Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study
Abstract
1. Introduction
2. Results
2.1. Genetic Diversity
2.2. Bottleneck Test and Effective Population Size Analysis
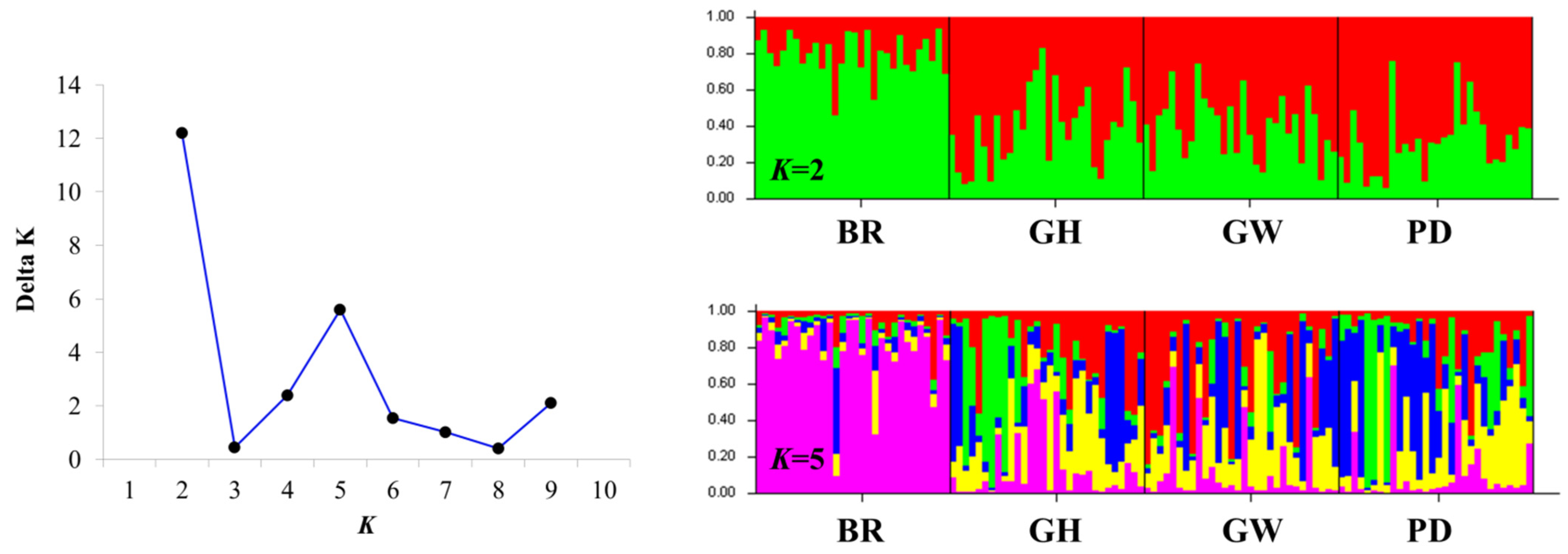
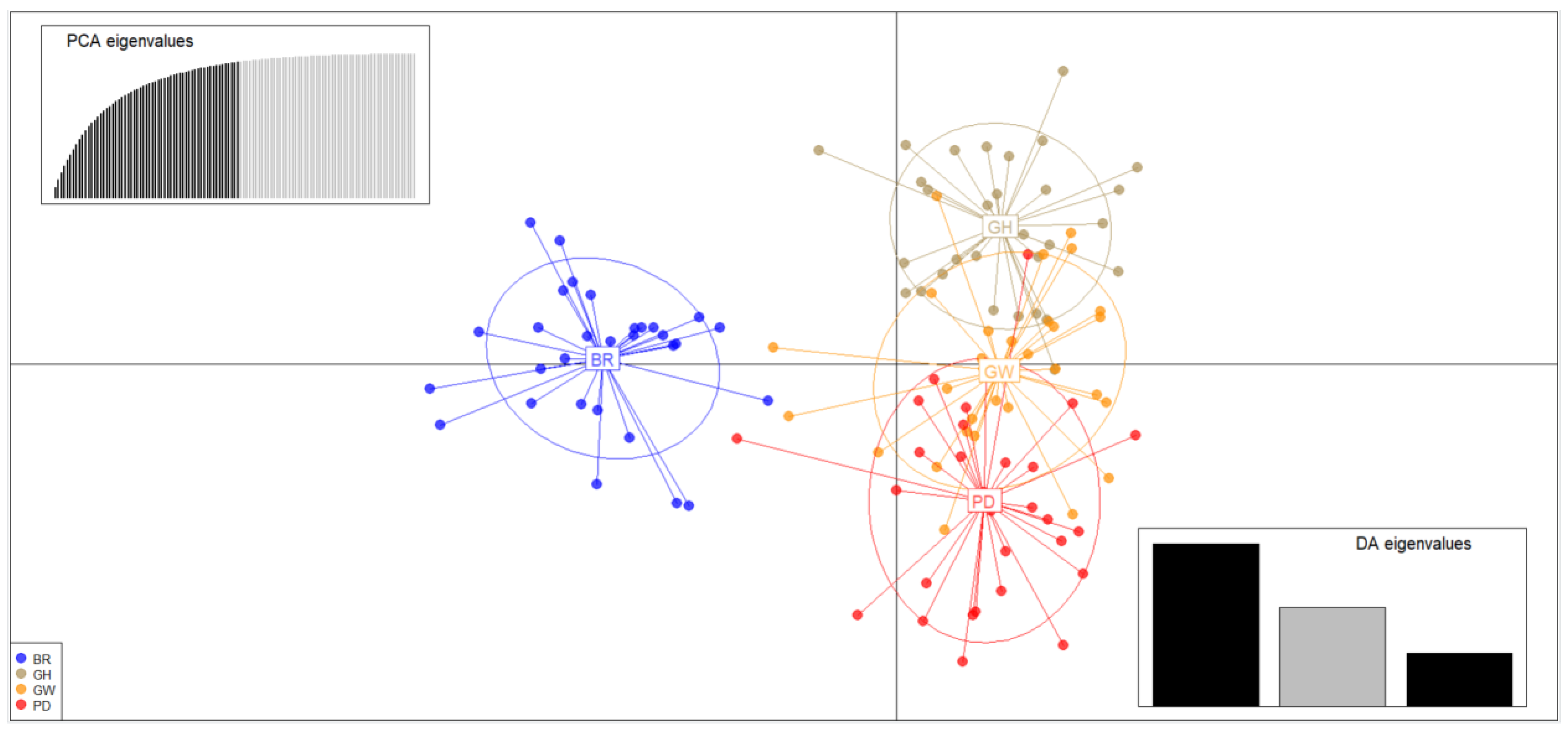
2.3. Population Structure and Genetic Differentiation Analyses
3. Discussion
3.1. Genetic Diversity
3.2. Genetic Structure
4. Materials and Methods
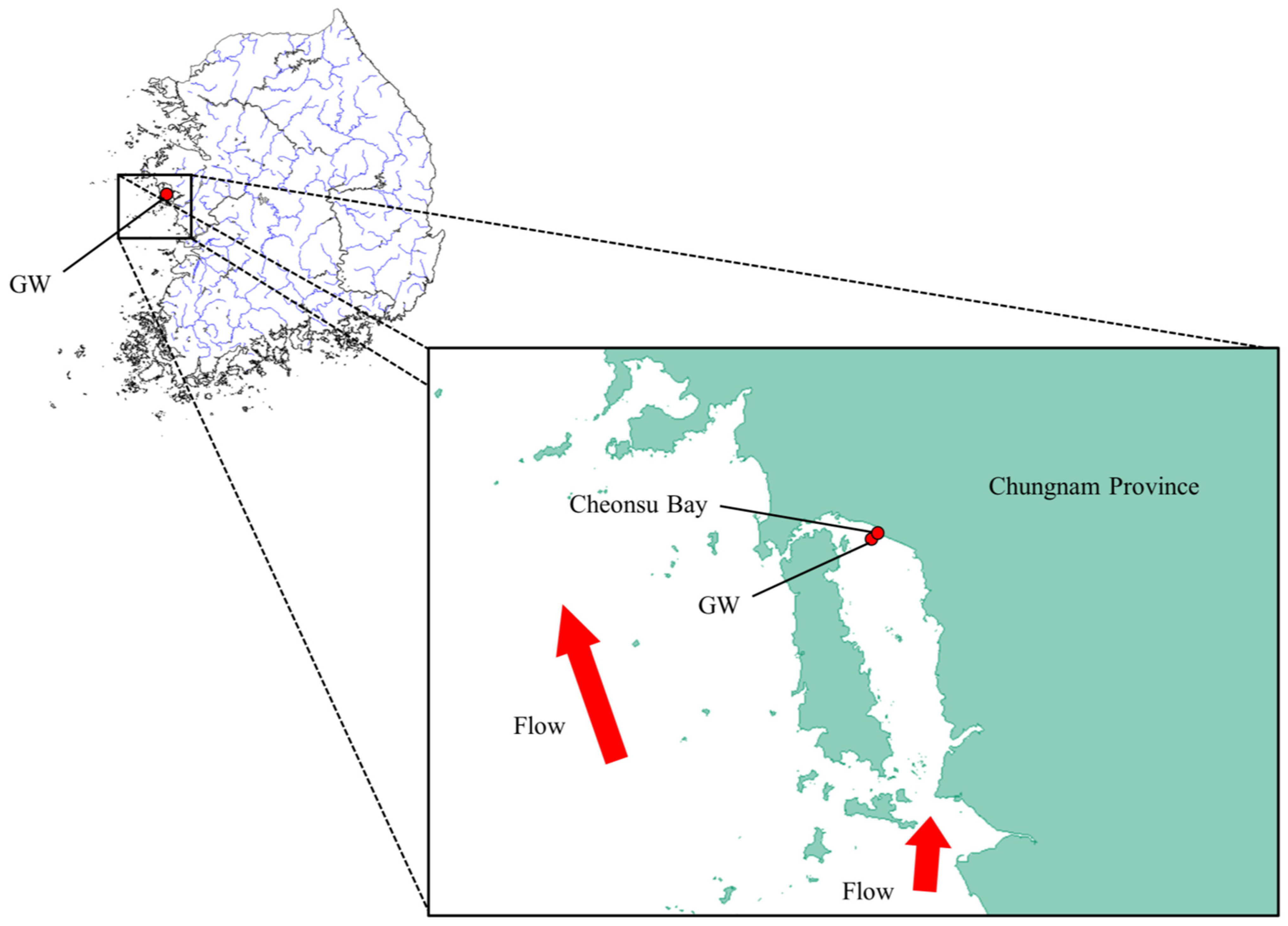
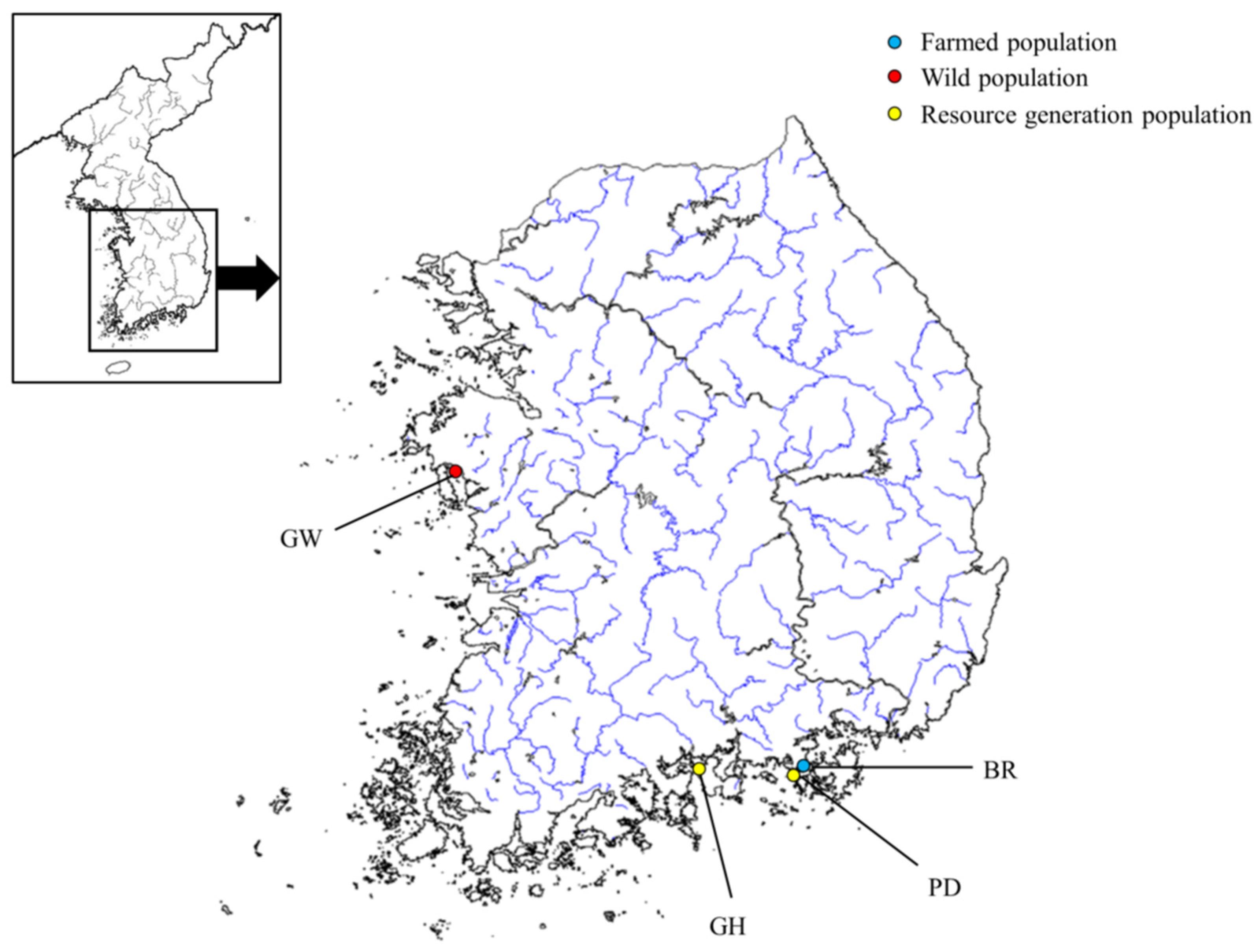
4.1. Sampling and DNA Extraction
4.2. Microsatellite Genotyping
4.3. Genetic Diversity Analysis
4.4. Population Genetic Structure Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Q.; Yu, H.; Yu, R. Genetic variability assessed by microsatellites in cultured populations of the Pacific oyster (Crassostrea gigas) in China. Aquaculture 2006, 259, 95–102. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Xu, C.; Li, Q. Comparative analysis of genetic diversity and structure among four shell color strains of the Pacific oyster Crassostrea gigas based on the mitochondrial COI gene and microsatellites. Aquaculture 2023, 563, 738990. [Google Scholar] [CrossRef]
- An, H.S.; Lee, J.W.; Kim, W.J.; Lim, H.J.; Kim, E.M.; Byun, S.G.; An, C.M. Comparative genetic diversity of wild and hatchery-produced Pacific oyster (Crassostrea gigas) populations in Korea using multiplex PCR assays with nine polymorphic microsatellite markers. Genes Genom. 2013, 35, 805–815. [Google Scholar] [CrossRef]
