rDNA Copy Number Variation and Methylation in Human and Mouse Sperm
Abstract
1. Introduction
2. Results
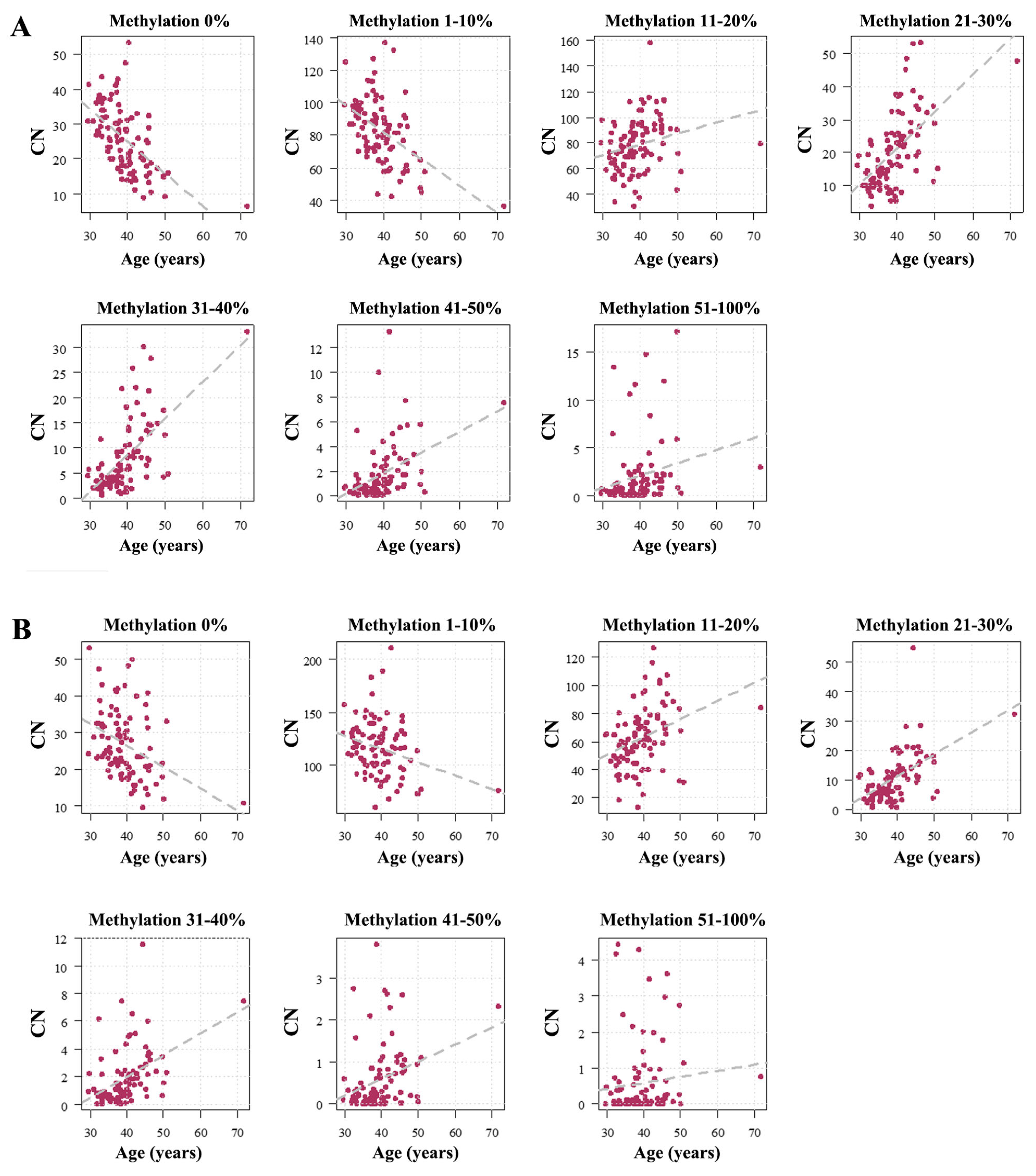
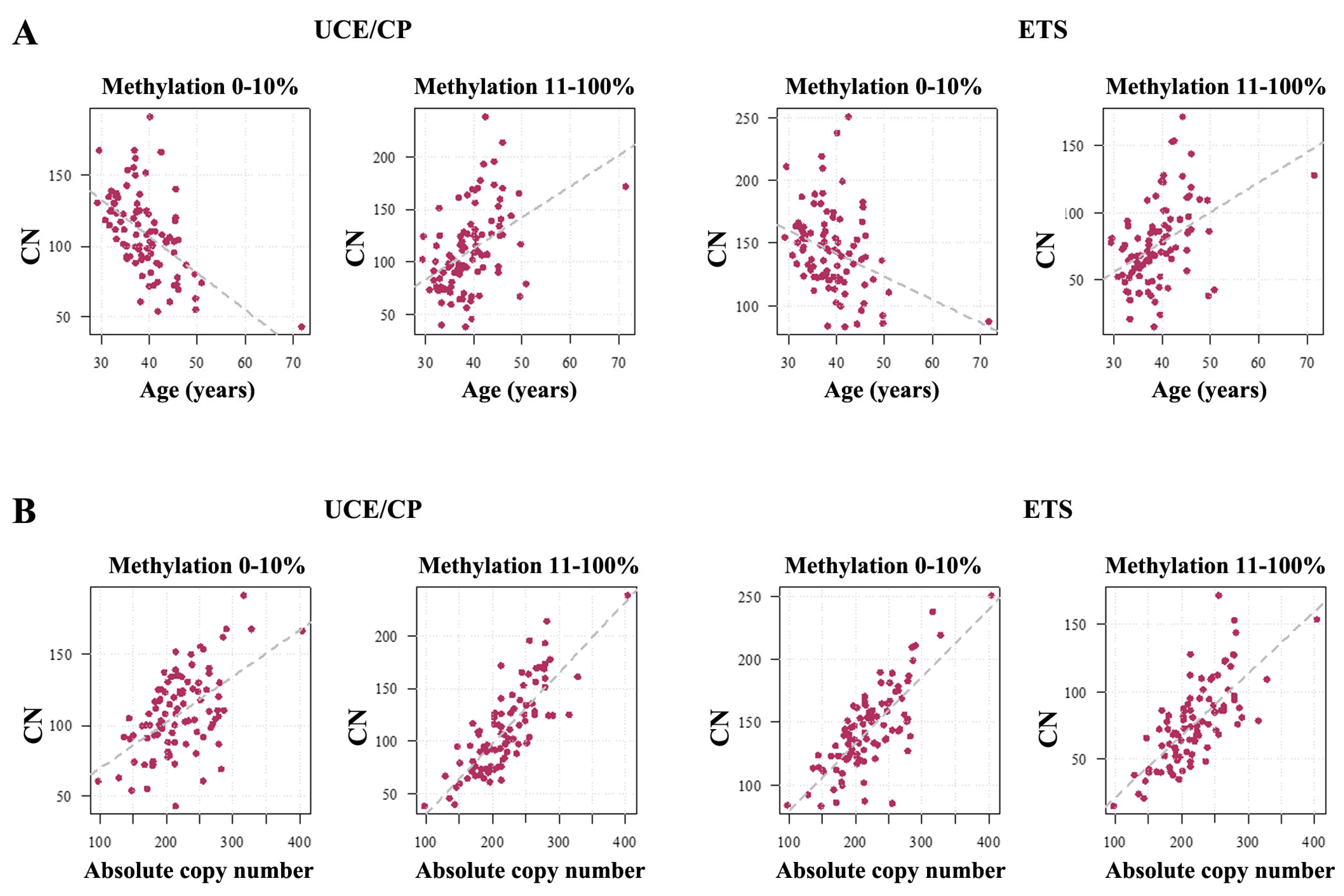
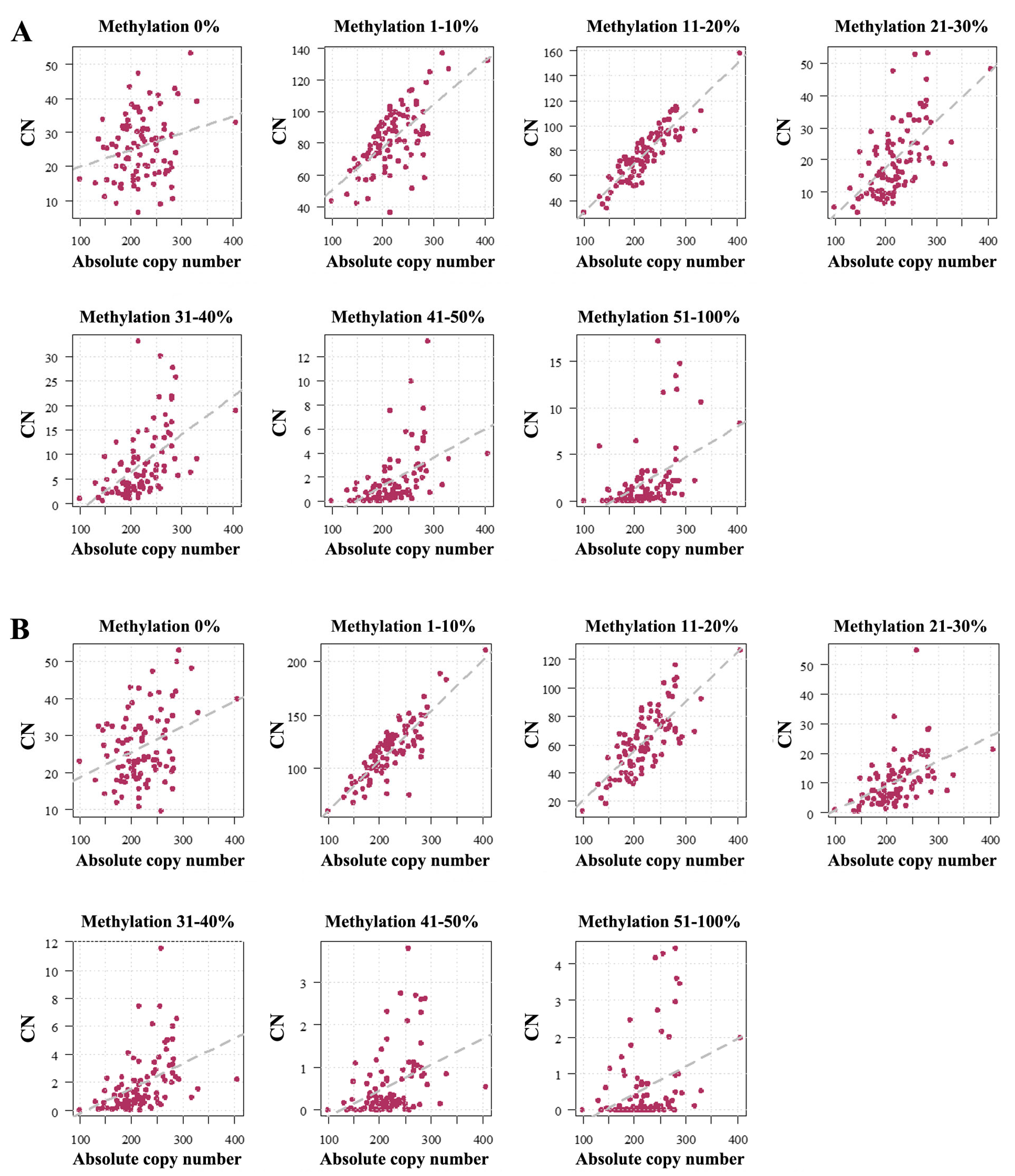
2.1. Human Sperm Samples
2.2. Mouse Sperm Samples
3. Discussion
3.1. Absolute rDNA CN and Methylation in the Mammalian Germline
3.2. rDNA Promoter Methylation Differences in the Human and Mouse Germline
3.3. The Paternal Age Effect on Sperm rDNA Methylation in Humans and Mice
3.4. Limitations
4. Materials and Methods
4.1. Study Samples
4.2. Droplet Digital PCR
4.3. Deep Bisulfite Sequencing
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CN | copy number |
| CP | core promoter |
| ddPCR | droplet digital polymerase chain reaction |
| EGA | embryonic genome activation |
| ETS | external transcribed spacer |
| r | ribosomal |
| SP | spacer promoter |
| TU | transcription unit |
| UCE | upstream control element |
References
- Potabattula, R.; Dittrich, M.; Böck, J.; Haertle, L.; Müller, T.; Hahn, T.; Schorsch, M.; Hajj, N.E.; Haaf, T. Allele-specific methylation of imprinted genes in fetal cord blood is influenced by cis-acting genetic variants and parental factors. Epigenomics 2018, 10, 1315–1326. [Google Scholar] [CrossRef]
- Potabattula, R.; Zacchini, F.; Ptak, G.E.; Dittrich, M.; Müller, T.; El Hajj, N.; Hahn, T.; Drummer, C.; Behr, R.; Lucas-Hahn, A.; et al. Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline. Aging Cell 2020, 19, e13181. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Zhou, Y.; Liu, S.; Jiang, J.; Bickhart, D.M.; Null, D.J.; Li, B.; Schroeder, S.G.; Rosen, B.D.; Cole, J.B.; et al. Comparative analyses of sperm DNA methylomes among human, mouse and cattle provide insights into epigenomic evolution and complex traits. Epigenetics 2019, 14, 260–276. [Google Scholar] [CrossRef]
