Stevia Leaf Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T Displays Anticancer Activity to Pancreatic Cancer PANC-1 Cell Line
Abstract
1. Introduction
2. Results and Discussion
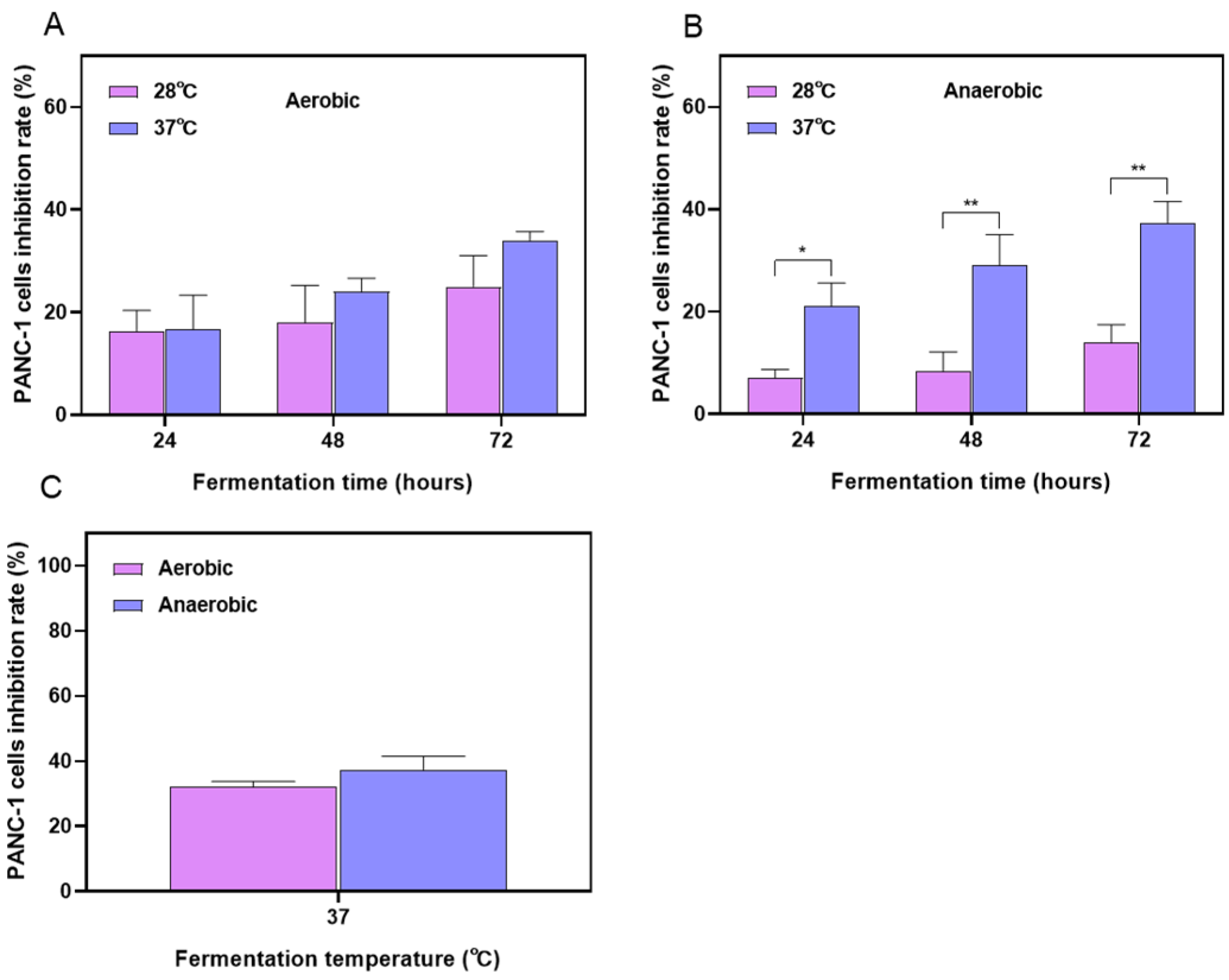
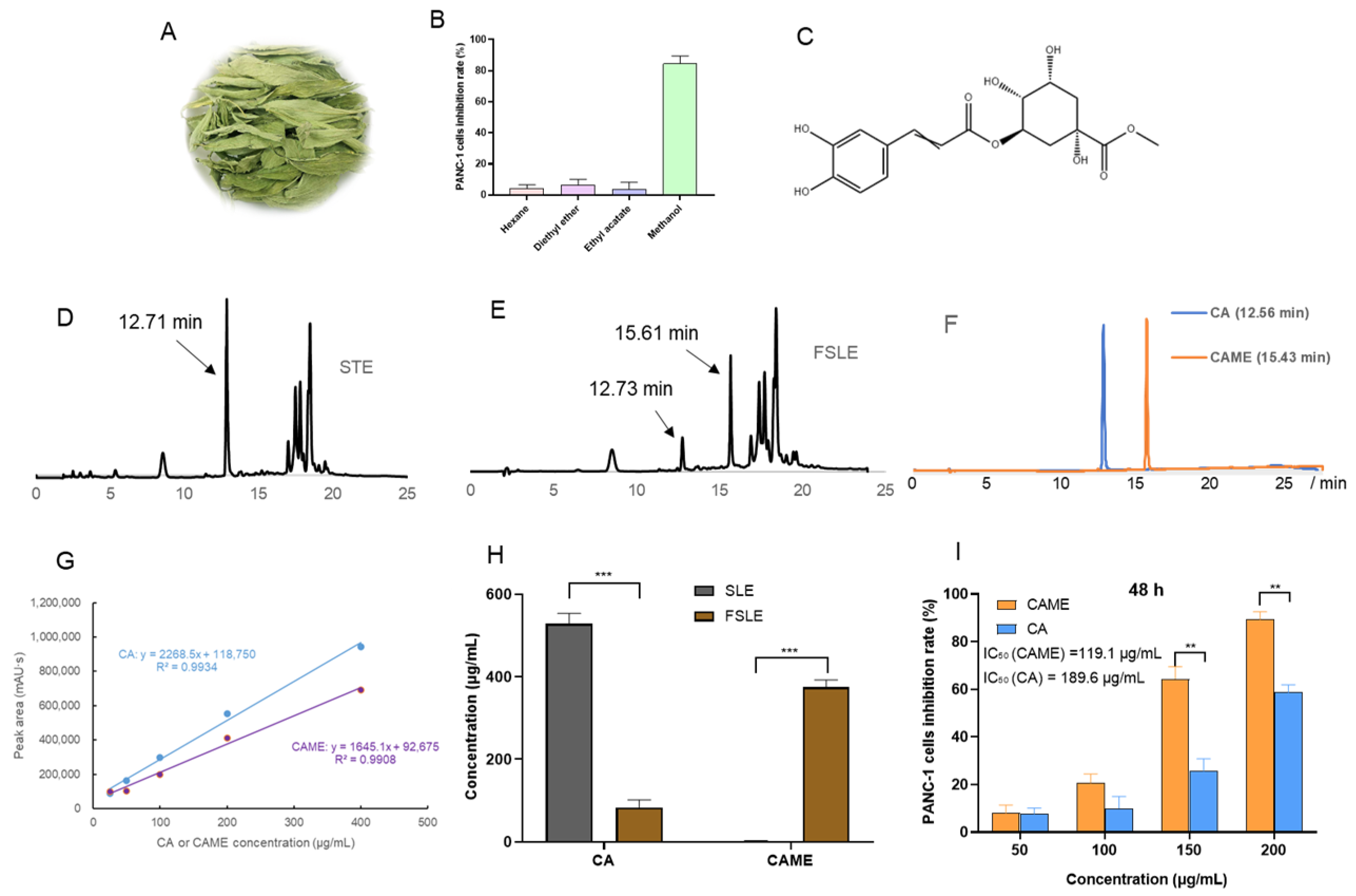
2.1. Optimization Results of the Fermentation Process for Stevia Leaf Extract
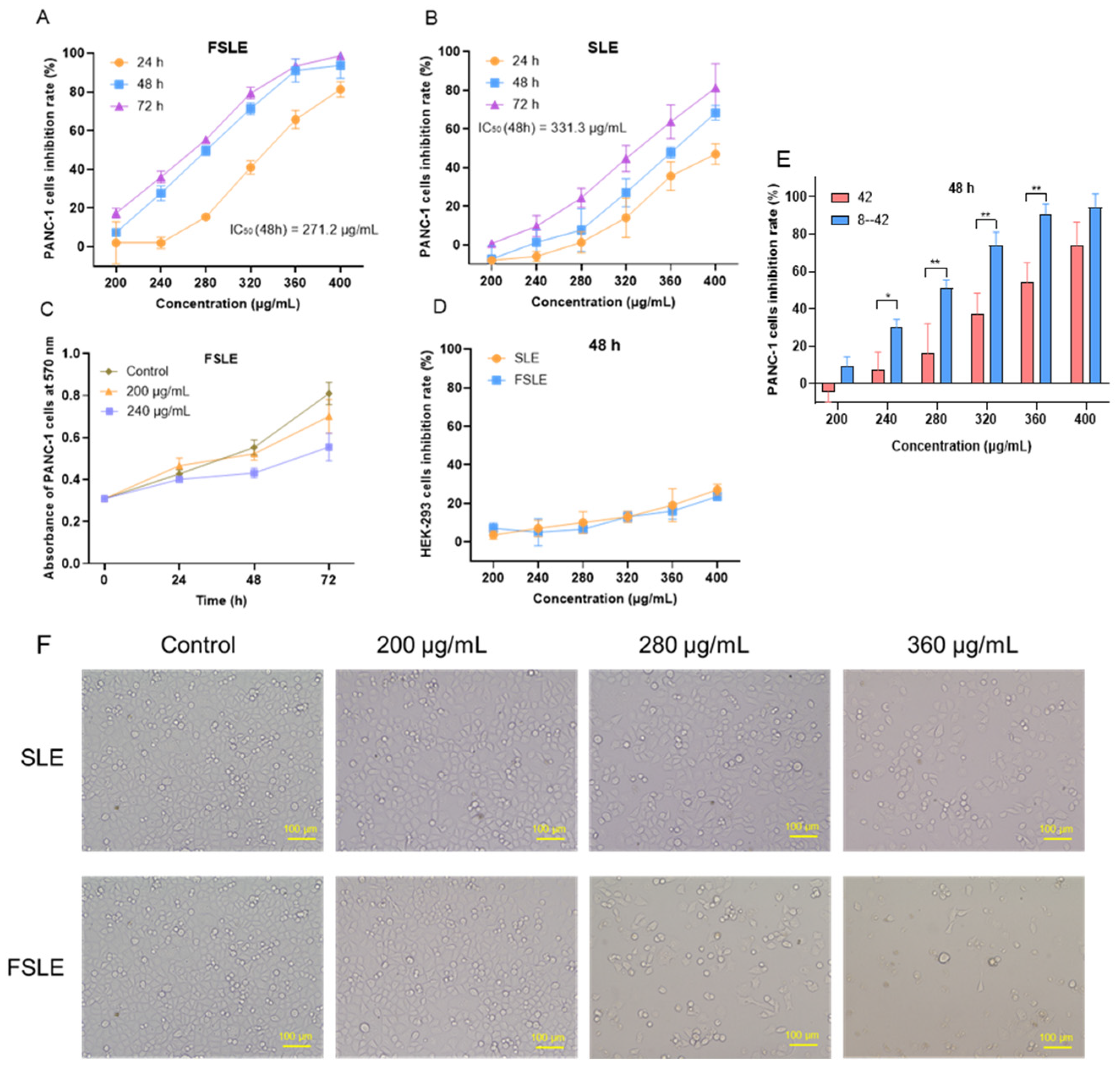
2.2. Cytotoxicity Evaluation In Vitro
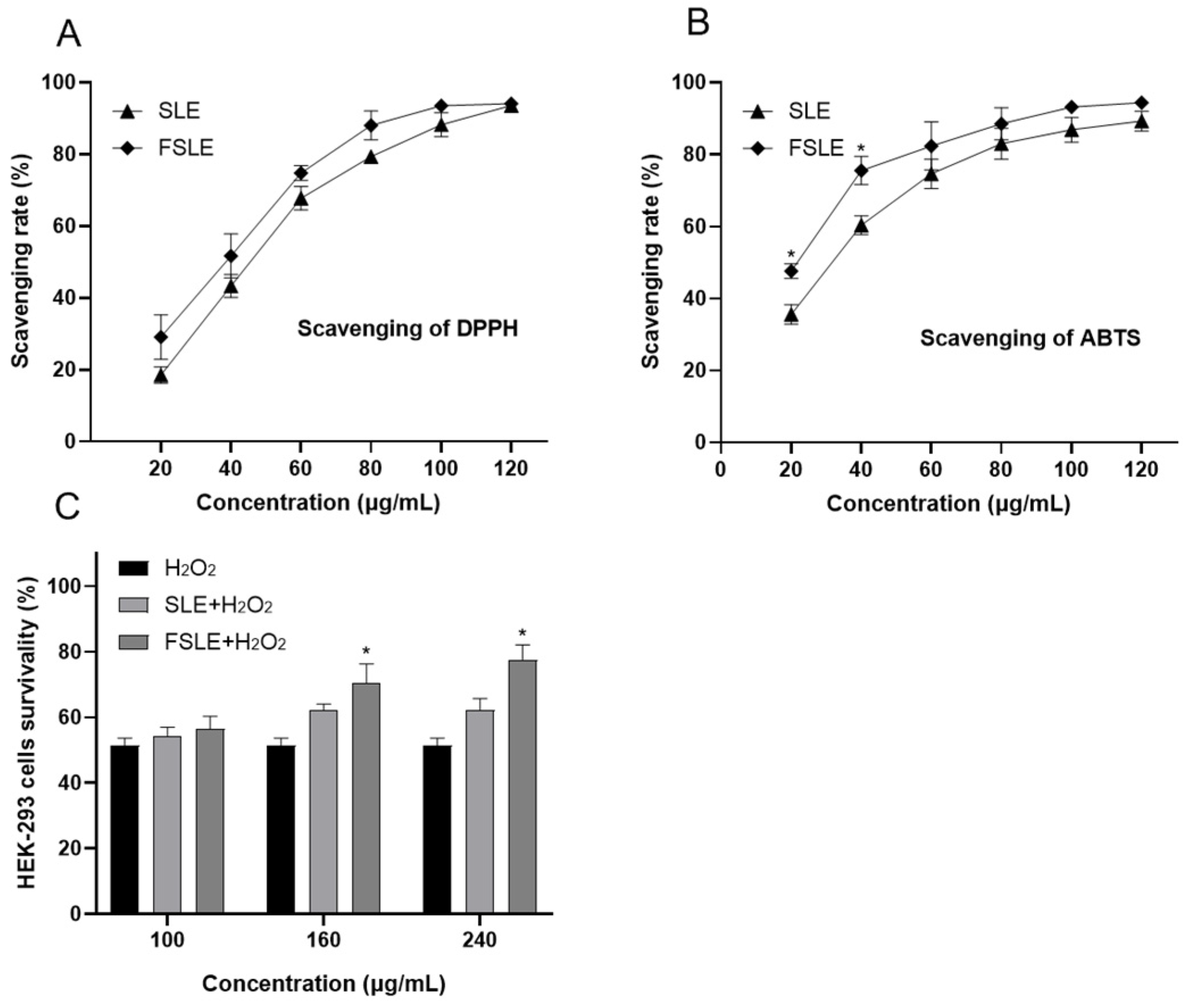
2.3. In Vitro Determination of Antioxidant Capacity
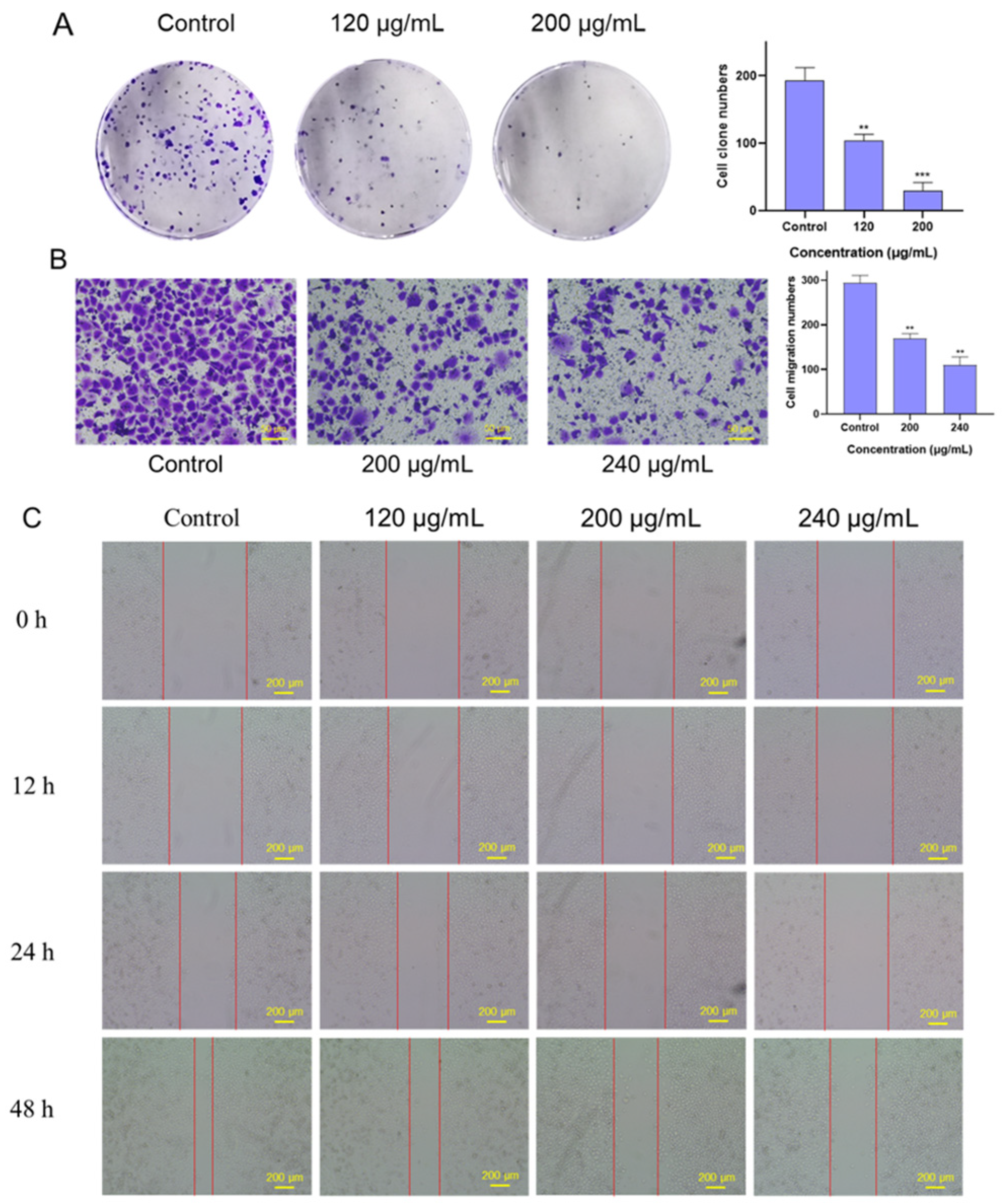
2.4. Effects of FSLE on the Proliferation and Migration of PANC-1 Cells
2.5. Analysis of Antitumor Active Components in FSLE
2.6. Effects of CAME on Cell Cycle and Apoptosis in PANC-1 Cells
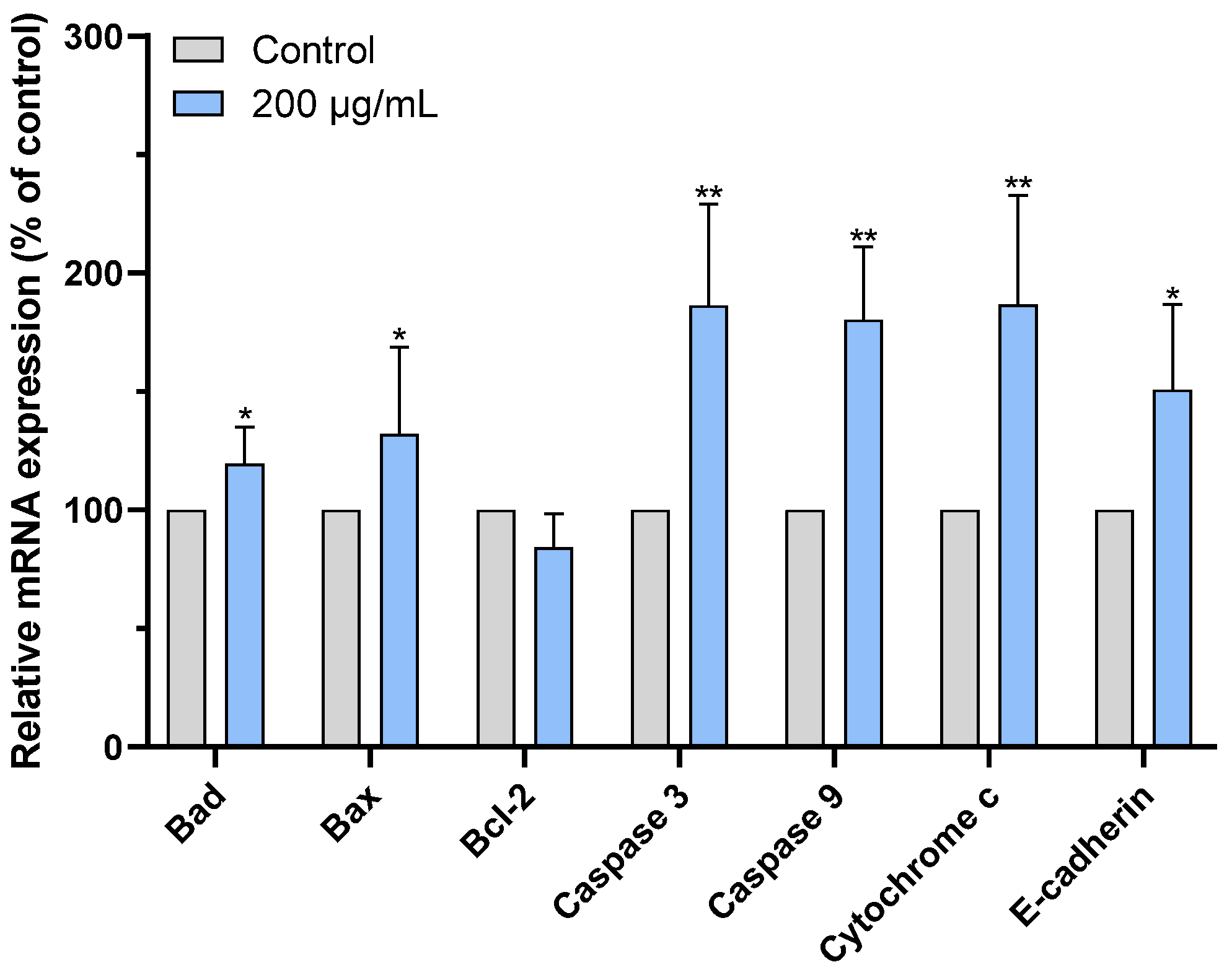
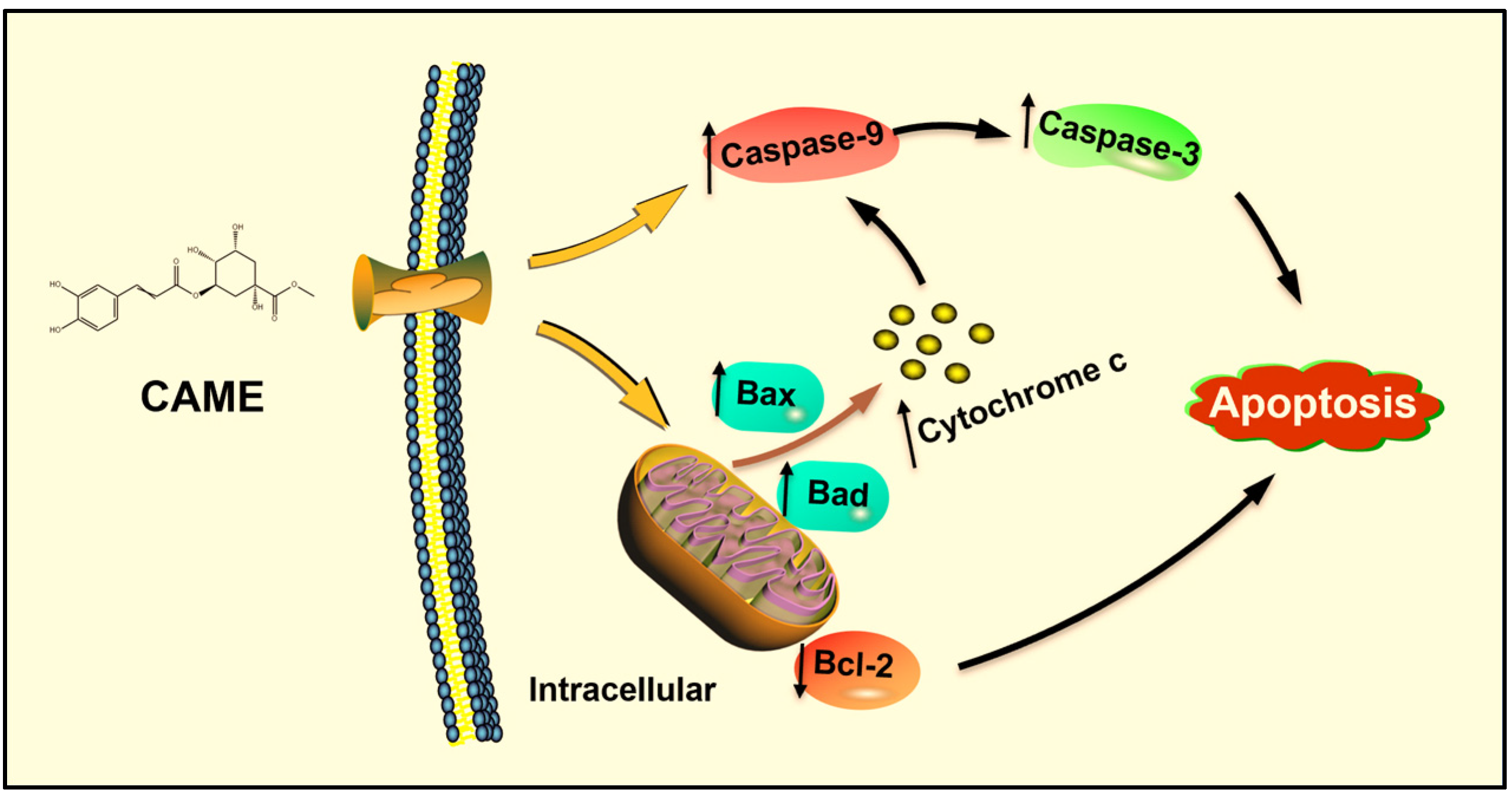
2.7. CAME Pro-Apoptotic Effects by Regulating the Expression of Apoptosis-Related Genes
3. Conclusions
4. Materials and Methods
4.1. Bacterial Cultivation and Fermentation Conditions
4.2. Optimization of Fermentation Conditions for Stevia Leaf Extract
4.3. Cell Culture and Cell Growth Inhibition Assay
4.4. Antioxidant Testing
4.4.1. Scavenging DPPH Free Radicals
4.4.2. Scavenging ABTS Free Radicals
4.4.3. Antioxidant Activity of HEK-293 Cells
4.5. Colony Formation Assay
4.6. Wound-Healing Assay
4.7. Migration Assay
4.8. Extraction and Identification of Active Compounds Produced in FSLE
4.9. Flow Cytometry Analysis
4.9.1. Cell Cycle Analysis
4.9.2. Cell Apoptosis Analysis
4.10. RNA Extraction and qRT-PCR Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Lyssiotis, C.A.; Pasca di Magliano, M.; Maitra, A. Pancreatic cancer: Advances and challenges. Cell 2023, 186, 1729–1754. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Cronin, K.A.; Scott, S.; Firth, A.U.; Sung, H.; Henley, S.J.; Sherman, R.L.; Siegel, R.L.; Anderson, R.N.; Kohler, B.A.; Benard, V.B.; et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 2022, 128, 4251–4284. [Google Scholar] [CrossRef]
