Genome-Wide Identification and Functional Analysis of the Norcoclaurine Synthase Gene Family in Aristolochia contorta
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of 15 AcNCS Genes in A. contorta
2.2. Phylogenetic Relationships of AcNCS Proteins
2.3. Conserved Domain and Motifs of AcNCS Proteins
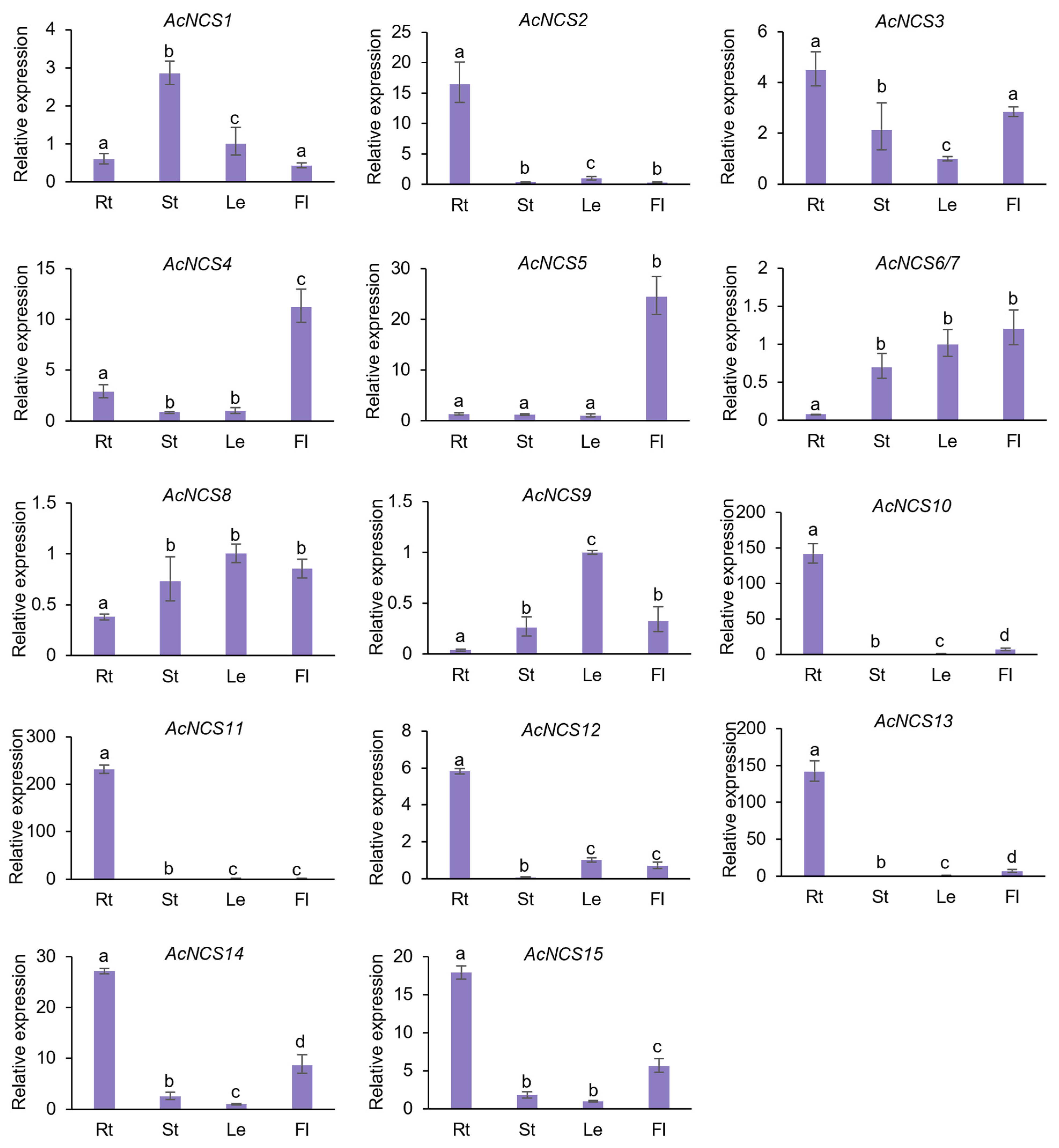
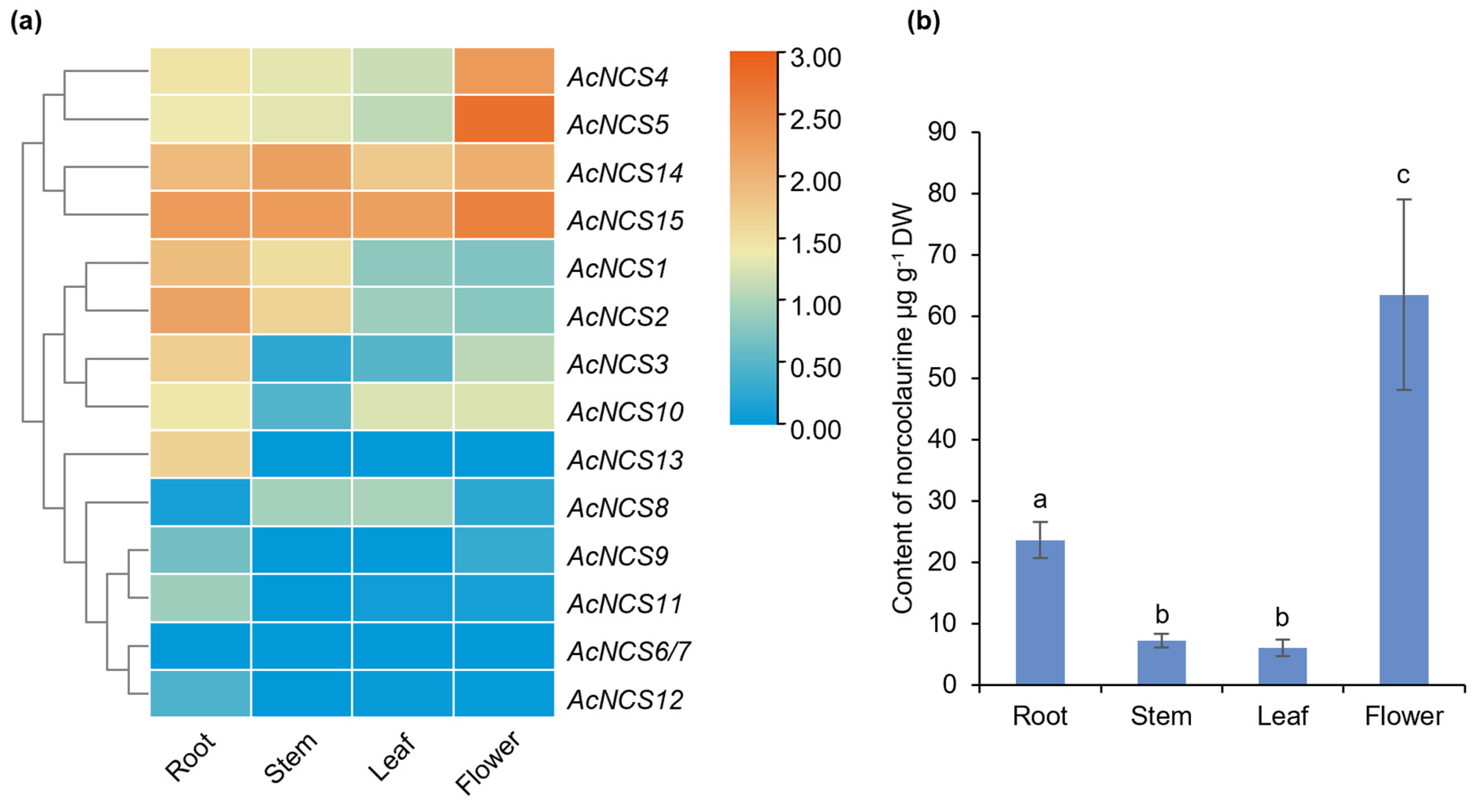
2.4. Differential Expression of AcNCS Genes in A. contorta
2.5. Accumulation of Norcoclaurine in Roots and Flowers of A. contorta
2.6. AcNCS Proteins Catalyze the Formation of Norcoclaurine
2.7. Affinity of AcNCSs Toward Dopamine and 4-HPPA
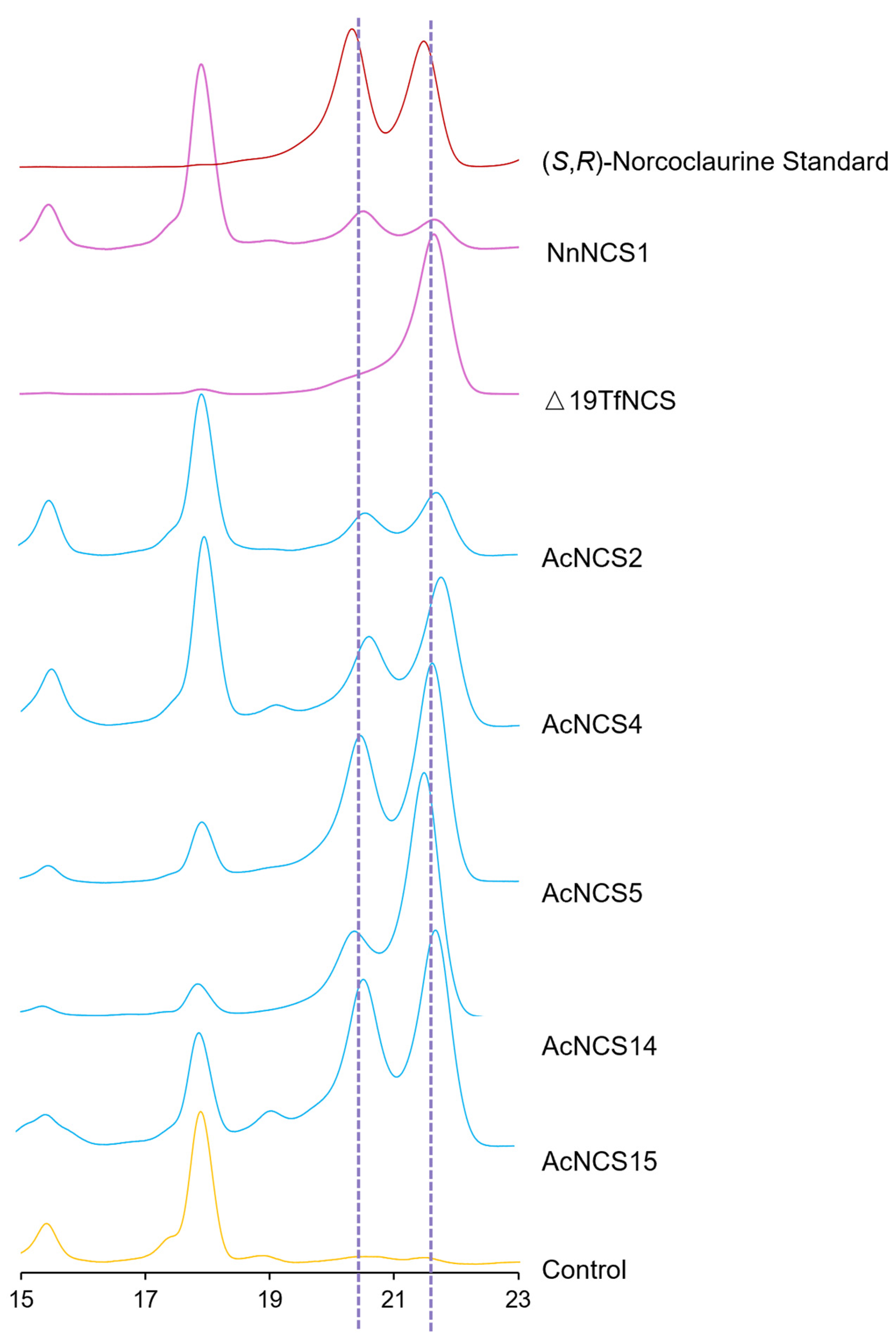
2.8. Existence of (S)- and (R)-Norcoclaurine in AcNCS-Catalyzed Products
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Chemicals
4.2. AcNCS Gene Identification
4.3. Analyses of Sequence Features, Chromosomal Locations, and Gene Structures
4.4. Phylogenetic Tree Construction
4.5. Quantitative Real-Time Reverse Transcription-PCR (qRT-PCR)
4.6. Norcoclaurine Content Determination
4.7. AcNCS Recombinant Protein Expression and Purification
4.8. In Vitro Enzymatic Activity Assay of AcNCS Recombinant Proteins
4.9. Kinetic Analysis of AcNCS Recombinant Proteins
4.10. Chiral Analysis of the Configuration of Products
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuo, P.C.; Li, Y.C.; Wu, T.S. Chemical constituents and pharmacology of the Aristolochia species. J. Tradit. Complement. Med. 2012, 2, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Chawla, P.; Chawla, A.; Shah, G.; Baghel, U.S.; Dhawan, R.K. A review on pharmacognosy and biological activities of Aristolochia. Asian J. Res. Biol. Pharm. Sci. 2013, 1, 101–110. [Google Scholar]
- Comission, C.P. Pharmacopoeia of the People Republic of China; China Medical Science Press: Beijing, China, 2015; Volume 1, pp. 51–52. [Google Scholar]
- Zhang, C.Y.; Yu, J.; Wang, B. Analysis of volatile oil constituents in the fruits of Aristolochia contorta by GC-MS. Chin. J. Nat. Med. 2004, 2, 126–128. [Google Scholar]