- Moharrek, F.; Ingerslev, L.R.; Altıntaş, A.; Lundell, L.; Hansen, A.N.; Small, L.; Workman, C.T.; Barrès, R. Comparative analysis of sperm DNA methylation supports evolutionary acquired epigenetic plasticity for organ speciation. Epigenomics 2022, 14, 1305–1324. [Google Scholar] [CrossRef] [PubMed]
- Molaro, A.; Hodges, E.; Fang, F.; Song, Q.; McCombie, W.R.; Hannon, G.J.; Smith, A.D. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 2011, 146, 1029–1041. [Google Scholar] [CrossRef]
- Qu, J.; Hodges, E.; Molaro, A.; Gagneux, P.; Dean, M.D.; Hannon, G.J.; Smith, A.D. Evolutionary expansion of DNA hypomethylation in the mammalian germline genome. Genome Res. 2018, 28, 145–158. [Google Scholar] [CrossRef]
- Dittrich, M.; Bernhardt, L.; Penfold, C.A.; Boroviak, T.E.; Drummer, C.; Behr, R.; Müller, T.; Haaf, T. Age-related and species-specific methylation changes in the protein-coding marmoset sperm epigenome. Aging Cell 2024, 23, e14200. [Google Scholar] [CrossRef]
- Jenkins, T.G.; Carrell, D.T. The sperm epigenome and potential implications for the developing embryo. Reproduction 2012, 143, 727–734. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef]
- Teperek, M.; Simeone, A.; Gaggioli, V.; Miyamoto, K.; Allen, G.E.; Erkek, S.; Kwon, T.; Marcotte, E.M.; Zegerman, P.; Bradshaw, C.R.; et al. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016, 26, 1034–1046. [Google Scholar] [CrossRef]
- Martin, G.M. Geroscience: Addressing the mismatch between its exciting research opportunities, its economic imperative and its current funding crisis. Exp. Gerontol. 2017, 94, 46–51. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Geisen, A.B.C.; Santana Acevedo, N.; Oshima, J.; Dittrich, M.; Potabattula, R.; Haaf, T. rDNA copy number variation and methylation during normal and premature aging. Aging Cell 2025, 24, e14497. [Google Scholar] [CrossRef]