- Groot, V.P.; Rezaee, N.; Wu, W.; Cameron, J.L.; Fishman, E.K.; Hruban, R.H.; Weiss, M.J.; Zheng, L.; Wolfgang, C.L.; He, J. Patterns, timing, and predictors of recurrence following pancreatectomy for pancreatic ductal adenocarcinoma. Ann. Surg. 2018, 267, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Farrell, J.J.; Elsaleh, H.; Garcia, M.; Lai, R.; Ammar, A.; Regine, W.F.; Abrams, R.; Benson, A.B.; Macdonald, J.; Cass, C.E.; et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology 2009, 136, 187–195. [Google Scholar] [CrossRef]
- Iatridis, N.; Kougioumtzi, A.; Vlataki, K.; Papadaki, S.; Magklara, A. Anti-cancer properties of stevia rebaudiana; more than a sweetener. Molecules 2022, 27, 1362. [Google Scholar] [CrossRef]
- Kasti, A.N.; Nikolaki, M.D.; Synodinou, K.D.; Katsas, K.N.; Petsis, K.; Lambrinou, S.; Pyrousis, I.A.; Triantafyllou, K. The effects of stevia consumption on gut bacteria: Friend or foe? Microorganisms 2022, 10, 744. [Google Scholar] [CrossRef]
- Martinez-Rojo, E.; Carino-Cortes, R.; Berumen, L.C.; Garcia-Alcocer, G.; Escobar-Cabrera, J. Stevia eupatoria and stevia pilosa extracts inhibit the proliferation and migration of prostate cancer cells. Medicina 2020, 56, 90. [Google Scholar] [CrossRef]
- Lopez, V.; Perez, S.; Vinuesa, A.; Zorzetto, C.; Abian, O. Stevia rebaudiana ethanolic extract exerts better antioxidant properties and antiproliferative effects in tumour cells than its diterpene glycoside stevioside. Food Funct. 2016, 7, 2107–2113. [Google Scholar] [CrossRef] [PubMed]
- Khare, N.; Chandra, S. Stevioside mediated chemosensitization studies and cytotoxicity assay on breast cancer cell lines MDA-MB-231 and SKBR3. Saudi J. Biol. Sci. 2019, 26, 1596–1601. [Google Scholar] [CrossRef] [PubMed]
