Integration of Bioinformatics and Machine Learning Strategies Identifies Ferroptosis and Immune Infiltration Signatures in Peri-Implantitis
Abstract
1. Introduction
2. Results
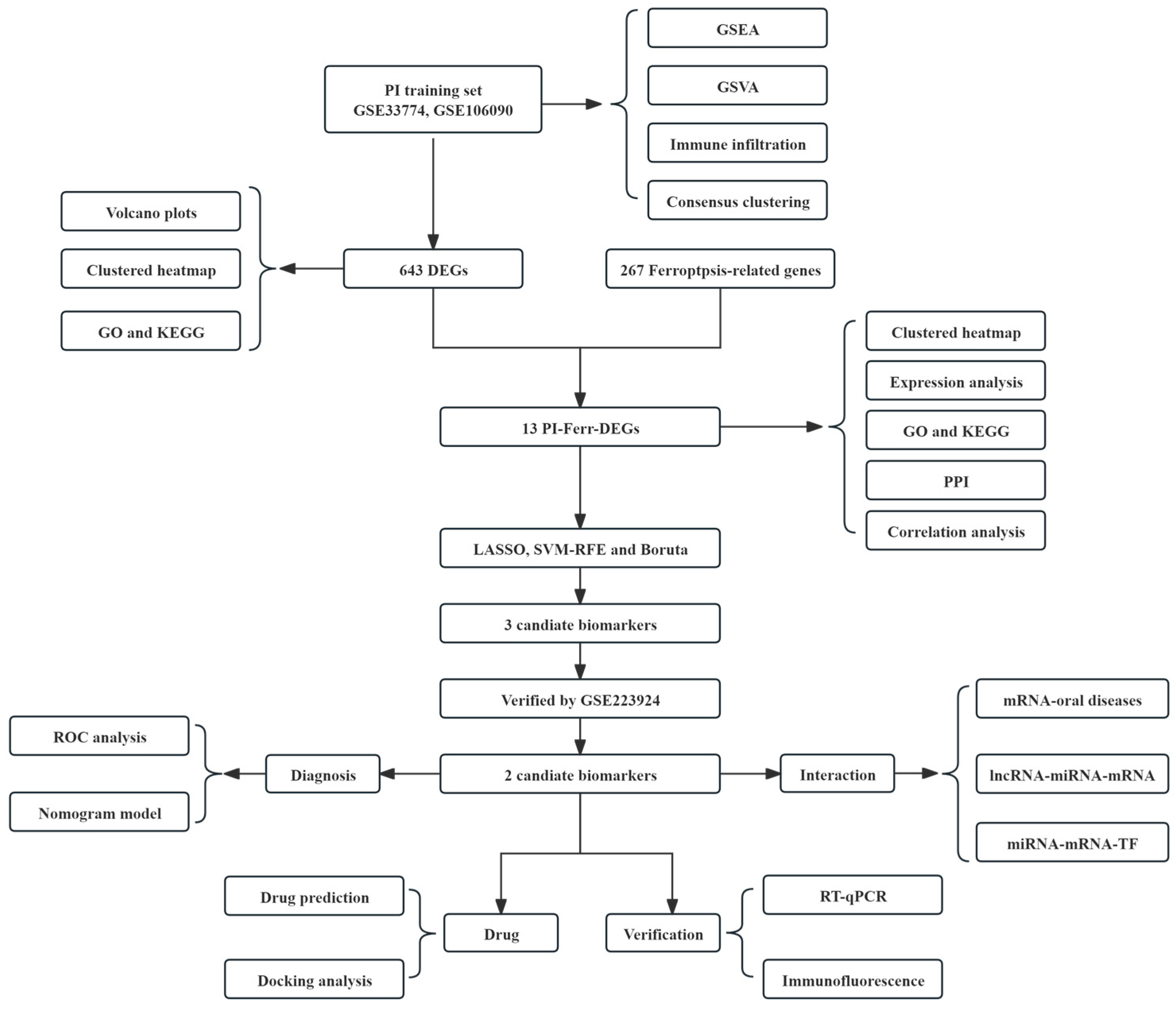
2.1. Data Collection and Preprocessing
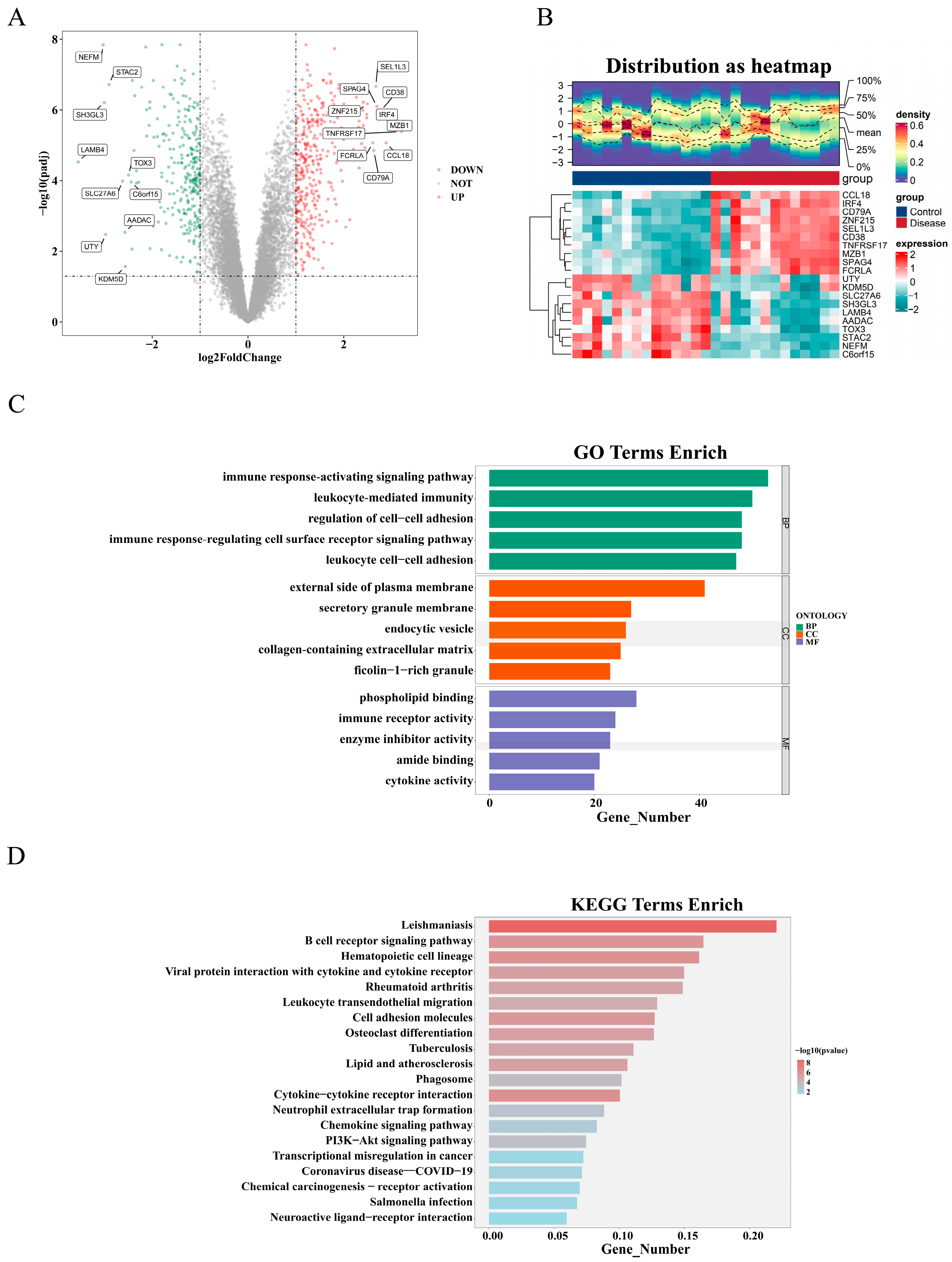
2.2. Identification and Functional Analysis of DEGs
2.3. Comprehensive Functional Analysis of PI-Ferr-DEGs
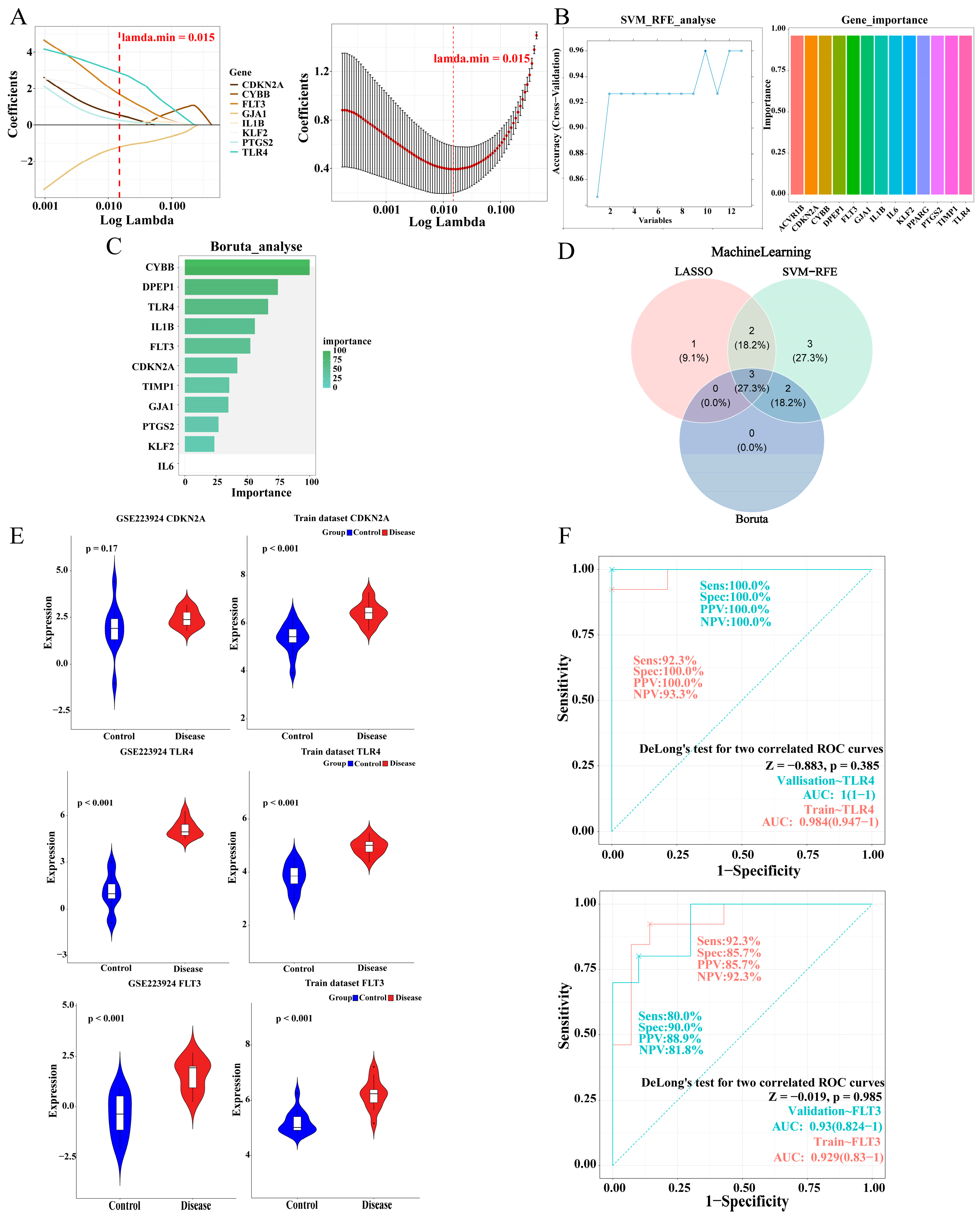
2.4. The Identification and Validation of TLR4 and FLT3
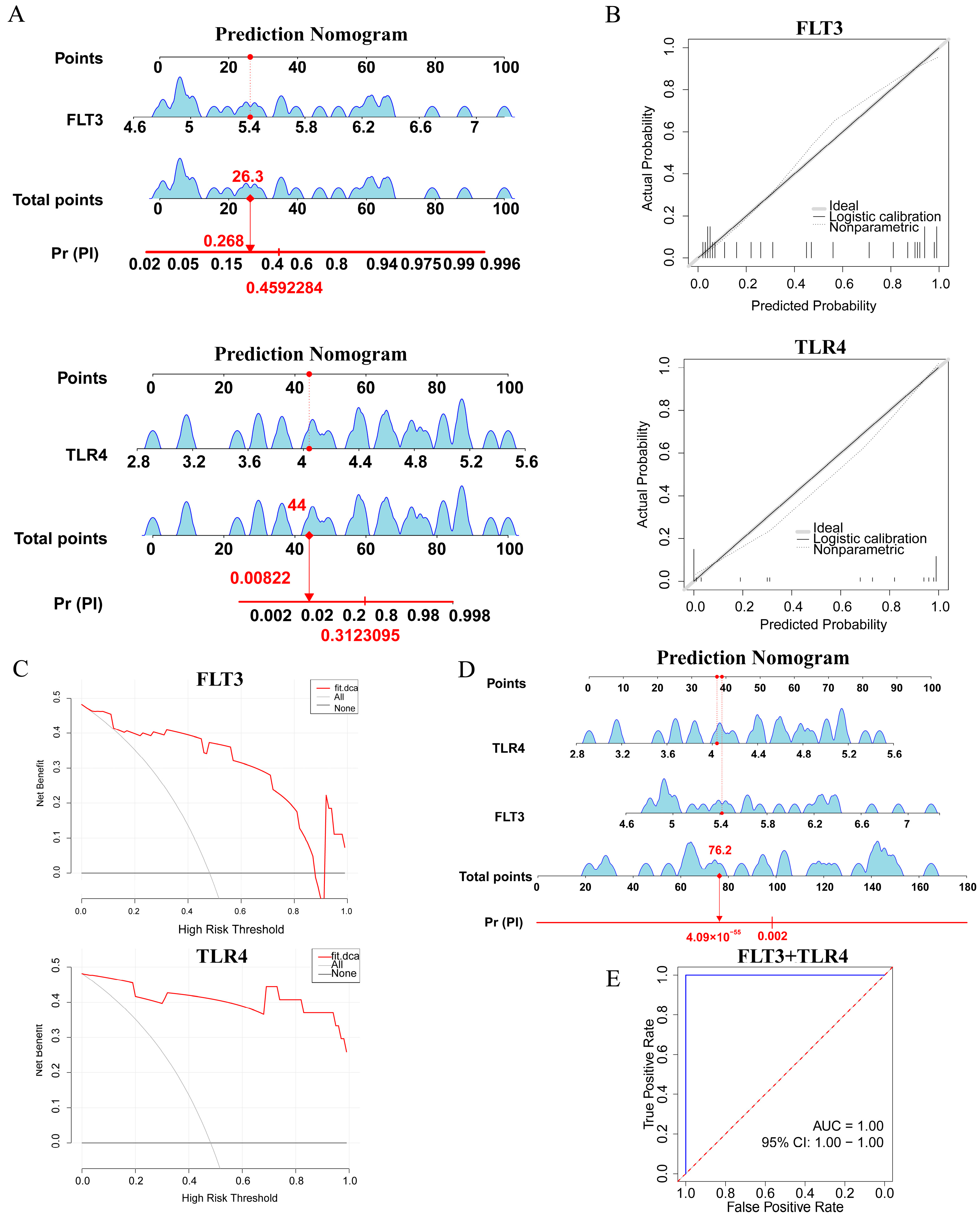
2.5. Construction and Assessment of a Nomogram for PI Diagnosis
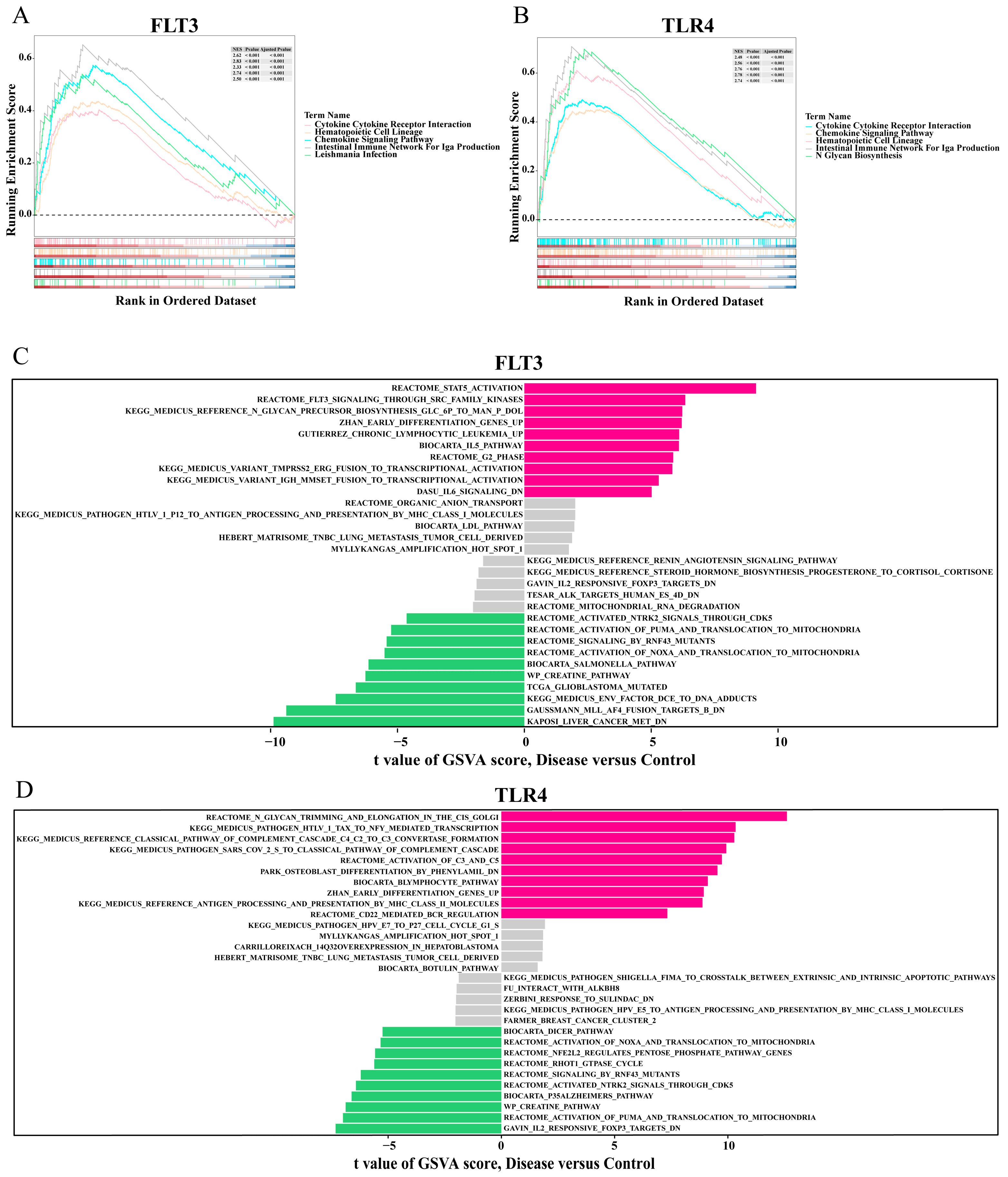
2.6. GSEA and GSVA of the Biomarkers
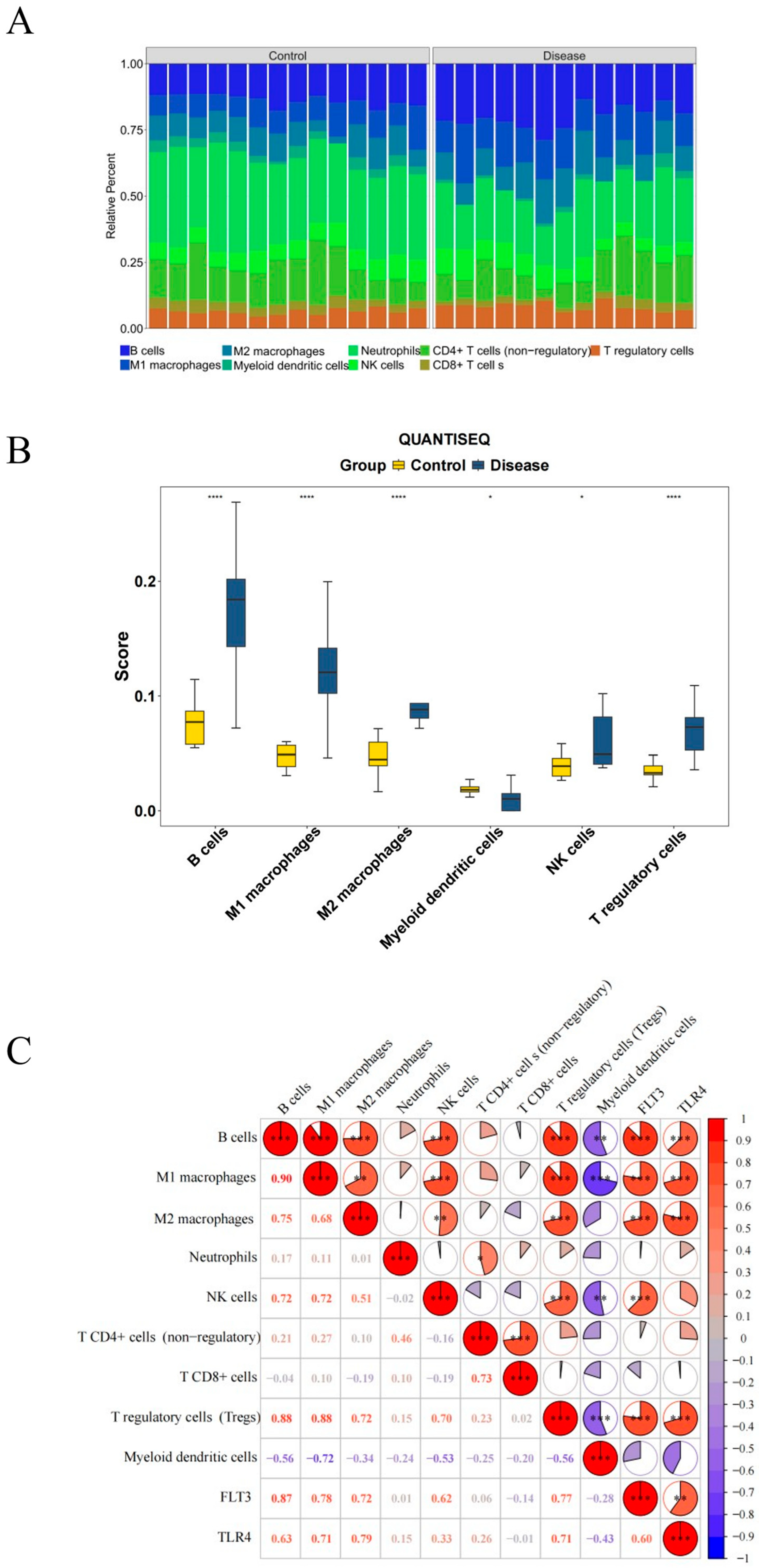
2.7. Immune Infiltration in PI
2.8. Network Construction for ‘Biomarkers-Oral Diseases’, ‘lncRNA-miRNA-mRNA’, and ‘miRNA-mRNA-TF’ Relationships
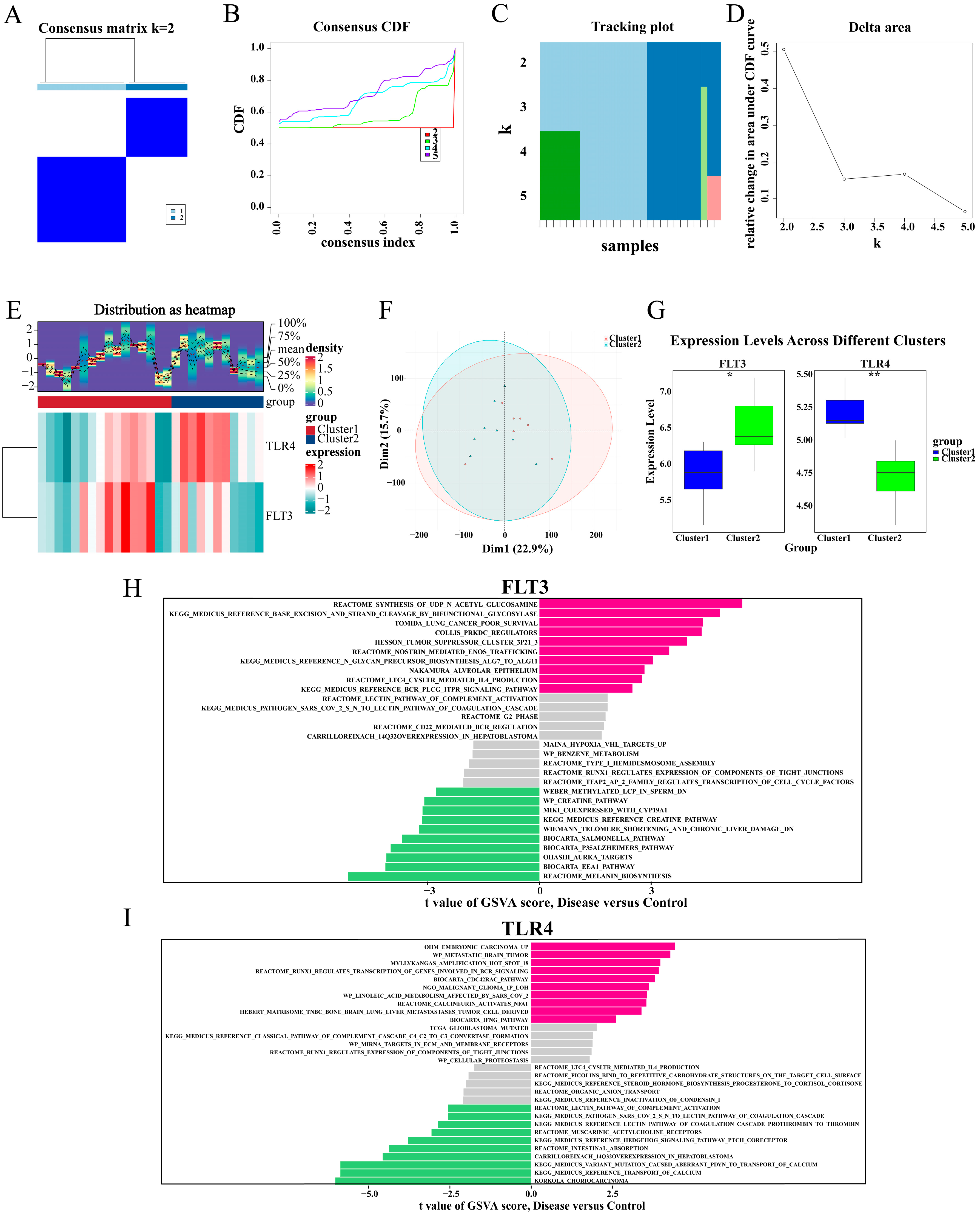
2.9. TLR4 and FLT3 Function Varied Significantly Between the Different Subtypes
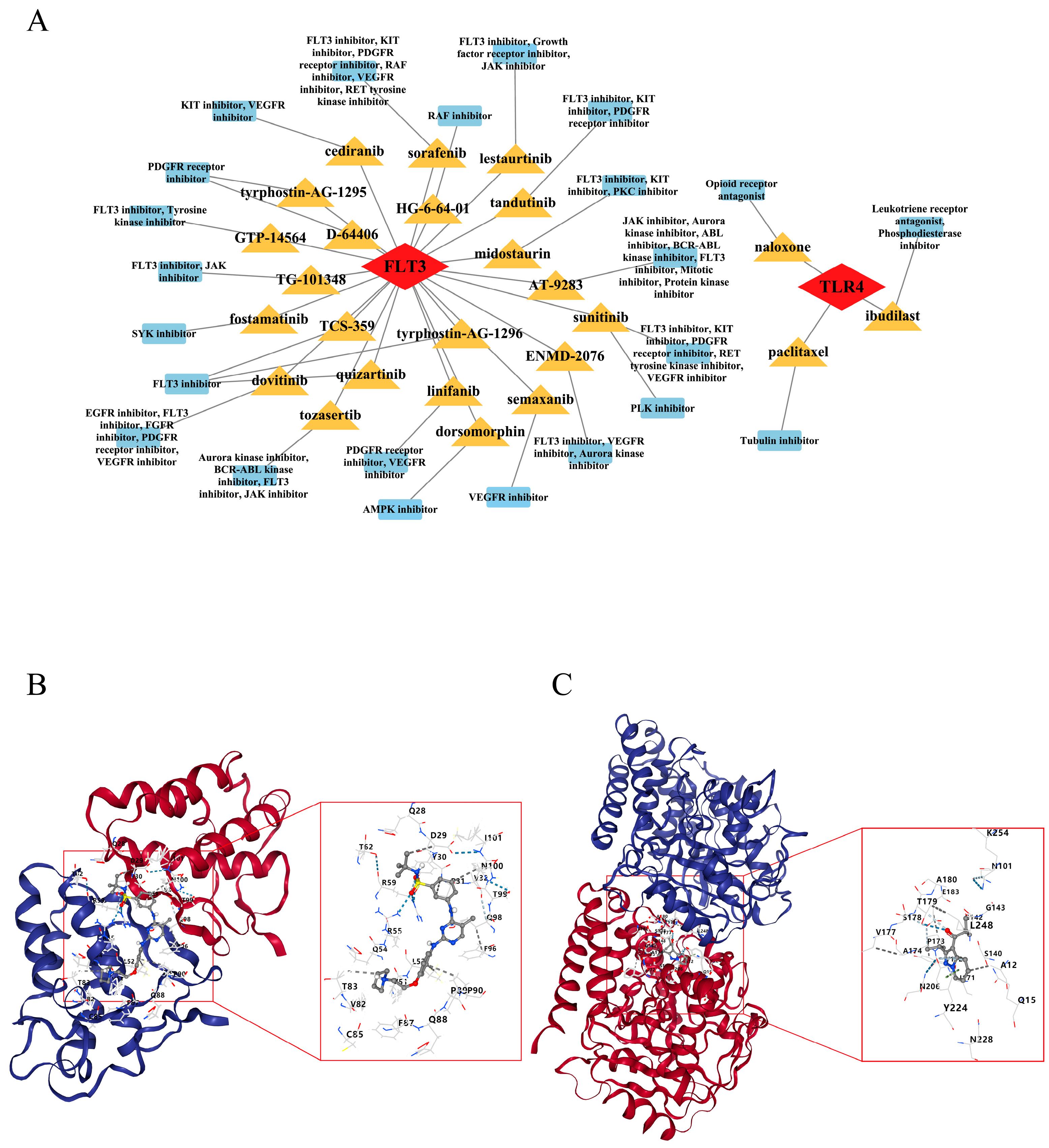
2.10. Fedratinib and Ibudilast Might Represent Potential Drugs for Treating PI
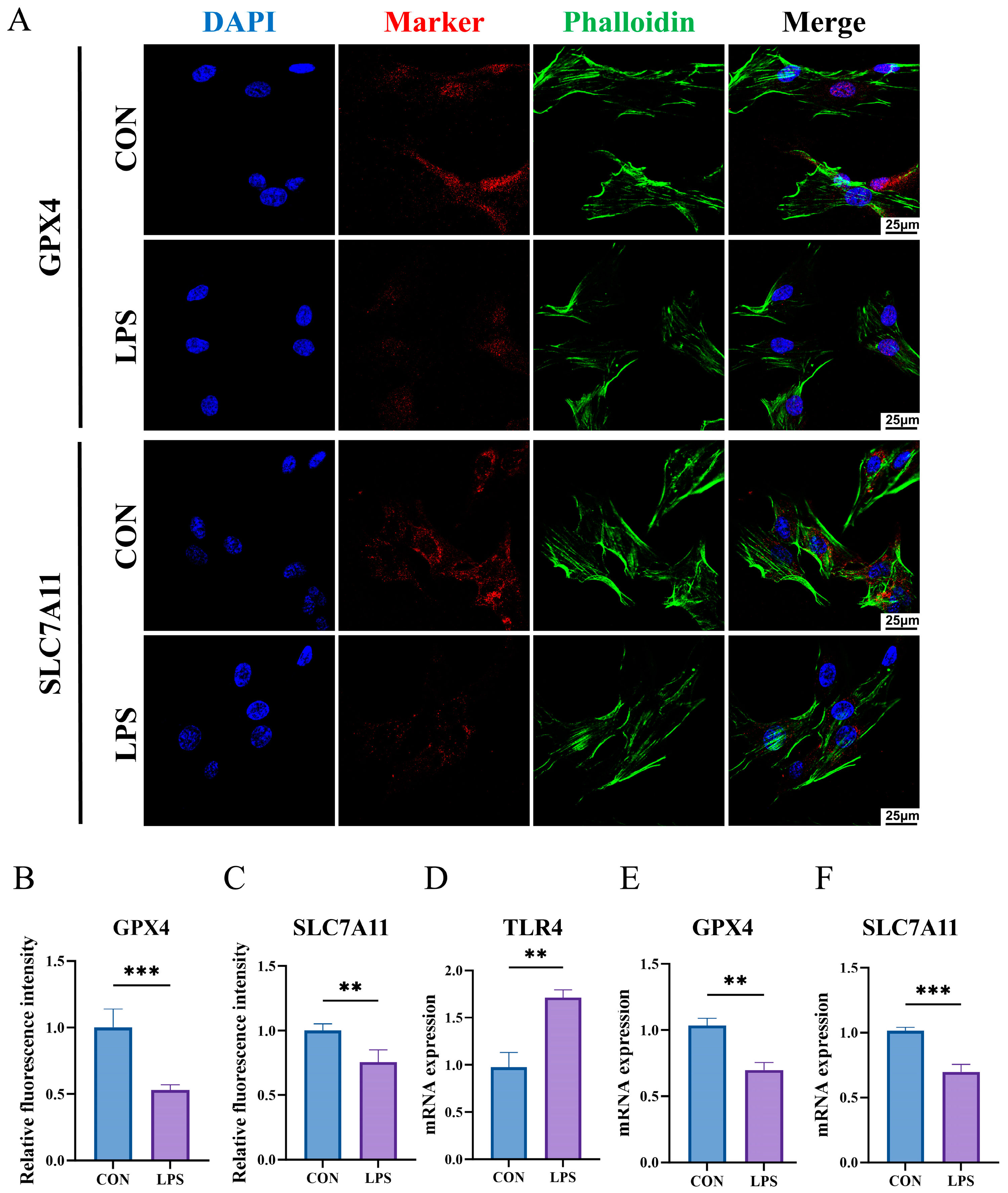
2.11. The Construction of an Inflammatory State Model of HGFs and Detection of Key Biomarkers
3. Discussion
4. Materials and Methods
4.1. Data Preparation
4.2. Normalization of the Obtained Data
4.3. Differential Expression Analysis and Function Analysis
4.4. Identification, Function Analysis, Protein–Protein Interaction (PPI) Network, and Correlation Analysis of PI-Ferr-DEGs
4.5. Machine Learning Screening for Identifying Candidate Biomarkers
4.6. Validation of Biomarkers by Expression Levels, Receiver Operating Characteristic (ROC), and Nomogram
4.7. Nomogram of Biomarkers’ Combination
4.8. Gene Set Enrichment Analysis (GSEA)
4.9. Gene Set Variation Analysis (GSVA)
4.10. Immune Infiltration Analysis
4.11. Gene-Disease Network Analysis and Molecular Regulatory Network
4.12. Comprehensive Analysis of Biomarkers Between Different Subtypes
4.13. Drug Prediction and Molecular Docking
4.14. Cell Culture
4.15. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
4.16. Immunofluorescence Staining (IF)
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berglundh, T.; Armitage, G.; Araujo, M.G.; Avila-Ortiz, G.; Blanco, J.; Camargo, P.M.; Chen, S.; Cochran, D.; Derks, J.; Figuero, E.; et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. S20), S286–S291. [Google Scholar] [CrossRef]