- Meistertzheim, A.L.; Arnaud-Haond, S.; Boudry, P.; Thébault, M.T. Genetic structure of wild European populations of the invasive Pacific oyster Crassostrea gigas due to aquaculture practices. Mar. Biol. 2013, 160, 453–463. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Frankham, R.; Bradshaw, C.J.A.; Brook, B.W. Genetics in conservation management: Revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol. Conserv. 2014, 170, 56–63. [Google Scholar] [CrossRef]
- Kim, K.R.; Kim, K.Y.; Song, H.Y. Genetic Structure and Diversity of Hatchery and Wild Populations of Yellow Catfish Tachysurus fulvidraco (Siluriformes: Bagridae) from Korea. Int. J. Mol. Sci. 2024, 25, 3923. [Google Scholar] [CrossRef] [PubMed]
- Appleyard, S.A.; Ward, R.D. Genetic diversity and effective population size in mass selection lines of Pacific oyster (Crassostrea gigas). Aquaculture 2006, 254, 148–159. [Google Scholar] [CrossRef]
- Yu, H.; Li, Q. Genetic variation of wild and hatchery populations of the Pacific oyster Crassostrea gigas assessed by microsatellite markers. J. Genet. Genom. 2007, 34, 1114–1122. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, C.; Li, Q. Genetic diversity in a genetically improved line of the Pacific oyster Crassostrea gigas with orange shell based on microsatellites and mtDNA data. Aquaculture 2022, 549, 737791. [Google Scholar] [CrossRef]
