Efficient Agrobacterium-Mediated Methods for Transient and Stable Transformation in Common and Tartary Buckwheat
Abstract
1. Introduction
2. Results
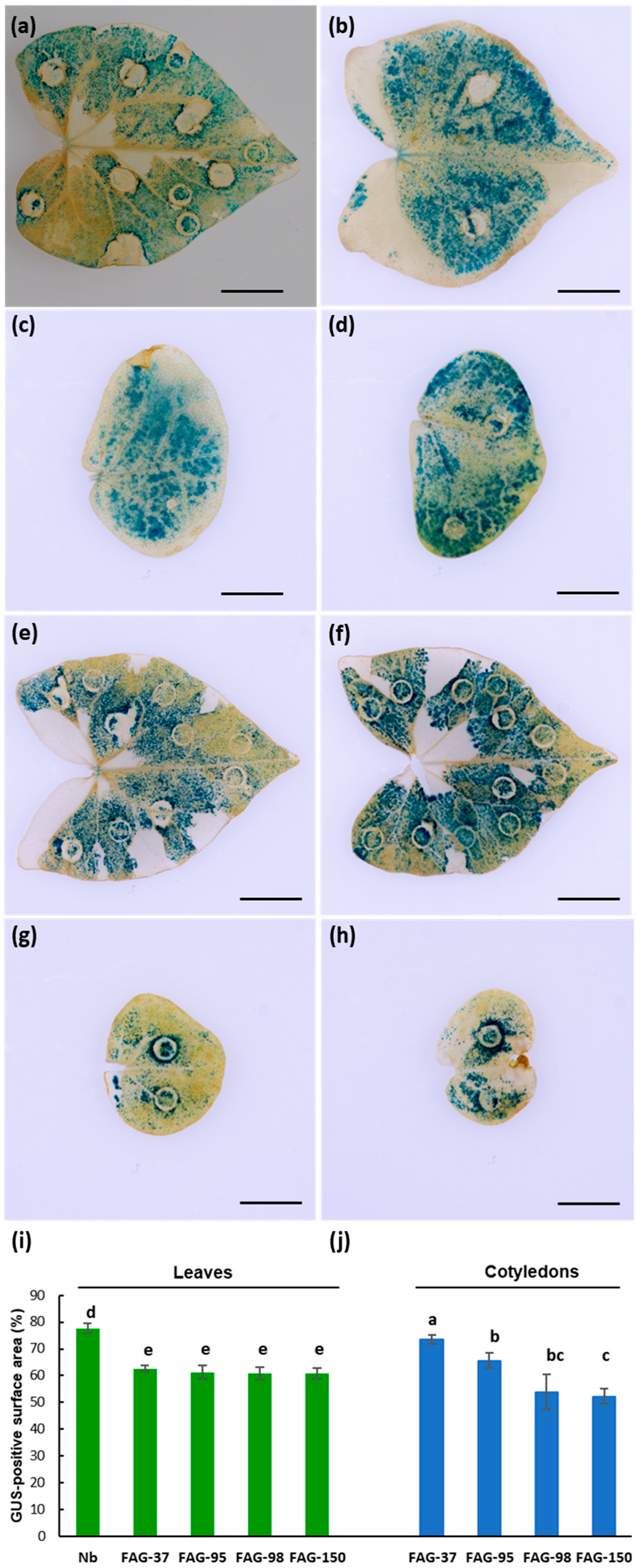
2.1. Transient Expression of Reporter Genes
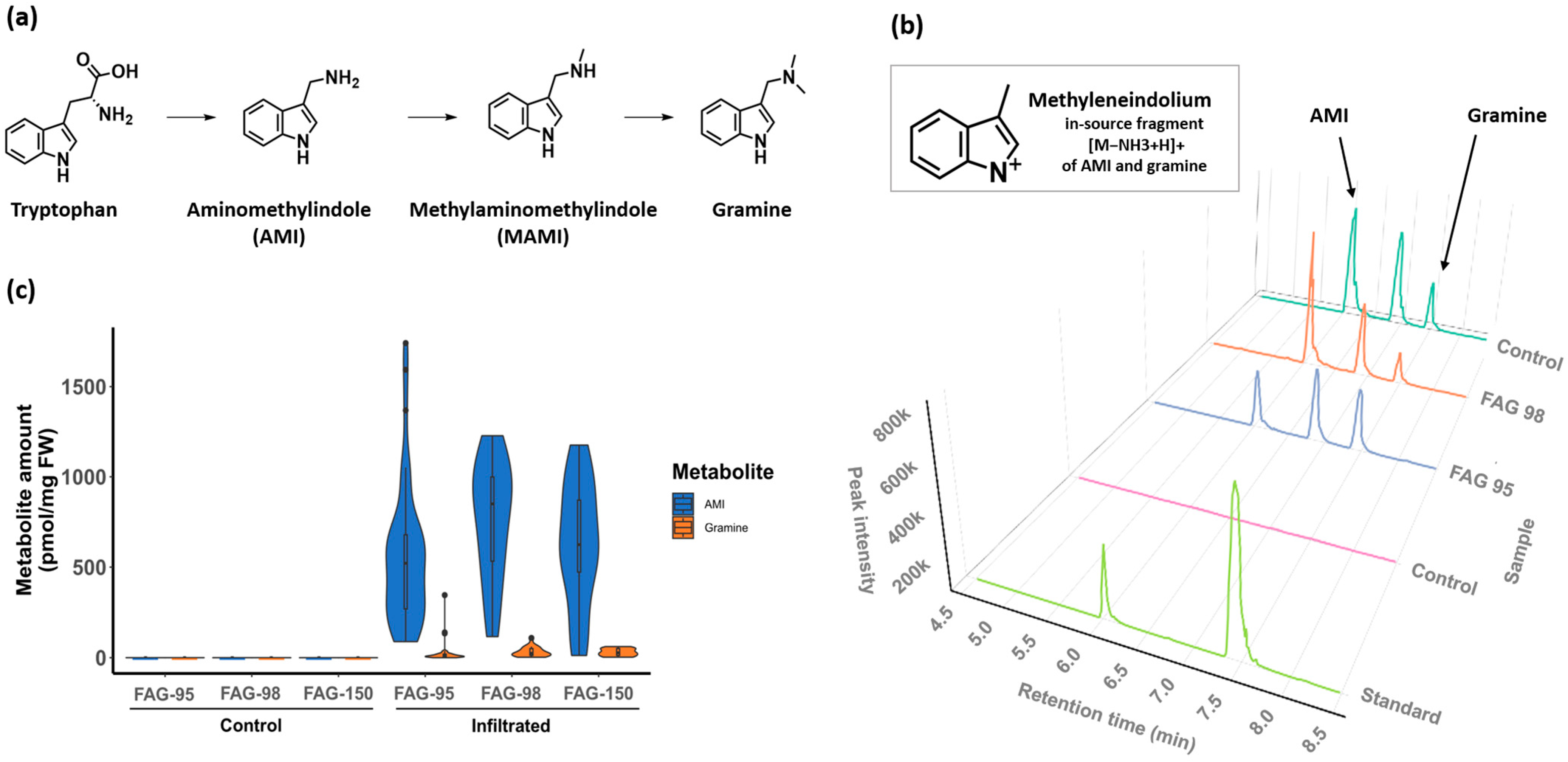
2.2. Assembly of the Gramine Biosynthesis Pathway
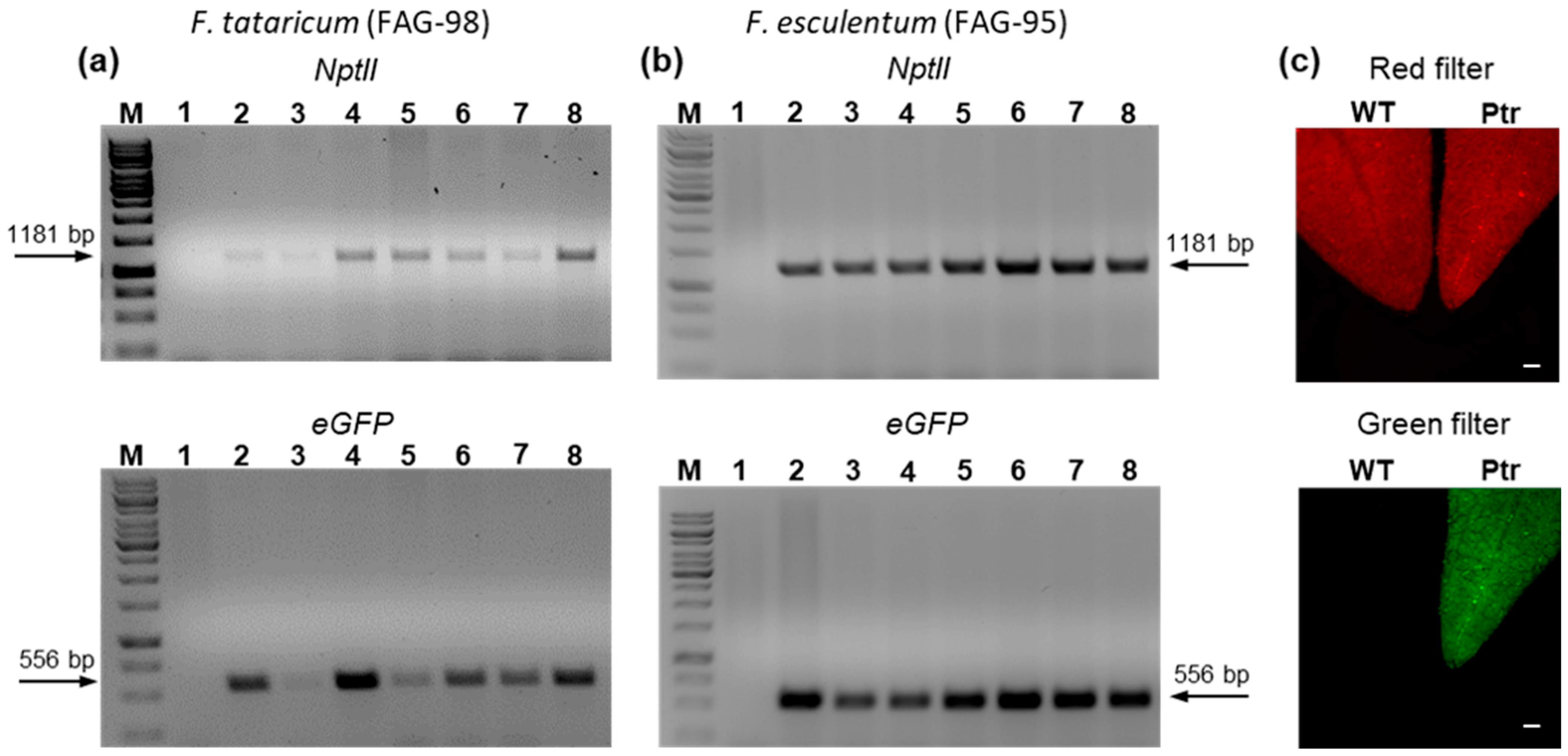
2.3. Stable Genetic Transformation of Common and Tartary Buckwheat
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Plant Material
4.3. GUS Staining
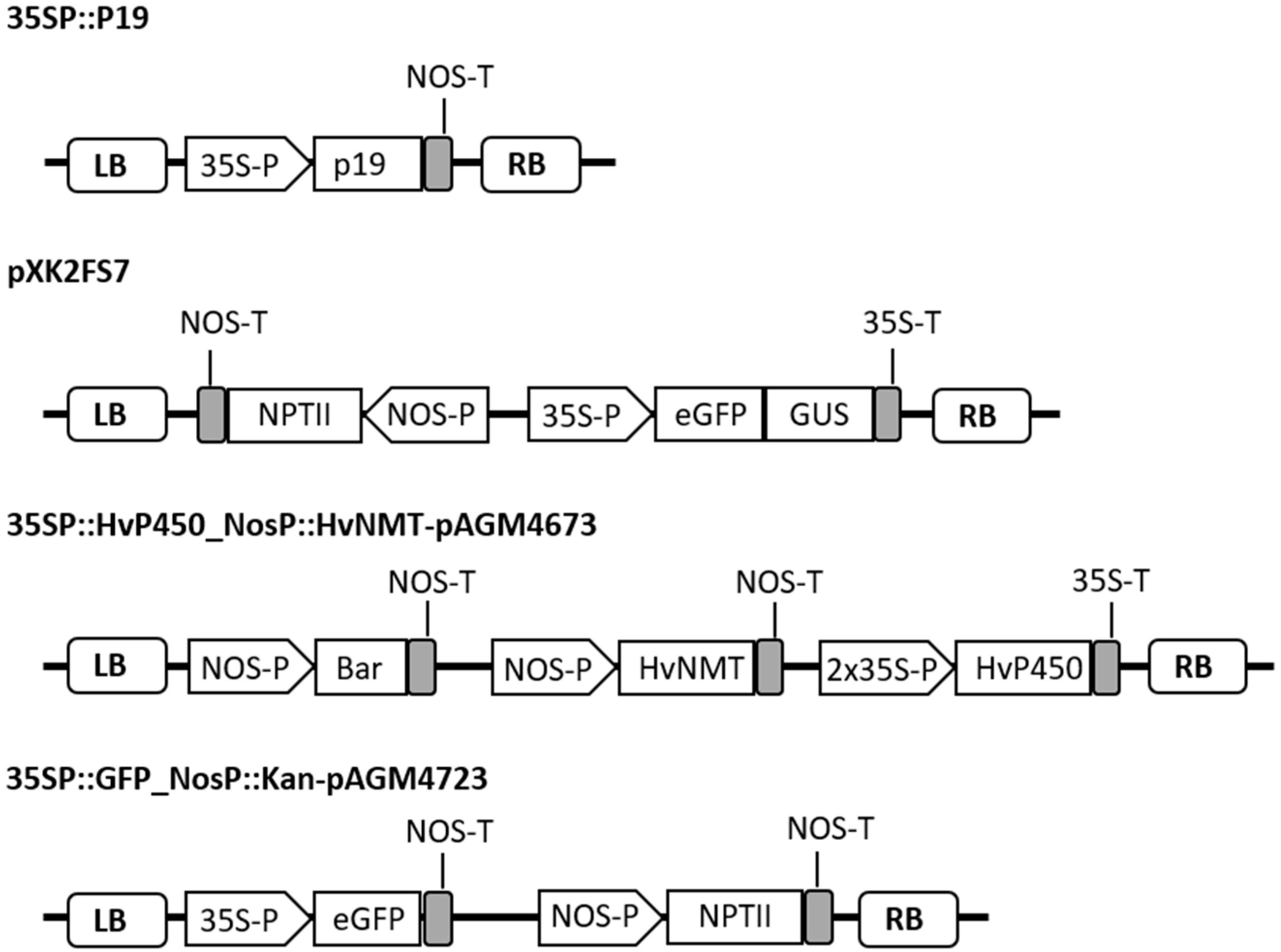
4.4. Plasmids
4.5. Transient Transformation by Agroinfiltration
4.6. Stable Transformation by Vacuum Infiltration
4.7. Selection and PCR Analysis of Stably Transformed Buckwheat Plants
4.8. Detection of GFP by Fluorescence Microscopy
4.9. Protein Extraction and Western Blot Analysis
4.10. Buckwheat Leaf Collection for Chemical Analysis
4.11. Buckwheat Sample Preparation for Chemical Analysis
4.12. Instrumentation and Conditions
4.13. Standards and Calibration Curves
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohsako, T.; Li, C. Classification and systematics of the Fagopyrum species. Breed. Sci. 2020, 70, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bonafaccia, G.; Gambelli, L.; Fabjan, N.; Kreft, I. Trace elements in flour and bran from common and tartary buckwheat. Food Chem. 2003, 83, 1–5. [Google Scholar] [CrossRef]
- Skřivan, P.; Chrpová, D.; Klitschová, B.; Švec, I.; Sluková, M. Buckwheat Flour (Fagopyrum esculentum Moench)—A Contemporary View on the Problems of Its Production for Human Nutrition. Foods 2023, 12, 3055. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Yamaguchi, H.; Degi, K. The Contribution of Polyphenols to Antioxidative Activity in Common Buckwheat and Tartary Buckwheat Grain. Plant Prod. Sci. 2007, 10, 99–104. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Zhou, M.-L.; Tang, Y.; Li, F.-L.; Tang, Y.-X.; Shao, J.-R.; Xue, W.-T.; Wu, Y.-M. Bioactive compounds in functional buckwheat food. Food Res. Int. 2012, 49, 389–395. [Google Scholar] [CrossRef]