- Yuan, J.; Nie, L.; Zeng, D.; Luo, X.; Tang, F.; Ding, L.; Liu, Q.; Guo, M.; Yao, S. Simultaneous determination of nine aristolochic acid analogues in medicinal plants and preparations by high-performance liquid chromatography. Talanta 2007, 73, 644–650. [Google Scholar] [CrossRef]
- Tian, H.M.; Cheng, X.M.; Wang, C.H.; Wang, Z.T. Simultaneous determination of four constituents in Aristolochia debilis by HPLC. Chin. Tradit. Pat. Med. 2016, 38, 560–565. [Google Scholar]
- Wu, P.L.; Su, G.C.; Wu, T.S. Constituents from the stems of Aristolochia manshuriensis. J. Nat. Prod. 2003, 66, 996–998. [Google Scholar] [CrossRef]
- Shi, L.S.; Kuo, P.C.; Tsai, Y.L.; Damu, A.G.; Wu, T.S. The alkaloids and other constituents from the root and stem of Aristolochia elegans. Bioorg. Med. Chem. 2004, 12, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.L.R.; de Pascoli, I.C.; Nascimento, I.R.; Zukerman- Schpector, J.; Lopes, L.M.X. Aporphine and bisaporphine alkaloids from Aristolochia lagesiana var. intermedia. Phytochemistry 2010, 71, 469–478. [Google Scholar] [CrossRef]
- Mao, W.W.; Gao, W.; Liang, Z.T.; Li, P.; Zhao, Z.Z.; Li, H.J. Characterization and quantitation of aristolochic acid analogs in different parts of Aristolochiae fructus using UHPLC-Q/TOF-MS and UHPLC-QqQ-MS. Chin. J. Nat. Med. 2017, 15, 392–400. [Google Scholar] [CrossRef]
- Cui, X.; Meng, F.; Pan, X.; Qiu, X.; Zhang, S.; Li, C.; Lu, S. Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Hortic. Res. 2022, 9, uhac005. [Google Scholar] [CrossRef]
- Meng, F.; Zhang, S.; Su, J.; Zhu, B.; Pan, X.; Qiu, X.; Cui, X.; Wang, C.; Niu, L.; Li, C.; et al. Characterization of two CYP80 enzymes provides insights into aporphine alkaloid skeleton formation in Aristolochia contorta. Plant J. 2024, 118, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, P.; Wei, F.; Lin, R.; Khan, I.A.; Pasco, D.S. Structure activity relationships of aristolochic acid analogues: Toxicity in cultured renal epithelial cells. Kidney Int. 2005, 67, 1797–1805. [Google Scholar] [CrossRef]