- Smeets, R.; Henningsen, A.; Jung, O.; Heiland, M.; Hammächer, C.; Stein, J.M. Definition, etiology, prevention and treatment of peri-implantitis—A review. Head Face Med. 2014, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Buser, D.; Sennerby, L.; De Bruyn, H. Modern implant dentistry based on osseointegration: 50 years of progress, current trends and open questions. Periodontology 2000 2017, 73, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Karoussis, I.K.; Salvi, G.E.; Heitz-Mayfield, L.J.; Brägger, U.; Hämmerle, C.H.; Lang, N.P. Long-term implant prognosis in patients with and without a history of chronic periodontitis: A 10-year prospective cohort study of the ITI Dental Implant System. Clin. Oral Implants Res. 2003, 14, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Vilela, N.; Gurgel, B.C.V.; Bruzos, C.; Duarte, W.R.; da Silva, H.D.P.; Pannuti, C.M.; Duarte, P.M. Preloading peri-implant crestal bone loss: A retrospective study of incidence and related factors. J. Periodontol. 2024, 95, 963–976. [Google Scholar] [CrossRef]
- Mauland, E.K.; Sørensen, K.; Aarbu, N.; Verket, A.; Ellingsen, S.A.; Bull, V.H.; Gaarden, R.T.; Lie, S.A.; Bunæs, D.F. A Cross-Sectional Study of Peri-Implant Diseases in a Random Norwegian Population: Prevalence, Risk Indicators, and Clinical Validation of Patient-Reported Outcomes. Clin. Oral Implants Res. 2025, 36, 153–165. [Google Scholar] [CrossRef]
- Josephraj, F.; Kumar, N.A.; Nandini, V.V.; Sujatha, S.; Karthik, V. Performance evaluation of carbon quantum dots impregnated glass ionomer cement to avoid peri-implant disease. Biomed. Mater. 2024, 19, 035040. [Google Scholar] [CrossRef]
- Di Spirito, F.; Giordano, F.; Di Palo, M.P.; D’Ambrosio, F.; Scognamiglio, B.; Sangiovanni, G.; Caggiano, M.; Gasparro, R. Microbiota of Peri-Implant Healthy Tissues, Peri-Implant Mucositis, and Peri-Implantitis: A Comprehensive Review. Microorganisms 2024, 12, 1137. [Google Scholar] [CrossRef]
- Ericsson, I.; Lindhe, J. Probing depth at implants and teeth. An experimental study in the dog. J. Clin. Periodontol. 1993, 20, 623–627. [Google Scholar] [CrossRef]
- Hermann, J.S.; Buser, D.; Schenk, R.K.; Schoolfield, J.D.; Cochran, D.L. Biologic Width around one- and two-piece titanium implants. Clin. Oral Implants Res. 2001, 12, 559–571. [Google Scholar] [CrossRef]
- Ericsson, I.; Berglundh, T.; Marinello, C.; Liljenberg, B.; Lindhe, J. Long-standing plaque and gingivitis at implants and teeth in the dog. Clin. Oral Implants Res. 1992, 3, 99–103. [Google Scholar] [CrossRef]
- Wickert, A.; Schwantes, A.; Fuhrmann, D.C.; Brüne, B. Inflammation in a ferroptotic environment. Front. Pharmacol. 2024, 15, 1474285. [Google Scholar] [CrossRef]
- Li, J.; Ye, L.J.; Dai, Y.W.; Wang, H.W.; Gao, J.; Shen, Y.H.; Wang, F.; Dai, Q.G.; Wu, Y.Q. Single-cell analysis reveals a unique microenvironment in peri-implantitis. J. Clin. Periodontol. 2024, 51, 1665–1676. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, G.; Wu, H.; Huang, X.; Xu, R.; Deng, F.; Li, Y. Ebselen restores peri-implantitis-induced osteogenic inhibition via suppressing BMSCs ferroptosis. Exp. Cell Res. 2023, 427, 113612. [Google Scholar] [CrossRef]