- Gharat, S.A.; Tamhane, V.A.; Giri, A.P.; Aharoni, A. Navigating the challenges of engineering composite specialized metabolite pathways in plants. Plant J. 2025, 121, e70100. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Liu, R.; Du, W.; Ma, M.; Han, Y.; Li, H.; Liu, L.; Hou, S. Comparative transcriptomic analysis reveals the regulatory mechanism of the gibberellic acid pathway of Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.) dwarf mutants. BMC Plant Biol. 2021, 21, 206. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Takeshima, R.; Kikuchi, S.; Yazaki, E.; Katsube-Tanaka, T.; Dong, Y.; Li, M.; Hunt, H.V.; Jones, M.K.; Lister, D.L.; et al. Genome sequencing reveals the genetic architecture of heterostyly and domestication history of common buckwheat. Nat. Plants 2023, 9, 1236–1251. [Google Scholar] [CrossRef]
- Guo, H.; Qin, Z.; Ren, W.; Feng, H.; Chen, W.; Liu, L.; Sun, Z. Ethyl Methanesulfonate Mutant Library Construction in Tartary Buckwheat with Agronomic Trait and Flavonoid Screening for Germplasm Innovation. Agronomy 2024, 14, 547. [Google Scholar] [CrossRef]
- Yasui, Y. History of the progressive development of genetic marker systems for common buckwheat. Breed. Sci. 2020, 70, 13–18. [Google Scholar] [CrossRef]
- Yasui, Y.; Hirakawa, H.; Ueno, M.; Matsui, K.; Katsube-Tanaka, T.; Yang, S.J.; Aii, J.; Sato, S.; Mori, M. Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res. 2016, 23, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; He, Y.; Lu, X.; Shi, Y.; Zhao, H.; Li, X.; Li, J.; Liu, Y.; Ouyang, Y.; Tang, Y.; et al. Comparative and population genomics of buckwheat species reveal key determinants of flavor and fertility. Mol. Plant 2023, 16, 1427–1444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Ma, B.; Gao, Q.; Du, H.; Han, Y.; Li, Y.; Cao, Y.; Qi, M.; Zhu, Y.; et al. The Tartary Buckwheat Genome Provides Insights into Rutin Biosynthesis and Abiotic Stress Tolerance. Mol. Plant 2017, 10, 1224–1237. [Google Scholar] [CrossRef]
- Yabe, S.; Iwata, H. Genomics-assisted breeding in minor and pseudo-cereals. Breed. Sci. 2020, 70, 19–31. [Google Scholar] [CrossRef]
- Meyers, B.; Zaltsman, A.; Lacroix, B.; Kozlovsky, S.V.; Krichevsky, A. Nuclear and plastid genetic engineering of plants: Comparison of opportunities and challenges. Biotechnol. Adv. 2010, 28, 747–756. [Google Scholar] [CrossRef]
