxtel, R. MutationalPatterns: The one stop shop for the analysis of mutational processes. BMC Genom. 2022, 23, 134. [Google Scholar] [CrossRef]
- Huang, M.N.; McPherson, J.R.; Cutcutache, I.; Teh, B.T.; Tan, P.; Rozen, S.G. MSIseq: Software for Assessing Microsatellite Instability from Catalogs of Somatic Mutations. Sci. Rep. 2015, 5, 13321. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://tnmplot.com/analysis/ (accessed on 7 April 2025).
- Available online: http://tisch.comp-genomics.org/home (accessed on 7 April 2025).
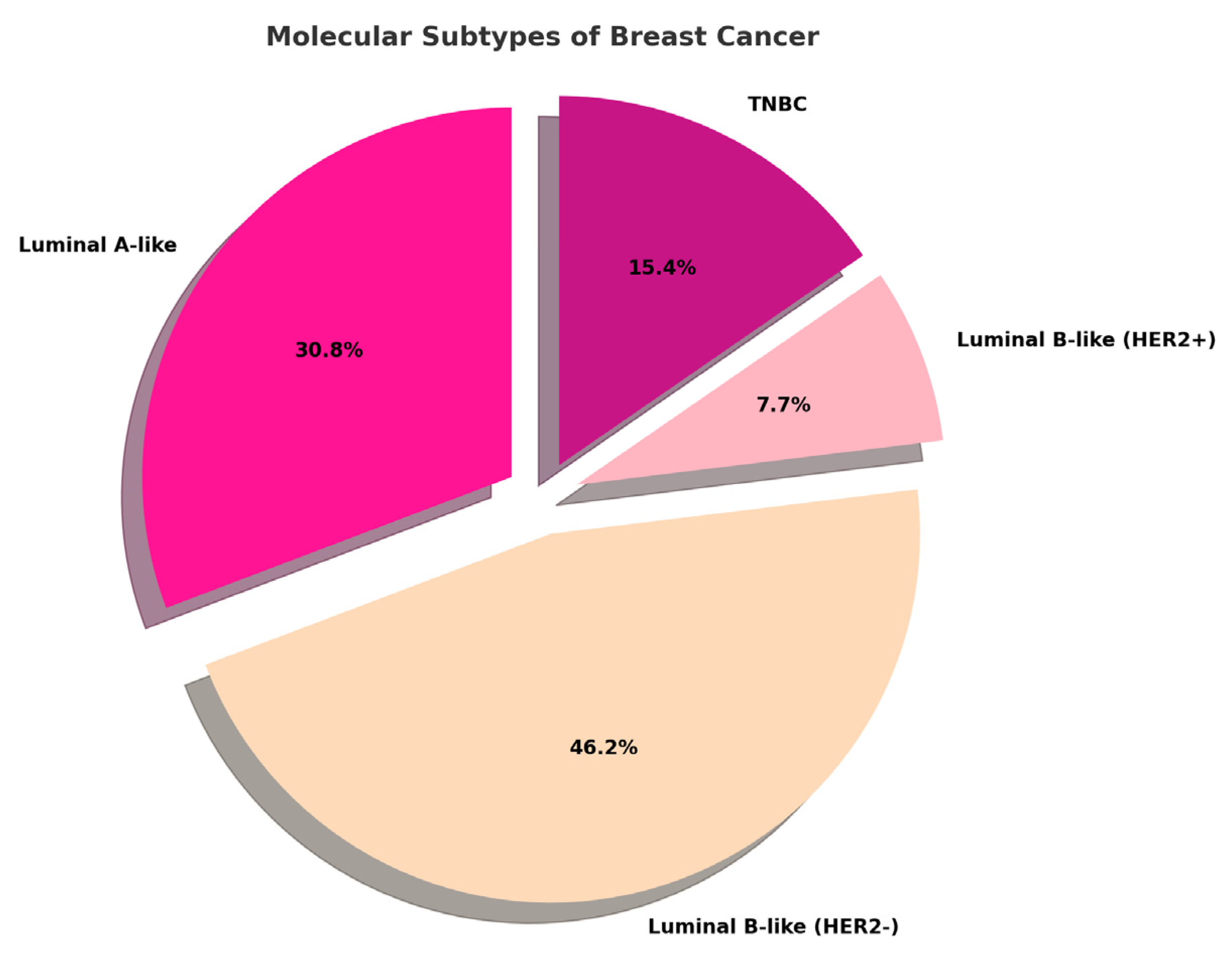
| Hg (mg/Kg) | AGE | Molecular Subtype | Histotype | Grade | T | N |
|---|---|---|---|---|---|---|
| 0.66 | 47 | Luminal-A-like | NST | G2 | pT1 | cN0(sn) |
| 0.5 | 46 | Luminal-B-like (HER2-) | NST | G3 | pT2(m) | N1a |
| 0.41 | 45 | TNBC | NST | G2 | pT2 | N0 |
| 0.35 | 44 | Luminal-B-like (HER2-) | NST | G2 | pT1c | Nx |
| 0.21 | 65 | Luminal-A-like | NST | G2 | pT1c(m) | N0 |
| 0.19 | 67 | Luminal-A-like | NST | G2 | pT1c | N0(sn) |
| 0.09 | 85 | Luminal-A-like | NST | G3 | pT2 | Nx |
| 0.07 | 46 | Luminal-B-like (HER2-) | NST | G3 | pT2 | N0(sn) |
| 0.06 | 71 | Luminal-B-like (HER2-) | NST | G2 | pT1c | N0 |
| 0.05 | 80 | TNBC | NST | G3 | pT2 | N0(sn) |
| 0.04 | 41 | Luminal-B-like (HER2+) | NST | G3 | pT3 | N0(sn) |
| 0.04 | 62 | TNBC | NST | G3 | pT3 | N3a |
| 0.04 | 72 | Luminal-A-like | NST | G2 | pT1c | N0 |
| 0.03 | 63 | Luminal-A-like | NST | G1 | pT1c | N1mi(sn) |
| 0.02 | 44 | TNBC | NST | G3 | pT3 | N3a |
| 0.02 | 81 | Luminal-B-like (HER2-) | NST | G3 | pT3 | N2a |
| 0.01 | 44 | Luminal-B-like (HER2-) | NST | G3 | pT1c | N1a |
| 0.01 | 78 | Luminal-A-like | NST | G2 | pT1 | cN0(sn) |
| 0 | 87 | Luminal-A-like | MIXED | G3/G1 | pT2(m) | Nx |
| 0 | 71 | Luminal-B-like (HER2+) | NST | G3 | pT1c | N1a |