- Mizushina, Y.; Akihisa, T.; Ukiya, M.; Hamasaki, Y.; Murakami-Nakai, C.; Kuriyama, I.; Takeuchi, T.; Sugawara, F.; Yoshida, H. Structural analysis of isosteviol and related compounds as DNA polymerase and DNA topoisomerase inhibitors. Life Sci. 2005, 77, 2127–2140. [Google Scholar] [CrossRef]
- Vasko, L.; Vaskova, J.; Fejercakova, A.; Mojzisova, G.; Poracova, J. Comparison of some antioxidant properties of plant extracts from Origanum vulgare, Salvia officinalis, Eleutherococcus senticosus and Stevia rebaudiana. In Vitro Cell Dev. Biol. Anim. 2014, 50, 614–622. [Google Scholar] [CrossRef]
- Chai, L.J.; Lan, T.; Cheng, Z.; Zhang, J.; Deng, Y.; Wang, Y.; Li, Y.; Wang, F.; Piao, M. Stevia rebaudiana leaves fermented by Lactobacillus plantarum exhibit resistance to microorganisms and cancer cell lines in vitro: A potential sausage preservative. Food Chem. 2024, 432, 137187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Guo, L.; Yu, D.; Du, X. New insights into immunomodulatory properties of lactic acid bacteria fermented herbal medicines. Front. Microbiol. 2022, 13, 1073922. [Google Scholar] [CrossRef]
- Fusco, V.; Fanelli, F.; Chieffi, D. Authenticity of probiotic foods and dietary supplements: A pivotal issue to address. Crit. Rev. Food Sci. Nutr. 2022, 62, 6854–6871. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, Z.; Ma, Y.; Du, K.; Sun, M.; Zhang, H.; Tu, H.; Jiang, X.; Lu, J.; Tu, L.; et al. Exopolysaccharide from Lactiplantibacillus plantarum YT013 and its apoptotic activity on gastric cancer AGS cells. Fermentation 2023, 9, 539. [Google Scholar] [CrossRef]
- Xiao, X.; Bai, J.; Zhang, J.; Wu, J.; Dong, Y. Inhibitory effect of fermented selected barley extracts with Lactobacillus plantarum dy-1 on the proliferation of human HT-29 Cells. J. Food Biochem. 2019, 43, e12989. [Google Scholar] [CrossRef]