- Özkan Karasu, Y.; Maden, O.; Çanakçı, C.F. Oxidative damage biomarkers and antioxidant enzymes in saliva of patients with peri-implant diseases. Int. J. Implant Dent. 2024, 10, 43. [Google Scholar] [CrossRef]
- Wang, J.; Hu, C.; Ma, X.; Zhang, Y.; Zhang, X.; Hong, X.; Chen, L.; Wang, Y.; Wang, J.; Chen, S.; et al. The role of oxidative stress biomarkers in the development of peri-implant disease: A systematic review and meta-analysis. J. Dent. 2024, 146, 105026. [Google Scholar] [CrossRef]
- Xie, L.; Zhu, G.; Long, S.; Wang, M.; Cheng, X.; Dong, Y.; Wang, C.; Wang, G. Identification of MORN3 and LLGL2 as novel diagnostic biomarkers for latent tuberculosis infection using machine learning strategies and experimental verification. Ann. Med. 2024, 56, 2380797. [Google Scholar] [CrossRef]
- Chen, J.; Ou, L.; Liu, W.; Gao, F. Exploring the molecular mechanisms of ferroptosis-related genes in periodontitis: A multi-dataset analysis. BMC Oral Health 2024, 24, 611. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, P.; Yu, C.; Song, X.; Wang, H.; Fang, J.; Zhu, C.; Li, Y. A LASSO-based model to predict central lymph node metastasis in preoperative patients with cN0 papillary thyroid cancer. Front. Oncol. 2023, 13, 1034047. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar]
- Hamidi, F.; Gilani, N.; Arabi Belaghi, R.; Yaghoobi, H.; Babaei, E.; Sarbakhsh, P.; Malakouti, J. Identifying potential circulating miRNA biomarkers for the diagnosis and prediction of ovarian cancer using machine-learning approach: Application of Boruta. Front. Digit. Health 2023, 5, 1187578. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.J.; Lai, Y.R.; Huang, Q.R.; Li, Y.R.; Zhang, Y.J.; Chen, R.Y.; Qian, S.J. Single-cell sequencing identifies inflammation-promoting fibroblast-neutrophil interaction in peri-implantitis. J. Clin. Periodontol. 2024, 51, 196–208. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Stavroullakis, A.; Brito, C.; Chen, H.Y.; Bajenova, E.; Prakki, A.; Nogueira-Filho, G. Dental implant surface treatments may modulate cytokine secretion in Porphyromonas gingivalis-stimulated human gingival fibroblasts: A comparative study. J. Biomed. Mater. Res. Part A 2015, 103, 1131–1140. [Google Scholar] [CrossRef]
- Pérez-Chaparro, P.J.; Duarte, P.M.; Shibli, J.A.; Montenegro, S.; Lacerda Heluy, S.; Figueiredo, L.C.; Faveri, M.; Feres, M. The Current Weight of Evidence of the Microbiologic Profile Associated With Peri-Implantitis: A Systematic Review. J. Periodontol. 2016, 87, 1295–1304. [Google Scholar] [CrossRef]