- Grosse, Y.; Baan, R.; Straif, K.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Benbrahim-Tallaa, L.; Guha, N.; Galichet, L.; Cogliano, V.; et al. A review of human carcinogens-Part A: Pharmaceuticals. Lancet Oncol. 2009, 10, 13–14. [Google Scholar] [CrossRef] [PubMed]
- Stiborová, M.; Arlt, V.M.; Schmeiser, H.H. DNA adducts formed by aristolochic acid are unique biomarkers of exposure and explain the initiation phase of upper urothelial cancer. Int. J. Mol. Sci. 2017, 18, 2144. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, C.; Xue, R.; Liu, M.; Bai, J.; Bao, J.; Wang, Y.; Jiang, N.; Li, Z.; Wang, W.; et al. Deep whole-genome analysis of 494 hepatocellular carcinomas. Nature 2024, 627, 586–593. [Google Scholar] [CrossRef]
- Sahib, S.; Yan, J.; Chen, T. Application of duplex sequencing to evaluate mutagenicity of aristolochic acid and methapyrilene in Fisher 344 rats. Food Chem. Toxicol. 2024, 185, 114512. [Google Scholar] [CrossRef]
- Senkin, S.; Moody, S.; Díaz-Gay, M.; Abedi-Ardekani, B.; Cattiaux, T.; Ferreiro-Iglesias, A.; Wang, J.; Fitzgerald, S.; Kazachkova, M.; Vangara, R.; et al. Geographic variation of mutagenic exposures in kidney cancer genomes. Nature 2024, 629, 910–918. [Google Scholar] [CrossRef]
- Liang, W.; Xu, Y.; Cui, X.; Li, C.; Lu, S. Genome-wide identification and characterization of miRNAs and natural antisense transcripts show the complexity of gene regulatory networks for secondary metabolism in Aristolochia contorta. Int. J. Mol. Sci. 2024, 25, 6043. [Google Scholar] [CrossRef]
- Samanani, N.; Alcantara, J.; Bourgault, R.; Zulak, K.G.; Facchini, P.J. The role of phloem sieve elements and laticifers in the biosynthesis and accumulation of alkaloids in opium poppy. Plant J. 2006, 47, 547–563. [Google Scholar] [CrossRef]
- Dastmalchi, M.; Park, M.R.; Morris, J.S.; Facchini, P. Family portraits: The enzymes behind benzylisoquinoline alkaloid diversity. Phytochem. Rev. 2018, 17, 249–277. [Google Scholar] [CrossRef]