- Lin, Y.-W.; Mou, Y.-C.; Su, C.-C.; Chiang, B.-H. Antihepatocarcinoma activity of lactic acid bacteria fermented Panax notoginseng. J. Agric. Food Chem. 2010, 58, 8528–8534. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.S.; Qi, S.S.; Wang, H.; Xiu, Z.L. Biotransformation of steriodal saponins in Dioscorea zingiberensis C. H. Wright to diosgenin by Trichoderma harzianum. Appl. Microbiol. Biotechnol. 2010, 85, 933–940. [Google Scholar] [CrossRef]
- Cardillo, A.B.; Perassolo, M.; Sartuqui, M.; Rodríguez Talou, J.; Giulietti, A.M. Production of tropane alkaloids by biotransformation using recombinant Escherichia coli whole cells. Biochem. Eng. J. 2017, 125, 180–189. [Google Scholar] [CrossRef]
- Dhiman, S.; Mukherjee, G.; Singh, A.K. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: A review. Int. Microbiol. 2018, 21, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Bodh, U.; Gupta, R. Isolation of a novel lipase producing fungal isolate Aspergillus fumigatus and production optimization of enzyme. Biocatal. Biotransform. 2018, 36, 450–457. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, Z.; Ma, Y.; Du, K.; Sun, M.; Zhang, H.; Tu, H.; Jiang, X.; Lu, J.; Tu, L.; et al. Production of the exopolysaccharide from Lactiplantibacillus plantarum YT013 under different growth conditions: Optimum parameters and mathematical analysis. Int. J. Food Prop. 2023, 26, 1941–1952. [Google Scholar] [CrossRef]
- Jiang, S.L.; Liu, H.J.; Liu, Z.C.; Liu, N.; Liu, R.; Kang, Y.R.; Ji, J.G.; Zhang, C.; Hua, B.J.; Kang, S.J. Adjuvant effects of fermented red ginseng extract on advanced non-small cell lung cancer patients treated with chemotherapy. Chin. J. Integr. Med. 2017, 23, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Le, B.; Ngoc, A.P.T.; Yang, S.H. Synbiotic fermented soymilk with Weissella cibaria FB069 and xylooligosaccharides prevents proliferation in human colon cancer cells. J. Appl. Microbiol. 2020, 128, 1486–1496. [Google Scholar] [CrossRef]