- Santoro, R.; Grummt, I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 2001, 8, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef]
- Potapova, T.; Kostos, P.; McKinney, S.; Borchers, M.; Haug, J.; Guarracino, A.; Solar, S.; Gogol, M.; Monfort Anez, G.; de Lima, L.G.; et al. Epigenetic control and inheritance of rDNA arrays. bioRxiv 2024. preprint. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Shao, F.; Liu, X.; Zhang, X.; Wang, Q.; Wang, W. Methylation of 45S ribosomal DNA (rDNA) is associated with cancer and aging in humans. Int. J. Genom. 2021, 2021, 8818007. [Google Scholar] [CrossRef]
- Yang, F.; Guo, X.; Bao, Y.; Li, R. The role of ribosomal DNA methylation in embryonic development, aging and diseases. Epigenet. Chromatin 2024, 17, 23. [Google Scholar] [CrossRef]
- Tiku, V.; Jain, C.; Raz, Y.; Nakamura, S.; Heestand, B.; Liu, W.; Späth, M.; Suchiman, H.E.D.; Müller, R.U.; Slagboom, P.E.; et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017, 8, 16083. [Google Scholar] [CrossRef]
- Buchwalter, A.; Hetzer, M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017, 8, 328. [Google Scholar] [CrossRef]
- D’Aquila, P.; Montesanto, A.; Mandalà, M.; Garasto, S.; Mari, V.; Corsonello, A.; Bellizzi, D.; Passarino, G. Methylation of the ribosomal RNA gene promoter is associated with aging and age-related decline. Aging Cell 2017, 16, 966–975. [Google Scholar] [CrossRef]
- Flunkert, J.; Maierhofer, A.; Dittrich, M.; Müller, T.; Horvath, S.; Nanda, I.; Haaf, T. Genetic and epigenetic changes in clonal descendants of irradiated human fibroblasts. Exp. Cell Res. 2018, 370, 322–332. [Google Scholar] [CrossRef]
- Oakes, C.C.; Smiraglia, D.J.; Plass, C.; Trasler, J.M.; Robaire, B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc. Natl. Acad. Sci. USA 2003, 100, 1775–1780. [Google Scholar] [CrossRef]