- Lee, E.J.; Facchini, P. Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell 2010, 22, 3489–3503. [Google Scholar] [CrossRef] [PubMed]
- Berkner, H.; Seutter von Loetzen, C.; Hartl, M.; Randow, S.; Gubesch, M.; Vogel, L.; Husslik, F.; Reuter, A.; Lidholm, J.; Ballmer-Weber, B.; et al. Enlarging the toolbox for allergen epitope definition with an allergen-type model protein. PLoS ONE 2014, 9, e11169. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Q.; Jiao, X.; Tang, H.; Cheng, Y.; Ma, Y.; Cui, G.; Tang, J.; Chen, Y.; Guo, J.; et al. Phylogenetic analysis and functional characterization of norcoclaurine synthase involved in benzylisoquinoline alkaloids biosynthesis in Stephania tetrandra. J. Cell. Physiol. 2024, 239, e31065. [Google Scholar] [CrossRef]
- Samanani, N.; Facchini, P.J. Purification and characterization of norcoclaurine synthase. J. Biol. Chem. 2002, 277, 33878–33883. [Google Scholar] [CrossRef]
- Samanani, N.; Liscombe, D.K.; Facchini, P.J. Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyz ing the first committed step in benzylisoquinoline alkaloid bio synthesis. Plant J. 2004, 40, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Liscombe, D.K.; MacLeod, B.P.; Loukanina, N.; Nandi, O.I.; Facchini, P.J. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 2005, 66, 2501–2520. [Google Scholar] [CrossRef]
- Minami, H.; Dubouzet, E.; Iwasa, K.; Sato, F. Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 2007, 282, 6274–6282. [Google Scholar] [CrossRef]
- Pesnot, T.; Gershater, M.C.; Ward, J.M.; Hailes, H.C. The catalytic potential of Coptis japonica NCS2 revealed-development and utilisation of a fluorescamine-based assay. Adv. Synth. Catal. 2012, 354, 2997–3008. [Google Scholar] [CrossRef]
- Marques, J.V.; Dalisay, D.S.; Yang, H.; Lee, C.; Davin, L.B.; Lewis, N.G. A multi-omics strategy resolves the elusive nature of alkaloids in Podophyllum species. Mol. Biosyst. 2014, 10, 2838–2849. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Deng, X.; Owiti, A.; Meelaph, T.; Ogutu, C.; Han, Y. Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants. Sci. Rep. 2016, 6, 26323. [Google Scholar] [CrossRef]