- Lizardo, R.C.M.; Cho, H.D.; Lee, J.H.; Won, Y.S.; Seo, K.I. Extracts of Elaeagnus multiflora Thunb. fruit fermented by lactic acid bacteria inhibit SW480 human colon adenocarcinoma via induction of cell cycle arrest and suppression of metastatic potential. J. Food Sci. 2020, 85, 2565–2577. [Google Scholar] [CrossRef]
- Retamozo, M.H.; Silva, C.C.; Tamayose, C.I.; Carvalho, J.C.S.; Romoff, P.; Fávero, O.A.; Ferreira, M.J.P. Chemical constituents from leaves of Baccharis sphenophylla (Asteraceae) and their antioxidant effects. Plants 2023, 12, 1262. [Google Scholar] [CrossRef]
- Latha, S.; Chaudhary, S.; Ray, R.S. Hydroalcoholic extract of Stevia rebaudiana bert. leaves and stevioside ameliorates lipopolysaccharide induced acute liver injury in rats. Biomed. Pharmacother. 2017, 95, 1040–1050. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, M.; Gao, L.; Bao, Y.; Li, P.; Wang, M.; Shao, T.; Wang, G.; Liu, C. Optimal extraction of polysaccharides from Stevia rebaudiana roots for protection against hydrogen peroxide-induced oxidative damage in RAW264.7 cells. Nat. Prod. Res. 2023, 38, 3865–3869. [Google Scholar] [CrossRef] [PubMed]
- Entschladen, F.; Drell, T.L.; Lang, K.; Joseph, J.; Zaenker, K.S. Tumour-cell migration, invasion, and metastasis: Navigation by neurotransmitters. Lancet Oncol. 2004, 5, 254–258. [Google Scholar] [CrossRef]
- Mostafa, E.M.; Mohammed, H.A.; Musa, A.; Abdelgawad, M.A.; Al-Sanea, M.M.; Almahmoud, S.A.; Ghoneim, M.M.; Gomaa, H.A.M.; Rahman, F.; Shalaby, K.; et al. In vitro anti-proliferative, and kinase inhibitory activity of phenanthroindolizidine alkaloids isolated from tylophora indica. Plants 2022, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Xiong, T.; Peng, Z.; Xiao, Y.S.; Liu, Z.G.; Hu, M.; Xie, M.Y. Genomic analysis revealed adaptive mechanism to plant-related fermentation of Lactobacillus plantarum NCU116 and Lactobacillus spp. Genomics 2020, 112, 703–711. [Google Scholar] [CrossRef]