- Yutoku, M.; Fujita, K.; Chiba, N.; Tada, R.; Ohnishi, T.; Sugimura, M.; Matsuguchi, T. Early Growth Response 1 Plays an Essential Role in Proinflammatory and Osteoclastogenic Activities of Lipopolysaccharide-Stimulated Osteoblasts. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2025, 39, e70532. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, J.; Fan, X.; Miao, R.; Wang, Y. HSP90AA1 promotes the inflammation in human gingival fibroblasts induced by Porphyromonas gingivalis lipopolysaccharide via regulating of autophagy. BMC Oral Health 2022, 22, 366. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, S.B.; Hakki, S.S.; Hakki, E.E.; Durak, Y.; Kantarci, A. Porphyromonas gingivalis Lipopolysaccharide Induces a Pro-inflammatory Human Gingival Fibroblast Phenotype. Inflammation 2017, 40, 144–153. [Google Scholar] [CrossRef]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef]
- Esposito, M.; Grusovin, M.G.; Worthington, H.V. Interventions for replacing missing teeth: Treatment of peri-implantitis. Cochrane Database Syst. Rev. 2012, 1, Cd004970. [Google Scholar] [CrossRef]
- Majzoub, J.; Chen, Z.; Saleh, I.; Askar, H.; Wang, H.L. Influence of restorative design on the progression of peri-implant bone loss: A retrospective study. J. Periodontol. 2021, 92, 536–546. [Google Scholar] [CrossRef]
- Ozawa, R.; Saita, M.; Sakaue, S.; Okada, R.; Sato, T.; Kawamata, R.; Sakurai, T.; Hamada, N.; Kimoto, K.; Nagasaki, Y. Redox injectable gel protects osteoblastic function against oxidative stress and suppresses alveolar bone loss in a rat peri-implantitis model. Acta Biomater. 2020, 110, 82–94. [Google Scholar] [CrossRef]
- Kos, M.; Bojarski, K.; Mertowska, P.; Mertowski, S.; Tomaka, P.; Dziki, Ł.; Grywalska, E. Immunological Strategies in Gastric Cancer: How Toll-like Receptors 2, -3, -4, and -9 on Monocytes and Dendritic Cells Depend on Patient Factors? Cells 2024, 13, 1708. [Google Scholar] [CrossRef]
- Ma, M.; Jiang, W.; Zhou, R. DAMPs and DAMP-sensing receptors in inflammation and diseases. Immunity 2024, 57, 752–771. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Ferroptosis in infection, inflammation, and immunity. J. Exp. Med. 2021, 218, e20210518. [Google Scholar] [CrossRef]
- Yamashiro, K.; Ideguchi, H.; Aoyagi, H.; Yoshihara-Hirata, C.; Hirai, A.; Suzuki-Kyoshima, R.; Zhang, Y.; Wake, H.; Nishibori, M.; Yamamoto, T.; et al. High Mobility Group Box 1 Expression in Oral Inflammation and Regeneration. Front. Immunol. 2020, 11, 1461. [Google Scholar] [CrossRef]
- Deng, S.; Hu, Y.; Zhou, J.; Wang, Y.; Wang, Y.; Li, S.; Huang, G.; Peng, C.; Hu, A.; Yu, Q.; et al. TLR4 mediates alveolar bone resorption in experimental peri-implantitis through regulation of CD45(+) cell infiltration, RANKL/OPG ratio, and inflammatory cytokine production. J. Periodontol. 2020, 91, 671–682. [Google Scholar] [CrossRef]
- Wu, X.; Qiao, S.; Wang, W.; Zhang, Y.; Shi, J.; Zhang, X.; Gu, W.; Zhang, X.; Li, Y.; Ding, X.; et al. Melatonin prevents peri-implantitis via suppression of TLR4/NF-κB. Acta Biomater. 2021, 134, 325–336. [Google Scholar] [CrossRef]