- Eidenberger, L.; Kogelmann, B.; Steinkellner, H. Plant-based biopharmaceutical engineering. Nat. Rev. Bioeng. 2023, 1, 426–439. [Google Scholar] [CrossRef]
- Peyret, H.; Lomonossoff, G.P. When plant virology met Agrobacterium: The rise of the deconstructed clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef]
- Cardi, T. Cisgenesis and genome editing: Combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 2016, 135, 139–147. [Google Scholar] [CrossRef]
- Manghwar, H.; Lindsey, K.; Zhang, X.; Jin, S. CRISPR/Cas System: Recent Advances and Future Prospects for Genome Editing. Trends Plant Sci. 2019, 24, 1102–1125. [Google Scholar] [CrossRef]
- Platani, M.; Sokefun, O.; Bassil, E.; Apidianakis, Y. Genetic engineering and genome editing in plants, animals and humans: Facts and myths. Gene 2023, 856, 147141. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 2005, 23, 2042–2048. [Google Scholar] [CrossRef] [PubMed]
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminium-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Ma, J.F. Two MATE Transporters with Different Subcellular Localization are Involved in Al Tolerance in Buckwheat. Plant Cell Physiol. 2017, 58, 2179–2189. [Google Scholar] [CrossRef]
- Sakamoto, S.; Matsui, K.; Oshima, Y.; Mitsuda, N. Efficient transient gene expression system using buckwheat hypocotyl protoplasts for large-scale experiments. Breed. Sci. 2020, 70, 128–134. [Google Scholar] [CrossRef]
- Li, X.; Sathasivam, R.; Park, N.I.; Wu, Q.; Park, S.U. Enhancement of phenylpropanoid accumulation in tartary buckwheat hairy roots by overexpression of MYB transcription factors. Ind. Crops Prod. 2020, 156, 112887. [Google Scholar] [CrossRef]
- Lee, S.Y.; Cho, S.I.; Park, M.H.; Kim, Y.K.; Choi, J.E.; Park, S.U. Growth and rutin production in hairy root cultures of buckwheat (Fagopyrum esculentum M.). Prep. Biochem. Biotechnol. 2007, 37, 239–246. [Google Scholar] [CrossRef]
- Kim, Y.K.; Li, X.; Xu, H.; Il Park, N.; Uddin, M.R.; Pyon, J.Y.; Park, S.U. Production of phenolic compounds in hairy root culture of tartary buckwheat (Fagopyrum tataricum Gaertn). J. Crop Sci. Biotechnol. 2009, 12, 53–57. [Google Scholar] [CrossRef]