- Lechner, H.; Soriano, P.; Poschner, R.; Hailes, H.C.; Ward, J.M.; Kroutil, W. Library of norcoclaurine synthases and their immobilization for biocatalytic transformations. Biotechnol. J. 2018, 13, 1700542. [Google Scholar]
- Liu, T.; Zhang, W.; Wang, S.; Tian, Y.; Wang, Y.; Gao, R.; Chen, S.; Sun, W.; Ma, W.; Xu, Z. Metabolome and transcriptome association study reveals biosynthesis of specialized benzylisoquinoline alkaloids in Phellodendron amurense. Chin. Herb. Med. 2024, 17, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.X.; Pan, Y.; Wang, Y.; Cui, Y.Z.; Zhang, Y.J.; Mo, R.Y.; Wu, X.L.; Tan, J.; Zhang, J.; Guo, L.A.; et al. The chromosome-level reference genome of Coptis chinensis provides insights into genomic evolution and berberine biosynthesis. Hortic. Res. 2021, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Mahdi, J.G.; Bowen, I.D. Pharmacological importance of stereochemical resolution of enantiomeric drugs. Drug Saf. 1997, 17, 149–165. [Google Scholar] [CrossRef]
- Wang, Y.; Ngo, V.A.; Wang, X. Stereoselective recognition of morphine enantiomers by μ-opioid receptor. Natl. Sci. Rev. 2024, 11, nwae029. [Google Scholar] [CrossRef] [PubMed]
- Winzer, T.; Kern, M.; King, A.J.; Larson, T.R.; Teodor, R.I.; Donninger, S.L.; Li, Y.; Dowle, A.A.; Cartwright, J.; Bates, R.; et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 2015, 349, 309–312. [Google Scholar] [CrossRef]
- Farrow, S.C.; Hagel, J.M.; Beaudoin, G.A.W.; Burns, D.C.; Facchini, P.J. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 2015, 11, 728–732. [Google Scholar] [CrossRef]
- Guo, L.; Winzer, T.; Yang, X.; Li, Y.; Ning, Z.; He, Z.; Teodor, R.; Lu, Y.; Bowser, T.A.; Graham, I.A.; et al. The opium poppy genome and morphinan production. Science 2018, 362, 343–347. [Google Scholar] [CrossRef]
- Catania, T.; Li, Y.; Winzer, T.; Harvey, D.; Meade, F.; Caridi, A.; Leech, A.; Larson, T.R.; Ning, Z.; Chang, J.; et al. A functionally conserved STORR gene fusion in Papaver species that diverged 16.8 million years ago. Nat. Commun. 2022, 13, 3150. [Google Scholar] [CrossRef]