- Izquierdo, M.S.; Fernandez-Palacios, H.; Tacon, A.G.J. Effect of broodstock nutrition on reproductive performance of fish. Aquaculture 2022, 197, 25–42. [Google Scholar] [CrossRef]
- Doyle, R.M.; Perez-Enriquez, R.; Takagi, M.; Taniguchi, N. Selective recovery of founder genetic diversity in aquacultural broodstocks and captive, endangered fish populations. Genetica 2001, 111, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Van Doornik, D.M.; Kozfkay, C.C.; Waples, R.S. Genetic diversity in the Snake River sockeye salmon captive broodstock program as estimated from broodstock records. Conserv. Genet. 2012, 13, 1183–1193. [Google Scholar] [CrossRef]
- Teterina, A.A.; Willis, J.H.; Lukac, M.; Jovelin, R.; Cutter, A.D.; Phillips, P.C. Genomic diversity landscapes in outcrossing and selfing Caenorhabditis nematodes. PLoS Genet. 2023, 19, e1010879. [Google Scholar] [CrossRef]
- Glemin, S.; Galtier, N. Genome evolution in outcrossing versus selfing versus asexual species. Evol. Genom. 2012, 1, 311–335. [Google Scholar] [CrossRef]
- Neaves, L.E.; Eales, J.; Whitlock, R.; Hollingsworth, P.M.; Burke, T.; Pullin, A.S. The fitness consequences of inbreeding in natural populations and their implications for species conservation–a systematic map. Environ. Evid. 2015, 4, 5. [Google Scholar] [CrossRef]
- Geletu, T.T.; Zhao, J. Genetic resources of Nile tilapia (Oreochromis niloticus Linnaeus, 1758) in its native range and aquaculture. Hydrobiologia 2023, 850, 2425–2445. [Google Scholar] [CrossRef]