- Leifheit, M.E.; Johnson, G.; Kuzel, T.M.; Schneider, J.R.; Barker, E.; Yun, H.D.; Ustun, C.; Goldufsky, J.W.; Gupta, K.; Marzo, A.L. Enhancing Therapeutic Efficacy of FLT3 Inhibitors with Combination Therapy for Treatment of Acute Myeloid Leukemia. Int. J. Mol. Sci. 2024, 25, 9448. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 2019, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Tiziani, S.; Park, G.; Kaul, M.; Paternostro, G. Cellular protection using Flt3 and PI3Kα inhibitors demonstrates multiple mechanisms of oxidative glutamate toxicity. Nat. Commun. 2014, 5, 3672. [Google Scholar] [CrossRef]
- Ramos, M.I.; Tak, P.P.; Lebre, M.C. Fms-like tyrosine kinase 3 ligand-dependent dendritic cells in autoimmune inflammation. Autoimmun. Rev. 2014, 13, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, L.; Zhao, D.; Gao, F.; Yan, J. Classical dendritic cells regulate acute lung inflammation and injury in mice with lipopolysaccharide-induced acute respiratory distress syndrome. Int. J. Mol. Med. 2019, 44, 617–629. [Google Scholar] [CrossRef]
- Li, Y.J.; Zhang, C.; Martincuks, A.; Herrmann, A.; Yu, H. STAT proteins in cancer: Orchestration of metabolism. Nat. Rev. Cancer 2023, 23, 115–134. [Google Scholar] [CrossRef]
- Villalobos, V.; Silva, I.; Morales, D.; Canelo, J.; Garrido, M.; Carreño, L.J.; Cavalla, F.; Dutzan, N.; Caceres, M. Topological insight of immune-vascular distribution in peri-implantitis lesions. Oral Dis. 2024, 30, 5305–5314. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Guo, D.; Meng, L.; Feng, X.; Zhang, Y.; Pan, S. Immune dysregulation and macrophage polarization in peri-implantitis. Front. Bioeng. Biotechnol. 2024, 12, 1291880. [Google Scholar] [CrossRef]
- Zhao, X.; Peng, T.; Cao, X.; Hou, Y.; Li, R.; Han, T.; Fan, Z.; Zhao, M.; Chang, Y.; Chen, H.; et al. In vivo G-CSF treatment activates the GR-SOCS1 axis to suppress IFN-γ secretion by natural killer cells. Cell Rep. 2022, 40, 111342. [Google Scholar] [CrossRef]
- Wanderley, C.W.; Colón, D.F.; Luiz, J.P.M.; Oliveira, F.F.; Viacava, P.R.; Leite, C.A.; Pereira, J.A.; Silva, C.M.; Silva, C.R.; Silva, R.L.; et al. Paclitaxel Reduces Tumor Growth by Reprogramming Tumor-Associated Macrophages to an M1 Profile in a TLR4-Dependent Manner. Cancer Res. 2018, 78, 5891–5900. [Google Scholar] [CrossRef]
- Li, Y.; Adamek, P.; Zhang, H.; Tatsui, C.E.; Rhines, L.D.; Mrozkova, P.; Li, Q.; Kosturakis, A.K.; Cassidy, R.M.; Harrison, D.S.; et al. The Cancer Chemotherapeutic Paclitaxel Increases Human and Rodent Sensory Neuron Responses to TRPV1 by Activation of TLR4. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 13487–13500. [Google Scholar] [CrossRef] [PubMed]