- Thwe, A.; Valan Arasu, M.; Li, X.; Park, C.H.; Kim, S.J.; Al-Dhabi, N.A.; Park, S.U. Effect of Different Agrobacterium rhizogenes Strains on Hairy Root Induction and Phenylpropanoid Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum Gaertn). Front. Microbiol. 2016, 7, 318. [Google Scholar] [CrossRef]
- Wen, D.; Wu, L.; Wang, M.; Yang, W.; Wang, X.; Ma, W.; Sun, W.; Chen, S.; Xiang, L.; Shi, Y. CRISPR/Cas9-Mediated Targeted Mutagenesis of FtMYB45 Promotes Flavonoid Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum). Front. Plant Sci. 2022, 13, 879390. [Google Scholar] [CrossRef]
- Pinski, A.; Betekhtin, A. Efficient Agrobacterium-mediated transformation and genome editing of Fagopyrum tataricum. Front. Plant Sci. 2023, 14, 1270150. [Google Scholar] [CrossRef] [PubMed]
- Miljuš-Djukić, J.; Nešković, M.; Ninković, S.; Crkvenjakov, R. Agrobacterium-mediated transformation and plant regeneration of buckwheat (Fagopyrum esculentum Moench.). Plant Cell Tissue Organ Cult. 1992, 29, 101–108. [Google Scholar] [CrossRef]
- Kim, H.-S.; Kang, H.-J.; Lee, Y.-T.; Lee, S.-Y.; Ko, J.; Rha, E.-S. Direct regeneration of transgenic buckwheat from hypocotyl segment by agrobacterium-mediated transformation. Korean J. Crop Sci. 2001, 46, 375–379. [Google Scholar]
- Chen, L.H.; Zhang, B.; Xu, Z.Q. Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res. 2008, 17, 121–132. [Google Scholar] [CrossRef]
- Bélanger, J.G.; Copley, T.R.; Hoyos-Villegas, V.; Charron, J.B.; O’Donoughue, L. A comprehensive review of in planta stable transformation strategies. Plant Methods 2024, 20, 79. [Google Scholar] [CrossRef]
- Sebiani-Calvo, A.; Hernández-Soto, A.; Hensel, G.; Gatica-Arias, A. Crop genome editing through tissue-culture-independent transformation methods. Front. Genome Ed. 2024, 6, 1490295. [Google Scholar] [CrossRef]
- Bechtold, N.; Bouchez, D. In Planta Agrobacterium-Mediated Transformation of Adult Arabidopsis thaliana Plants by Vacuum Infiltration. In Gene Transfer to Plants; Potrykus, I., Spangenberg, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1995; pp. 19–23. [Google Scholar]