- Diaz-Bárcena, A.; Fernandez-Pacios, L.; Giraldo, P. Structural characterization and molecular dynamics study of the REPI fusion protein from Papaver somniferum L. Biomolecules 2023, 14, 2. [Google Scholar] [CrossRef]
- Luk, L.Y.; Bunn, S.; Liscombe, D.K.; Facchini, P.J.; Tanner, M.E. Mechanistic studies on norcoclaurine synthase of benzylisoquinoline alkaloid biosynthesis: An enzymatic Pictet-Spengler reaction. Biochemistry 2007, 46, 10153–10161. [Google Scholar] [CrossRef]
- Leng, L.; Xu, Z.; Hong, B.; Zhao, B.; Tian, Y.; Wang, C.; Yang, L.; Zou, Z.; Li, L.; Liu, K.; et al. Cepharanthine analogs mining and genomes of Stephania accelerate anti-coronavirus drug discovery. Nat. Commun. 2024, 15, 1537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Liao, L.; Tang, H.; Wang, W.; Yin, F.; Han, L.; Zhu, K.; Liu, Y.; Xu, D.; et al. Discovery, structure, and mechanism of the (R, S)-norcoclaurine synthase for the chiral synthesis of benzylisoquinoline alkaloids. ACS Catal. 2023, 13, 15164–15174. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ilari, A.; Franceschini, S.; Bonamore, A.; Arenghi, F.; Botta, B.; Macone, A.; Pasquo, A.; Bellucci, L.; Boffi, A. Structural basis of enzymatic (S)-norcoclaurine biosynthesis. J. Biol. Chem. 2009, 284, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Hu, Y.; Wang, J.; Wang, X.; Zhao, R.; Shan, H.; Li, K.; Xu, P.; Wu, H.; Yan, X.; et al. Insights into angiosperm evolution, floral development and chemical biosynthesis from the Aristolochia fimbriata genome. Nat. Plants 2021, 7, 1239–1253. [Google Scholar] [CrossRef]
- Li, J.; Lee, E.J.; Chang, L.; Facchini, P.J. Genes encoding norcoclaurine synthase occur as tandem fusions in the Papaveraceae. Sci. Rep. 2016, 6, 39256. [Google Scholar] [CrossRef] [PubMed]