- Farkas, A.; Lynch, M.J.; Westover, R.; Giles, J.; Siripong, N.; Nalatwad, A.; Pizon, A.F.; Martin-Gill, C. Pulmonary Complications of Opioid Overdose Treated With Naloxone. Ann. Emerg. Med. 2020, 75, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Djuric, V.J.; Wang, L.; Bienenstock, J.; Perdue, M.H. Naloxone Exacerbates Intestinal and Systemic Anaphylaxis in the Rat. Brain Behav. Immun. 1995, 9, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Kadota, N.; Yoshida, A.; Sawamoto, A.; Okuyama, S.; Nakajima, M. Ibudilast Reduces IL-6 Levels and Ameliorates Symptoms in Lipopolysaccharide-Induced Sepsis Mice. Biol. Pharm. Bull. 2022, 45, 1180–1184. [Google Scholar] [CrossRef]
- Lusardi, M.; Rapetti, F.; Spallarossa, A.; Brullo, C. PDE4D: A Multipurpose Pharmacological Target. Int. J. Mol. Sci. 2024, 25, 8052. [Google Scholar] [CrossRef]
- Crocetti, L.; Floresta, G.; Cilibrizzi, A.; Giovannoni, M.P. An Overview of PDE4 Inhibitors in Clinical Trials: 2010 to Early 2022. Molecules 2022, 27, 4964. [Google Scholar] [CrossRef]
- Chen, Y.; Wyatt, D.; Attanasio, M.; Thomas, M.; Thomas, M.; He, B.; Nishii, R.; Liu, L.; Shan, V.; Xue, Y.; et al. Relative bioavailability of fedratinib through various alternative oral administration methods in healthy adults. Cancer Chemother. Pharmacol. 2024, 93, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Yang, J.; Chen, H.; Liu, R.; Hu, R.; Cai, J.; Yang, S.; Zheng, B.; Guo, P.; Cai, Z.; et al. Macrophages hijack carbapenem-resistance hypervirulent Klebsiella pneumoniae by blocking SLC7A11/GSH-manipulated iron oxidative stress. Free Radic. Biol. Med. 2025, 230, 234–247. [Google Scholar] [CrossRef]
- Bruzzese, A.; Martino, E.A.; Labanca, C.; Mendicino, F.; Lucia, E.; Olivito, V.; Rossi, T.; Neri, A.; Morabito, F.; Vigna, E.; et al. Advances and Challenges in Quizartinib-Based FLT3 Inhibition for Acute Myeloid Leukemia: Mechanisms of Resistance and Prospective Combination Therapies. Eur. J. Haematol. 2025, 114, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Monogiou Belik, D.; Bernasconi, R.; Xu, L.; Della Verde, G.; Lorenz, V.; Grüterich, V.; Balzarolo, M.; Mochizuki, M.; Pfister, O.; Kuster, G.M. The Flt3-inhibitor quizartinib augments apoptosis and promotes maladaptive remodeling after myocardial infarction in mice. Apoptosis Int. J. Program. Cell Death 2024, 29, 357–371. [Google Scholar] [CrossRef]
- Ruan, H.; Luan, J.; Gao, S.; Li, S.; Jiang, Q.; Liu, R.; Liang, Q.; Zhang, R.; Zhang, F.; Li, X.; et al. Fedratinib Attenuates Bleomycin-Induced Pulmonary Fibrosis via the JAK2/STAT3 and TGF-β1 Signaling Pathway. Molecules 2021, 26, 4491. [Google Scholar] [CrossRef]
- Akcora, B.; Dathathri, E.; Ortiz-Perez, A.; Gabriël, A.V.; Storm, G.; Prakash, J.; Bansal, R. TG101348, a selective JAK2 antagonist, ameliorates hepatic fibrogenesis in vivo. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 9466–9475. [Google Scholar] [CrossRef]