- Bratic, A.; Majić, D.B.; Miljuš-Djukić, J.; Jovanović, Ž.; Maksimovic, V. In planta transformation of buckwheat (Fagopyrum esculentum Moench.). Arch. Biol. Sci. 2007, 59, 135–138. [Google Scholar] [CrossRef]
- Kojima, M.; Arai, Y.; Iwase, N.; Shirotori, K.; Shioiri, H.; Nozue, M. Development of a simple and efficient method for transformation of buckwheat plants (Fagopyrum esculentum) using Agrobacterium tumefaciens. Biosci. Biotechnol. Biochem. 2000, 64, 845–847. [Google Scholar] [CrossRef]
- Kojima, M.; Hihara, M.; Koyama, S.-i.; Shioiri, H.; Nozue, M.; Yamamoto, K.; Sasaki, T. Buckwheat transformed with cDNA of a rice MADS box gene is stimulated in branching. Plant Biotechnol. 2000, 17, 35–42. [Google Scholar] [CrossRef]
- Zhou, B.; Ding, F.; Yang, Z.; Song, Z.; Sun, J.; Wang, S.; Wang, X.; Liu, Z.-x.; Fang, Z.; Zhang, Y. Establishing a highly efficient Agrobacterium-mediated transformation system in sweet buckwheat. J. Consum. Prot. Food Saf. 2023, 18, 433–441. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, H.; Sun, L.; Wu, W.; Li, C.; Wu, Q. Genome-wide identification of MAPK gene family members in Fagopyrum tataricum and their expression during development and stress responses. BMC Genom. 2022, 23, 96. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, H.; Lin, J.; Ouyang, Y.; Li, C.; Li, H.; Wang, T.; Wu, Q.; Zhao, H. Establishment and application of novel transient cotyledon and seed transformation systems in Tartary buckwheat. Plant Cell Tissue Organ Cult. 2025, 160, 52. [Google Scholar] [CrossRef]
- Bally, J.; Jung, H.; Mortimer, C.; Naim, F.; Philips, J.G.; Hellens, R.; Bombarely, A.; Goodin, M.M.; Waterhouse, P.M. The Rise and Rise of Nicotiana benthamiana: A Plant for All Reasons. Annu. Rev. Phytopathol. 2018, 56, 405–426. [Google Scholar] [CrossRef]
- Kopertekh, L. Improving transient expression in N. benthamiana by suppression of the Nb-SABP2 and Nb-COI1 plant defence response related genes. Front Plant Sci. 2024, 15, 1453930. [Google Scholar] [CrossRef]
- Beritza, K.; Watts, E.C.; van der Hoorn, R.A.L. Improving transient protein expression in agroinfiltrated Nicotiana benthamiana. New Phytol. 2024, 243, 846–850. [Google Scholar] [CrossRef]