| 0 | 51 | Luminal-B-like (HER2-) | NST | G3 | pT1c | N0(sn) |
| 0 | 43 | Luminal-B-like (HER2-) | NST | G3 | pT1c | N0 |
| 0 | 41 | Luminal-B-like (HER2-) | NST | G3 | pT2 | N0(sn) |
| 0 | 65 | Luminal-B-like (HER2-) | NST | G3 | pT1c | N0(sn) |
| 0 | 66 | Luminal-B-like (HER2-) | NST | G3 | pT1c | N0(sn) |
| 0 | 51 | Luminal-B-like (HER2-) | LIC | G3 | pT3(m) | N3a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Servadei, F.; Bonfiglio, R.; Sisto, R.; Casciardi, S.; Giacobbi, E.; Scioli, M.P.; Palumbo, V.; Buonomo, C.O.; Melino, G.; Mauriello, A.; et al. Mercury Bioaccumulation in Female Breast Cancer Is Associated to CXCR4 Expression. Int. J. Mol. Sci. 2025, 26, 4427. https://doi.org/10.3390/ijms26094427
Servadei F, Bonfiglio R, Sisto R, Casciardi S, Giacobbi E, Scioli MP, Palumbo V, Buonomo CO, Melino G, Mauriello A, et al. Mercury Bioaccumulation in Female Breast Cancer Is Associated to CXCR4 Expression. International Journal of Molecular Sciences. 2025; 26(9):4427. https://doi.org/10.3390/ijms26094427
Chicago/Turabian StyleServadei, Francesca, Rita Bonfiglio, Renata Sisto, Stefano Casciardi, Erica Giacobbi, Maria Paola Scioli, Valeria Palumbo, Claudio Oreste Buonomo, Gerry Melino, Alessandro Mauriello, and et al. 2025. "Mercury Bioaccumulation in Female Breast Cancer Is Associated to CXCR4 Expression" International Journal of Molecular Sciences 26, no. 9: 4427. https://doi.org/10.3390/ijms26094427
APA StyleServadei, F., Bonfiglio, R., Sisto, R., Casciardi, S., Giacobbi, E., Scioli, M. P., Palumbo, V., Buonomo, C. O., Melino, G., Mauriello, A., & Scimeca, M. (2025). Mercury Bioaccumulation in Female Breast Cancer Is Associated to CXCR4 Expression. International Journal of Molecular Sciences, 26(9), 4427. https://doi.org/10.3390/ijms26094427