- Ginesin, O.; Mayer, Y.; Gabay, E.; Rotenberg, D.; Machtei, E.E.; Coyac, B.R.; Bar-On, Y.; Zigdon-Giladi, H. Revealing leukocyte populations in human peri-implantitis and periodontitis using flow cytometry. Clin. Oral Investig. 2023, 27, 5499–5508. [Google Scholar] [CrossRef]
- Yin, J.; Fang, Y.; Liao, Y.; Chen, Z.; Liu, S.; Zhu, H.; Song, K.; Shi, B. Bioinformatics investigation of adaptive immune-related genes in peri-implantitis and periodontitis: Characteristics and diagnostic values. Immun. Inflamm. Dis. 2024, 12, e1272. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Li, Z.; Acharya, A.; Chen, D.; Chen, Z.; Mattheos, N.; Chen, Z.; Huang, B. Long non-coding RNA and mRNA expression profiles in peri-implantitis vs periodontitis. J. Periodontal Res. 2020, 55, 342–353. [Google Scholar] [CrossRef]
- Becker, S.T.; Beck-Broichsitter, B.E.; Graetz, C.; Dörfer, C.E.; Wiltfang, J.; Häsler, R. Peri-implantitis versus periodontitis: Functional differences indicated by transcriptome profiling. Clin. Implant Dent. Relat. Res. 2014, 16, 401–411. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Oh, J.M.; Kim, Y.; Son, H.; Kim, Y.H.; Kim, H.J. Comparative transcriptome analysis of periodontitis and peri-implantitis in human subjects. J. Periodontol. 2024, 95, 337–349. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Cheng, Y.; Liu, K.; Zou, H.; You, Z. Identification of potential ferroptosis-related biomarkers and a pharmacological compound in diabetic retinopathy based on machine learning and molecular docking. Front. Endocrinol. 2022, 13, 988506. [Google Scholar] [CrossRef]
- He, J.; Zheng, Z.; Li, S.; Liao, C.; Li, Y. Identification and assessment of differentially expressed necroptosis long non-coding RNAs associated with periodontitis in human. BMC Oral Health 2023, 23, 632. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Hübschmann, D. Make Interactive Complex Heatmaps in R. Bioinformatics 2022, 38, 1460–1462. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Liu, P.; Xu, H.; Shi, Y.; Deng, L.; Chen, X. Potential Molecular Mechanisms of Plantain in the Treatment of Gout and Hyperuricemia Based on Network Pharmacology. Evid.-Based Complement. Altern. Med. eCAM 2020, 2020, 3023127. [Google Scholar] [CrossRef]
- Leyva-Moral, J.M.; Watson, C.; Granel, N.; Raij-Johansen, C.; Ayala, R.A. Cultural adaptation and validation of the caring behaviors assessment tool into Spanish. BMC Nurs. 2024, 23, 240. [Google Scholar] [CrossRef]
- Li, Y.; Lu, F.; Yin, Y. Applying logistic LASSO regression for the diagnosis of atypical Crohn’s disease. Sci. Rep. 2022, 12, 11340. [Google Scholar] [CrossRef]
- Cinelli, M.; Sun, Y.; Best, K.; Heather, J.M.; Reich-Zeliger, S.; Shifrut, E.; Friedman, N.; Shawe-Taylor, J.; Chain, B. Feature selection using a one dimensional naïve Bayes’ classifier increases the accuracy of support vector machine classification of CDR3 repertoires. Bioinformatics 2017, 33, 951–955. [Google Scholar] [CrossRef]