- Golubova, D.; Tansley, C.; Su, H.; Patron, N.J. Engineering Nicotiana benthamiana as a platform for natural product biosynthesis. Curr. Opin. Plant Biol. 2024, 81, 102611. [Google Scholar] [CrossRef]
- Leite Dias, S.; Chuang, L.; Liu, S.; Seligmann, B.; Brendel, F.L.; Chavez, B.G.; Hoffie, R.E.; Hoffie, I.; Kumlehn, J.; Bültemeier, A.; et al. Biosynthesis of the allelopathic alkaloid gramine in barley by a cryptic oxidative rearrangement. Science 2024, 383, 1448–1454. [Google Scholar] [CrossRef]
- Venkataraman, S.; Khan, I.; Habibi, P.; Le, M.; Lippert, R.; Hefferon, K. Recent advances in expression and purification strategies for plant made vaccines. Front. Plant Sci. 2023, 14, 1273958. [Google Scholar] [CrossRef]
- van Herpen, T.W.; Cankar, K.; Nogueira, M.; Bosch, D.; Bouwmeester, H.J.; Beekwilder, J. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE 2010, 5, e14222. [Google Scholar] [CrossRef]
- Shamloul, M.; Trusa, J.; Mett, V.; Yusibov, V. Optimization and utilization of Agrobacterium-mediated transient protein production in Nicotiana. J. Vis. Exp. 2014, 51204. [Google Scholar] [CrossRef]
- Drapal, M.; Enfissi, E.M.A.; Fraser, P.D. Metabolic effects of agro-infiltration on N. benthamiana accessions. Transgenic Res. 2021, 30, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.; Gkotsi, D.S.; Smith, D.R.M.; Goss, R.J.M.; Caputi, L.; O’Connor, S.E. Nicotiana benthamiana as a Transient Expression Host to Produce Auxin Analogs. Front. Plant Sci. 2020, 11, 581675. [Google Scholar] [CrossRef] [PubMed]
- Liu, C. Reconstitution of Metabolic Pathway in Nicotiana benthamiana. In Plant Metabolic Engineering: Methods and Protocols; Shulaev, V., Ed.; Springer: New York, NY, USA, 2022; pp. 29–33. [Google Scholar]
- Crocoll, C.; Mirza, N.; Reichelt, M.; Gershenzon, J.; Halkier, B.A. Optimization of Engineered Production of the Glucoraphanin Precursor Dihomomethionine in Nicotiana benthamiana. Front. Bioeng. Biotechnol. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; van der Krol, S.; Lugan, R.; Ilc, T.; et al. Correction: Corrigendum: The seco-iridoid pathway from Catharanthus roseus. Nature Comm. 2014, 5, 4175. [Google Scholar] [CrossRef]