- Potabattula, R.; Trapphoff, T.; Dittrich, M.; Fic, K.; Ptak, G.E.; Dieterle, S.; Haaf, T. Ribosomal DNA methylation in human and mouse oocytes increases with age. Aging 2022, 14, 1214–1232. [Google Scholar] [CrossRef]
- Perry, A.C.F.; Asami, M.; Lam, B.Y.H.; Yeo, G.S.H. The initiation of mammalian embryonic transcription: To begin at the beginning. Trends Cell Biol. 2023, 33, 365–373. [Google Scholar] [CrossRef]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.Y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E.; et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef]
- Chebrout, M.; Koné, M.C.; Jan, H.U.; Cournut, M.; Letheule, M.; Fleurot, R.; Aguirre-Lavin, T.; Peynot, N.; Jouneau, A.; Beaujean, N.; et al. Transcription of rRNA in early mouse embryos promotes chromatin reorganization and expression of major satellite repeats. J. Cell Sci. 2022, 135, jcs258798. [Google Scholar] [CrossRef]
- Koné, M.C.; Fleurot, R.; Chebrout, M.; Debey, P.; Beaujean, N.; Bonnet-Garnier, A. Three-dimensional distribution of UBF and Nopp140 in relationship to ribosomal DNA transcription during mouse preimplantation development. Biol. Reprod. 2016, 94, 95. [Google Scholar] [CrossRef]
- Kresoja-Rakic, J.; Santoro, R. Nucleolus and rRNA gene chromatin in early embryo development. Trends Genet. 2019, 35, 868–879. [Google Scholar] [CrossRef]
- Braude, P.; Bolton, V.; Moore, S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 1988, 332, 459–461. [Google Scholar] [CrossRef]
- Furuta, A.; Nakamura, T. DNA hypomethylation circuit of mouse rDNA repeats in the germ cell lineage. Biochem. Biophys. Res. Commun. 2017, 490, 429–433. [Google Scholar] [CrossRef]