- Michl, J.; Kite, G.C.; Wanke, S.; Zierau, O.; Vollmer, G.; Neinhuis, C.; Simmonds, M.S.; Heinrich, M. LC-MS- and 1H NMR-based metabolomic analysis and in vitro toxicological assessment of 43 Aristolochia species. J. Nat. Prod. 2016, 79, 30–37. [Google Scholar] [CrossRef]
- Pesnot, T.; Gershater, M.C.; Ward, J.M.; Hailes, H.C. Phosphate mediated biomimetic synthesis of tetrahydroisoquinoline alkaloids. Chem. Commun. 2011, 47, 3242–3244. [Google Scholar] [CrossRef]
- Ikezawa, N.; Iwasa, K.; Sato, F. Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 2008, 283, 8810–8821. [Google Scholar] [CrossRef]
- Menéndez-Perdomo, I.M.; Facchini, P.J. Elucidation of the (R)-enantiospecific benzylisoquinoline alkaloid biosynthetic pathways in sacred lotus (Nelumbo nucifera). Sci. Rep. 2023, 13, 2955. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
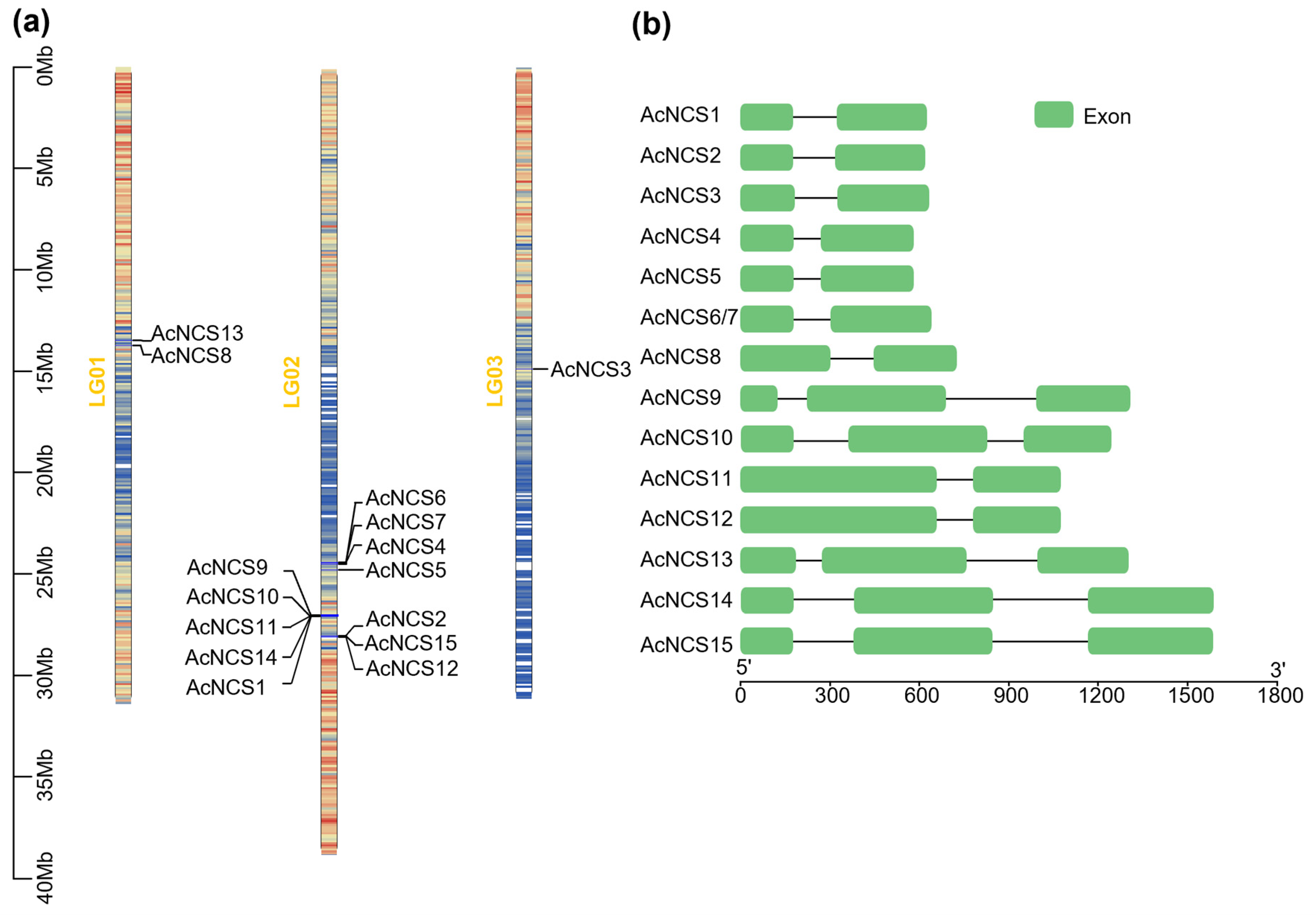
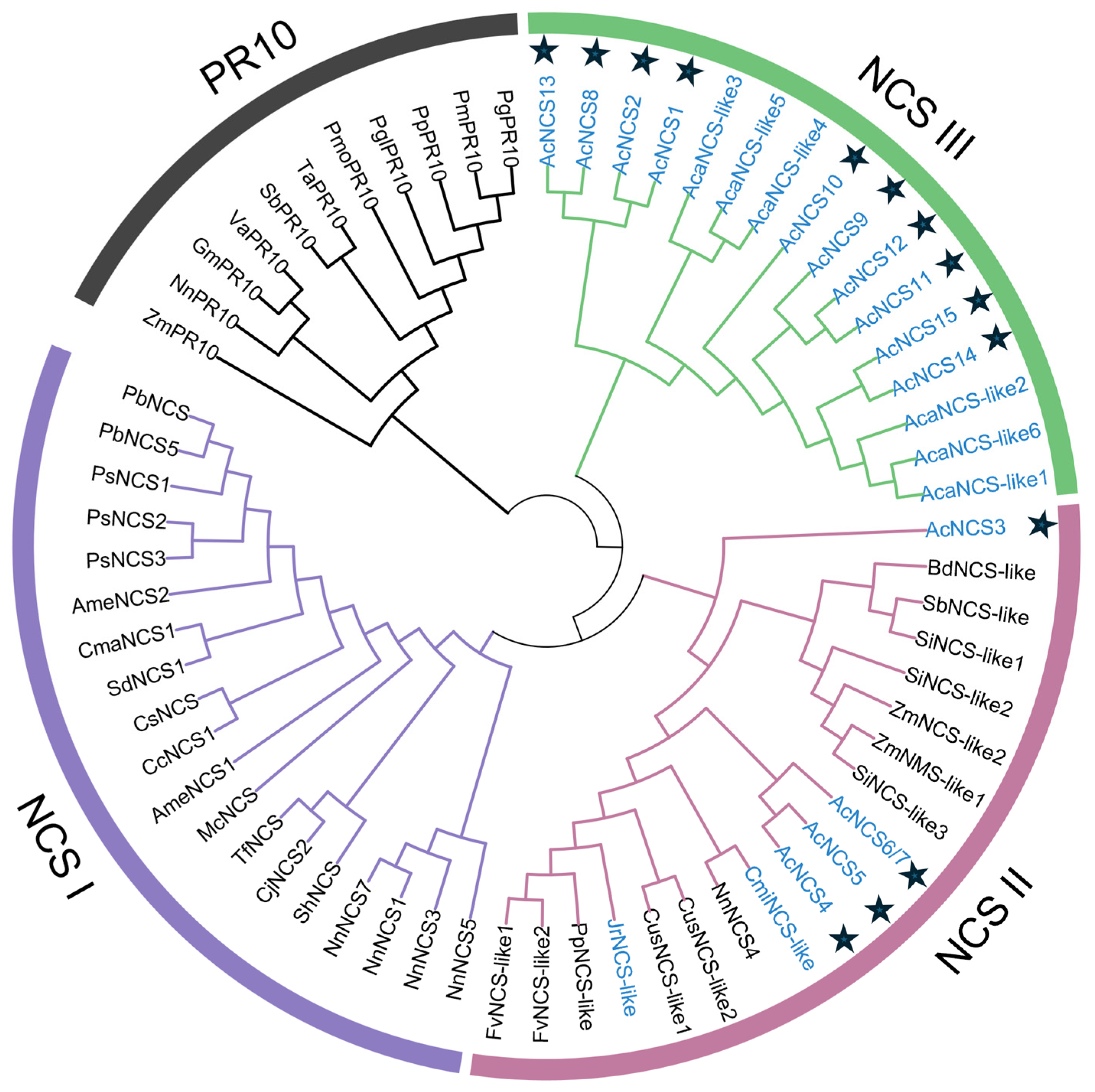
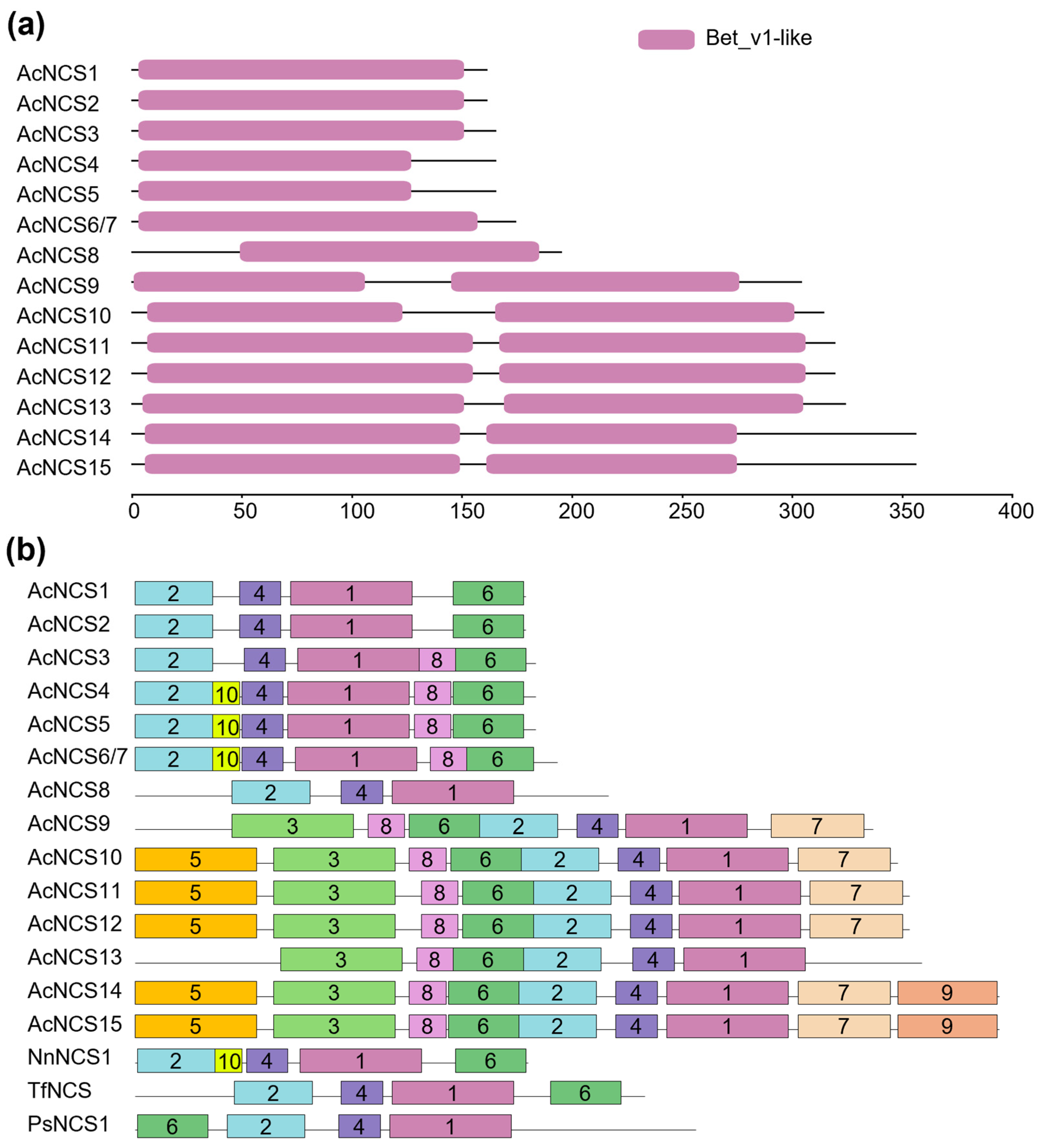
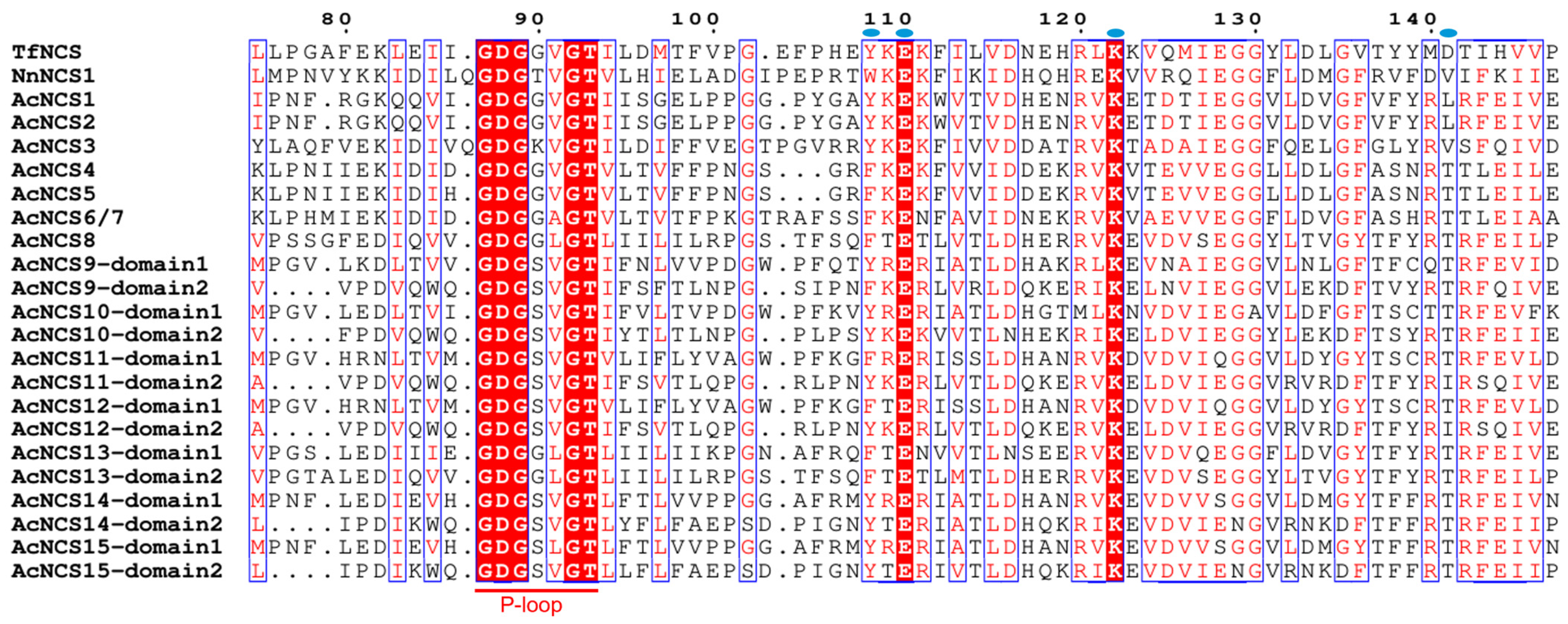

| Gene Name | Sequence ID | Number of Amino Acids | Molecular Weight (kDa) | Theoretical Isoelectric Point (pI) | Number of Introns | Chromosome Localization |
|---|---|---|---|---|---|---|
| AcNCS1 | EVM0011227.1 | 160 | 17.6 | 5.12 | 1 | LG02 |
| AcNCS2 | EVM0014712.1 | 160 | 17.6 | 4.99 | 1 | LG02 |
| AcNCS3 | EVM0011942.1 | 164 | 18.1 | 5.05 | 1 | LG03 |
| AcNCS4 | EVM0001864.1 | 164 | 17.9 | 4.91 | 1 | LG02 |
| AcNCS5 | EVM0010147.1 | 164 | 17.9 | 5.01 | 1 | LG02 |
| AcNCS6 | EVM0012460.1 | 173 | 18.6 | 5.05 | 1 | LG02 |
| AcNCS7 | EVM0000189.1 | 173 | 18.6 | 5.05 | 1 | LG02 |
| AcNCS8 | EVM0011298.1 | 194 | 21.3 | 5.24 | 1 | LG01 |
| AcNCS9 | EVM0016647.1 | 303 | 33.8 | 5.69 | 2 | LG02 |
| AcNCS10 | EVM0005050.1 | 313 | 34.6 | 5.00 | 2 | LG02 |
| AcNCS11 | EVM0007329.1 | 318 | 35.5 | 6.15 | 1 | LG02 |
| AcNCS12 | EVM0000274.1 | 318 | 35.5 | 6.15 | 1 | LG02 |
| AcNCS13 | EVM0013491.1 | 323 | 35.5 | 4.63 | 2 | LG01 |
| AcNCS14 | EVM0002685.1 | 355 | 39.0 | 5.40 | 2 | LG02 |
| AcNCS15 | EVM0014575.1 | 355 | 39.0 | 5.39 | 2 | LG02 |
| Protein | Vmax (µM min−1) | Km (µM) | Kcat (S−1) | Kcat/Km (M−1 S−1) |
|---|---|---|---|---|