- Chen, X.; Liu, B.; Tong, J.; Bo, J.; Feng, M.; Yin, L.; Lin, X. Chlorogenic acid inhibits proliferation, migration and invasion of pancreatic cancer cells via AKT/GSK-3β/β-catenin signaling pathway. Recent Pat. Anti-Cancer 2024, 19, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Meng, X.; Chen, S.; Huang, D.; Xia, Y.; Zhu, S. Antioxidant activities of chlorogenic acid derivatives with different acyl donor chain lengths and their stabilities during in vitro simulated gastrointestinal digestion. Food Chem. 2021, 357, 129904. [Google Scholar] [CrossRef]
- Phang, C.W.; Karsani, S.A.; Abd Malek, S.N. Induction of apoptosis and cell cycle arrest by flavokawain C on HT-29 human colon adenocarcinoma via enhancement of reactive oxygen species generation, upregulation of p21, p27, and GADD153, and inactivation of inhibitor of apoptosis proteins. Pharmacogn. Mag. 2017, 13 (Suppl. S2), S321–S328. [Google Scholar] [CrossRef]
- Lu, C.H.; Kuo, Y.Y.; Lin, G.B.; Chen, W.T.; Chao, C.Y. Application of non-invasive low-intensity pulsed electric field with thermal cycling-hyperthermia for synergistically enhanced anticancer effect of chlorogenic acid on PANC-1 cells. PLoS ONE 2020, 15, e0222126. [Google Scholar] [CrossRef]
- Kapoor, I.; Bodo, J.; Hill, B.T.; Hsi, E.D.; Almasan, A. Targeting BCL-2 in B-cell malignancies and overcoming therapeutic resistance. Cell Death Dis. 2020, 11, 941. [Google Scholar] [CrossRef]
- Antonsson, B. Bax and other pro-apoptotic Bcl-2 family “killer-proteins” and their victim the mitochondrion. Cell Tissue Res. 2001, 306, 347–361. [Google Scholar] [CrossRef]