- Boccia, M.; Grzech, D.; Lopes, A.A.; O’Connor, S.E.; Caputi, L. Directed Biosynthesis of New to Nature Alkaloids in a Heterologous Nicotiana benthamiana Expression Host. Front. Plant Sci. 2022, 13, 919443. [Google Scholar] [CrossRef]
- Polturak, G.; Breitel, D.; Grossman, N.; Sarrion-Perdigones, A.; Weithorn, E.; Pliner, M.; Orzaez, D.; Granell, A.; Rogachev, I.; Aharoni, A. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 2016, 210, 269–283. [Google Scholar] [CrossRef]
- Rajniak, J.; Barco, B.; Clay, N.K.; Sattely, E.S. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 2015, 525, 376–379. [Google Scholar] [CrossRef]
- Reed, J.; Osbourn, A. Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep. 2018, 37, 1431–1441. [Google Scholar] [CrossRef]
- Stephenson, M.J.; Reed, J.; Brouwer, B.; Osbourn, A. Transient Expression in Nicotiana benthamiana Leaves for Triterpene Production at a Preparative Scale. J. Vis. Exp. 2018, 138, e58169. [Google Scholar]
- Sanders, P.R.; Winter, J.A.; Barnason, A.R.; Rogers, S.G.; Fraley, R.T. Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants. Nucleic Acids Res. 1987, 15, 1543–1558. [Google Scholar] [CrossRef]
- Matsuura, H.N.; Malik, S.; de Costa, F.; Yousefzadi, M.; Mirjalili, M.H.; Arroo, R.; Bhambra, A.S.; Strnad, M.; Bonfill, M.; Fett-Neto, A.G. Specialized Plant Metabolism Characteristics and Impact on Target Molecule Biotechnological Production. Mol. Biotechnol. 2018, 60, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Martin, C.; Zhang, Y. Next-Generation Plant Metabolic Engineering, Inspired by an Ancient Chinese Irrigation System. Mol. Plant 2018, 11, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Anjanappa, R.B.; Gruissem, W. Current progress and challenges in crop genetic transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef]
- Us Camas, R.; Rivera-Solís, G.; Duarte, F.; De la Peña, C. In vitro culture: An epigenetic challenge for plants. Plant Cell Tissue Organ Cult. 2014, 118, 187–201. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Karimi, M.; Depicker, A.; Hilson, P. Recombinational cloning with plant gateway vectors. Plant Physiol. 2007, 145, 1144–1154. [Google Scholar] [CrossRef]