- Yue, S.; Li, S.; Huang, X.; Liu, J.; Hou, X.; Zhao, Y.; Niu, D.; Wang, Y.; Tan, W.; Wu, J. Machine learning for the prediction of acute kidney injury in patients with sepsis. J. Transl. Med. 2022, 20, 215. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Fang, K.; Song, X.; Bo, J. A nomogram to predict mortality in patients with severe fever with thrombocytopenia syndrome. Sci. Rep. 2024, 14, 10627. [Google Scholar] [CrossRef]
- Zhang, Z.; Rousson, V.; Lee, W.C.; Ferdynus, C.; Chen, M.; Qian, X.; Guo, Y. Decision curve analysis: A technical note. Ann. Transl. Med. 2018, 6, 308. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef]
- Orifjon, S.; Jammatov, J.; Sousa, C.; Barros, R.; Vasconcelos, O.; Rodrigues, P. Translation and Adaptation of the Adult Developmental Coordination Disorder/Dyspraxia Checklist (ADC) into Asian Uzbekistan. Sports 2023, 11, 135. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]


| Gene | Acc. No | Primer Sequence(5′-3′) |
|---|---|---|
| TLR4 | NM_003266.4 | F:AGACCTGTCCCTGAACCCTAT R:CGATGGACTTCTAAACCAGCCA |
| GPX4 | NM_001039847.3 | F:GAGGCAAGACCGAAGTAAACTAC R:CCGAACTGGTTACACGGGAA |
| SLC7A11 | NM_014331.4 | F:TCTCCAAAGGAGGTTACCTGC R: AGACTCCCCTCAGTAAAGTGAC |
| GAPDH | NM_001256799.3 | F: GGAGCGAGATCCCTCCAAAAT R: GGCTGTTGTCATACTTCTCATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zou, Y.; Deng, H.; Zha, J.; Pathak, J.L.; Chen, Y.; Ge, Q.; Wang, L. Integration of Bioinformatics and Machine Learning Strategies Identifies Ferroptosis and Immune Infiltration Signatures in Peri-Implantitis. Int. J. Mol. Sci. 2025, 26, 4306. https://doi.org/10.3390/ijms26094306
Huang J, Zou Y, Deng H, Zha J, Pathak JL, Chen Y, Ge Q, Wang L. Integration of Bioinformatics and Machine Learning Strategies Identifies Ferroptosis and Immune Infiltration Signatures in Peri-Implantitis. International Journal of Molecular Sciences. 2025; 26(9):4306. https://doi.org/10.3390/ijms26094306
Chicago/Turabian StyleHuang, Jieying, Yaokun Zou, Huizhi Deng, Jun Zha, Janak Lal Pathak, Yaxin Chen, Qing Ge, and Liping Wang. 2025. "Integration of Bioinformatics and Machine Learning Strategies Identifies Ferroptosis and Immune Infiltration Signatures in Peri-Implantitis" International Journal of Molecular Sciences 26, no. 9: 4306. https://doi.org/10.3390/ijms26094306
APA StyleHuang, J., Zou, Y., Deng, H., Zha, J., Pathak, J. L., Chen, Y., Ge, Q., & Wang, L. (2025). Integration of Bioinformatics and Machine Learning Strategies Identifies Ferroptosis and Immune Infiltration Signatures in Peri-Implantitis. International Journal of Molecular Sciences, 26(9), 4306. https://doi.org/10.3390/ijms26094306