- Frankham, R. Conservation genetics. In Encyclopedia of Genetics; Routledge: New York, NY, USA, 2014; pp. 910–914. [Google Scholar]
- Sui, L.; Zhang, F.; Wang, X.; Bossier, P.; Sorgeloos, P.; Hänfling, B. Genetic diversity and population structure of the Chinese mitten crab Eriocheir sinensis in its native range. Mar. Biol. 2009, 156, 1573–1583. [Google Scholar] [CrossRef]
- Ye, Y.; Ren, W.; Zhang, S.; Zhao, L.; Tang, J.; Hu, L.; Chen, X. Genetic diversity of fish in aquaculture and of common carp (Cyprinus carpio) in Traditional Rice–Fish Coculture. Agriculture 2022, 12, 997. [Google Scholar] [CrossRef]
- Chung, J.R.; Kwak, H.S. Growth of south and west coast pacific oyster spats (Crassostrea gigas) after cross-transplantation. Bull. Korean Fish. Soc. 1970, 3, 129–136. [Google Scholar]
- Lee, H.M.; Yoon, G.T. Comparison of Recruitment and Growth Patterns of Pacific Oysters (Crassostrea gigas) between a Natural Rocky Shore and Farming Substrate Within an Oil Spill Contaminated Area of Korea. Ocean Polar Res. 2014, 36, 145–156. [Google Scholar] [CrossRef]
- Chu, D.; Pan, H.P.; Li, X.C.; Guo, D.; Tao, Y.L.; Liu, B.M.; Zhang, Y.J. Spatial genetic heterogeneity in populations of a newly invasive whitefly in China revealed by a nation-wide field survey. PLoS ONE 2013, 8, e79997. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Santiago, E.; Caballero, A. Prediction and estimation of effective population size. Heredity 2016, 117, 193–206. [Google Scholar] [CrossRef]