- Angel, C.A.; Schoelz, J.E. A survey of resistance to Tomato bushy stunt virus in the genus Nicotiana reveals that the hypersensitive response is triggered by one of three different viral proteins. Mol. Plant Microbe Interact. 2013, 26, 240–248. [Google Scholar] [CrossRef]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.M.; Werner, S.; Jones, J.D.; Patron, N.J.; Marillonnet, S. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Leite Dias, S.; Garibay-Hernández, A.; Brendel, F.L.; Gabriel Chavez, B.; Brückner, E.; Mock, H.-P.; Franke, J.; D’Auria, J.C. A New Fluorescence Detection Method for Tryptophan- and Tyrosine-Derived Allelopathic Compounds in Barley and Lupin. Plants 2023, 12, 1930. [Google Scholar] [CrossRef] [PubMed]
- Vollheyde, K.; Dudley, Q.M.; Yang, T.; Oz, M.T.; Mancinotti, D.; Fedi, M.O.; Heavens, D.; Linsmith, G.; Chhetry, M.; Smedley, M.A.; et al. An improved Nicotiana benthamiana bioproduction chassis provides novel insights into nicotine biosynthesis. New Phytol. 2023, 240, 302–317. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leite Dias, S.; Rizzo, P.; D’Auria, J.C.; Kochevenko, A. Efficient Agrobacterium-Mediated Methods for Transient and Stable Transformation in Common and Tartary Buckwheat. Int. J. Mol. Sci. 2025, 26, 4425. https://doi.org/10.3390/ijms26094425
Leite Dias S, Rizzo P, D’Auria JC, Kochevenko A. Efficient Agrobacterium-Mediated Methods for Transient and Stable Transformation in Common and Tartary Buckwheat. International Journal of Molecular Sciences. 2025; 26(9):4425. https://doi.org/10.3390/ijms26094425
Chicago/Turabian StyleLeite Dias, Sara, Paride Rizzo, John Charles D’Auria, and Andriy Kochevenko. 2025. "Efficient Agrobacterium-Mediated Methods for Transient and Stable Transformation in Common and Tartary Buckwheat" International Journal of Molecular Sciences 26, no. 9: 4425. https://doi.org/10.3390/ijms26094425
APA StyleLeite Dias, S., Rizzo, P., D’Auria, J. C., & Kochevenko, A. (2025). Efficient Agrobacterium-Mediated Methods for Transient and Stable Transformation in Common and Tartary Buckwheat. International Journal of Molecular Sciences, 26(9), 4425. https://doi.org/10.3390/ijms26094425