- Gross, A.; McDonnell, J.M.; Korsmeyer, S.J. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999, 13, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L.; Korsmeyer, S.J. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem. Biophys. Res. Commun. 2003, 304, 437–444. [Google Scholar] [CrossRef]
- Hastak, K.; Gupta, S.; Ahmad, N.; Agarwal, M.K.; Agarwal, M.L.; Mukhtar, H. Role of p53 and NF-kappaB in epigallocatechin-3-gallate-induced apoptosis of LNCaP cells. Oncogene 2003, 22, 4851–4859. [Google Scholar] [CrossRef] [PubMed]
- Higashikawa, F.; Danshiitsoodol, N.; Kanno, K.; Ishida, R.; Tazuma, S.; Sugiyama, M. Lactobacillus plantarum SN13T cells improve hepatic dysfunction and fecal microbiota: A randomized pilot study. Arch. Clin. Biomed. Res. 2020, 4, 605–625. [Google Scholar] [CrossRef]
- Okamoto, T.; Sugimoto, S.; Noda, M.; Yokooji, T.; Danshiitsoodol, N.; Higashikawa, F.; Sugiyama, M. Interleukin-8 release inhibitors generated by fermentation of artemisia princeps pampanini herb extract with Lactobacillus plantarum SN13T. Front. Microbiol. 2020, 11, 1159. [Google Scholar] [CrossRef]
- Shakya, S.; Danshiitsoodol, N.; Noda, M.; Sugiyama, M. Role of phenolic acid metabolism in enhancing bioactivity of mentha extract fermented with plant-derived Lactobacillus plantarum SN13T. Probiotics Antimicrob. Proteins 2023, 16, 1052–1064. [Google Scholar] [CrossRef]
- Shakya, S.; Danshiitsoodol, N.; Sugimoto, S.; Noda, M.; Sugiyama, M. Anti-oxidant and anti-inflammatory substance generated newly in Paeoniae Radix alba extract fermented with plant-derived Lactobacillus brevis 174A. Antioxidants 2021, 10, 1071. [Google Scholar] [CrossRef]
- Benov, L. Improved formazan dissolution for bacterial MTT assay. Microbiol. Spectr. 2021, 9, e01637-21. [Google Scholar] [CrossRef]

| Genes | 5′-3′ Primer Pairs | Primer Sequences (5′-3′) |
|---|---|---|
| E-cadherin | Forward | GCCTCCTGAAAAGAGAGTGGAAG |
| Reverse | TGGCAGTGTCTCTCCAAATCCG | |
| Bcl-2 | Forward | ATCGCCCTGTGGATGACTGAGT |