| AcNCS2 | 1.6 ± 0.1 | 518.9 ± 109.9 | 20.3 ± 1.0 | 3.9× 104 |
| AcNCS4 | 1.6 ± 0.2 | 531.5 ± 86.5 | 21.4 ± 2.3 | 4.0 × 104 |
| AcNCS5 | 1.6 ± 0.2 | 534.9 ± 78.3 | 21.3 ± 2.1 | 4.0 × 104 |
| AcNCS14 | 2.3 ± 0.2 | 420.8 ± 45.9 | 22.1 ± 1.3 | 5.3 × 104 |
| AcNCS15 | 2.2 ± 0.2 | 502.9 ± 114.6 | 21.8 ± 1.4 | 4.3 × 104 |
| Protein | Vmax (µM min−1) | Km (µM) | Kcat (S−1) | Kcat/Km (M−1 S−1) |
|---|---|---|---|---|
| AcNCS2 | 1.3 ± 0.2 | 522.5 ± 72.6 | 13.1 ± 1.2 | 2.5 × 104 |
| AcNCS4 | 1.3 ± 0.2 | 530.3 ± 76.4 | 13.2 ± 2.0 | 2.5 × 104 |
| AcNCS5 | 1.4 ± 0.2 | 536.4 ± 78.6 | 13.2 ± 1.9 | 2.5 × 104 |
| AcNCS14 | 1.5 ± 0.2 | 495.3 ± 98.2 | 14.6 ± 1.2 | 2.9 × 104 |
| AcNCS15 | 1.5 ± 0.2 | 506.7 ± 63.8 | 14.2 ± 1.4 | 2.8 × 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zhang, S.; Meng, F.; Liang, W.; Peng, Y.; Zhu, B.; Niu, L.; Wang, C.; Li, C.; Lu, S. Genome-Wide Identification and Functional Analysis of the Norcoclaurine Synthase Gene Family in Aristolochia contorta. Int. J. Mol. Sci. 2025, 26, 4314. https://doi.org/10.3390/ijms26094314
Xu Y, Zhang S, Meng F, Liang W, Peng Y, Zhu B, Niu L, Wang C, Li C, Lu S. Genome-Wide Identification and Functional Analysis of the Norcoclaurine Synthase Gene Family in Aristolochia contorta. International Journal of Molecular Sciences. 2025; 26(9):4314. https://doi.org/10.3390/ijms26094314
Chicago/Turabian StyleXu, Yayun, Sixuan Zhang, Fanqi Meng, Wenjing Liang, Yunliang Peng, Butuo Zhu, Lili Niu, Chunling Wang, Caili Li, and Shanfa Lu. 2025. "Genome-Wide Identification and Functional Analysis of the Norcoclaurine Synthase Gene Family in Aristolochia contorta" International Journal of Molecular Sciences 26, no. 9: 4314. https://doi.org/10.3390/ijms26094314
APA StyleXu, Y., Zhang, S., Meng, F., Liang, W., Peng, Y., Zhu, B., Niu, L., Wang, C., Li, C., & Lu, S. (2025). Genome-Wide Identification and Functional Analysis of the Norcoclaurine Synthase Gene Family in Aristolochia contorta. International Journal of Molecular Sciences, 26(9), 4314. https://doi.org/10.3390/ijms26094314