- Demetrius, L. Of mice and men: When it comes to studying ageing and the means to slow it down, mice are not just small humans. EMBO Rep. 2005, 6, S39–S44. [Google Scholar] [CrossRef]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; WHO Press: Geneva, Switzerland, 2010; Available online: https://apps.who.int/iris/handle/10665/44261 (accessed on 1 April 2025).
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef]
- Salim, D.; Gerton, J.L. Ribosomal DNA instability and genome adaptability. Chromosome Res. 2019, 27, 73–87. [Google Scholar] [CrossRef]
- Babaian, A. Intra- and inter-individual variation in human ribosomal RNAs. bioRxiv 2017. preprint. [Google Scholar] [CrossRef]
- Leitao, E.; Beygo, J.; Zeschnigk, M.; Klein-Hitpass, L.; Bargull, M.; Rahmann, S.; Horsthemke, B. Locus-specific DNA methylation analysis by targeted deep bisulfite sequencing. Meth. Mol. Biol. 2018, 1767, 351–366. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potabattula, R.; Dittrich, M.; Hahn, T.; Schorsch, M.; Ptak, G.E.; Haaf, T. rDNA Copy Number Variation and Methylation in Human and Mouse Sperm. Int. J. Mol. Sci. 2025, 26, 4197. https://doi.org/10.3390/ijms26094197
Potabattula R, Dittrich M, Hahn T, Schorsch M, Ptak GE, Haaf T. rDNA Copy Number Variation and Methylation in Human and Mouse Sperm. International Journal of Molecular Sciences. 2025; 26(9):4197. https://doi.org/10.3390/ijms26094197
Chicago/Turabian StylePotabattula, Ramya, Marcus Dittrich, Thomas Hahn, Martin Schorsch, Grazyna Ewa Ptak, and Thomas Haaf. 2025. "rDNA Copy Number Variation and Methylation in Human and Mouse Sperm" International Journal of Molecular Sciences 26, no. 9: 4197. https://doi.org/10.3390/ijms26094197
APA StylePotabattula, R., Dittrich, M., Hahn, T., Schorsch, M., Ptak, G. E., & Haaf, T. (2025). rDNA Copy Number Variation and Methylation in Human and Mouse Sperm. International Journal of Molecular Sciences, 26(9), 4197. https://doi.org/10.3390/ijms26094197