| Reverse | GCCAGGAGAAATCAAACAGAGGC | |
| Bax | Forward | TCAGGATGCGTCCACCAAGAAG |
| Reverse | TGTGTCCACGGCGGCAATCATC | |
| Bad | Forward | CCAACCTCTGGGCAGCACAGC |
| Reverse | TTTGCCGCATCTGCGTTGCTGT | |
| Caspase-3 | Forward | GGAAGCGAATCAATGGACTCTGG |
| Reverse | GCATCGACATCTGTACCAGACC | |
| Caspase-9 | Forward | GTTTGAGGACCTTCGACCAGCT |
| Reverse | CAACGTACCAGGAGCCACTCTT | |
| GAPDH | Forward | GTCTCCTCTGACTTCAACAGCG |
| Reverse | ACCACCCTGTTGCTGTAGCCAA | |
| Cytochromec | Forward | AAGGGAGGCAAGCACAAGACTG |
| Reverse | CTCCATCAGTGTATCCTCTCCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Danshiitsoodol, N.; Noda, M.; Yonezawa, S.; Kanno, K.; Sugiyama, M. Stevia Leaf Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T Displays Anticancer Activity to Pancreatic Cancer PANC-1 Cell Line. Int. J. Mol. Sci. 2025, 26, 4186. https://doi.org/10.3390/ijms26094186
Zhang R, Danshiitsoodol N, Noda M, Yonezawa S, Kanno K, Sugiyama M. Stevia Leaf Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T Displays Anticancer Activity to Pancreatic Cancer PANC-1 Cell Line. International Journal of Molecular Sciences. 2025; 26(9):4186. https://doi.org/10.3390/ijms26094186
Chicago/Turabian StyleZhang, Rentao, Narandalai Danshiitsoodol, Masafumi Noda, Sayaka Yonezawa, Keishi Kanno, and Masanori Sugiyama. 2025. "Stevia Leaf Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T Displays Anticancer Activity to Pancreatic Cancer PANC-1 Cell Line" International Journal of Molecular Sciences 26, no. 9: 4186. https://doi.org/10.3390/ijms26094186
APA StyleZhang, R., Danshiitsoodol, N., Noda, M., Yonezawa, S., Kanno, K., & Sugiyama, M. (2025). Stevia Leaf Extract Fermented with Plant-Derived Lactobacillus plantarum SN13T Displays Anticancer Activity to Pancreatic Cancer PANC-1 Cell Line. International Journal of Molecular Sciences, 26(9), 4186. https://doi.org/10.3390/ijms26094186