- Mable, B.K. Conservation of adaptive potential and functional diversity: Integrating old and new approaches. Conserv. Genet. 2019, 20, 89–100. [Google Scholar] [CrossRef]
- Schumer, M.; Cui, R.; Rosenthal, G.G.; Andolfatto, P. Reproductive isolation of hybrid populations driven by genetic incompatibilities. PLoS Genet. 2015, 11, e1005041. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Song, K.; Li, C.; Wang, W.; Li, B.; Li, L.; Zhang, G. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas). PLoS ONE 2017, 12, e0174007. [Google Scholar] [CrossRef]
- Tournebize, R.; Chu, G.; Moorjani, P. Reconstructing the history of founder events using genome-wide patterns of allele sharing across individuals. PLoS Genet. 2022, 18, e1010243. [Google Scholar] [CrossRef]
- Dong, C.M.; Lee, H.M.; Lee, M.N.; Noh, E.S.; Nam, B.H.; Kim, Y.O.; Kim, E.M. Development of new Microsatellite DNA Markers Using Next-generation Sequencing in pacific oyster Crassostrea gigas. Korean J. Malacol. 2022, 38, 129–138. [Google Scholar]
- Li, G.; Hubert, S.; Bucklin, K.; Ribes, V.; Hedgecock, D. Characterization of 79 microsatellite DNA markers in the Pacific oyster Crassostrea gigas. Mol. Ecol. Notes 2003, 3, 228–232. [Google Scholar] [CrossRef]
- Currie-Fraser, E.; Shah, P.; True, S. Data analysis using GeneMapper® v4. 1: Comparing the newest generation of GeneMapper software to legacy Genescan® and Genotyper® Software. J. Biomol. Tech. 2010, 21, S31. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.B.K.E.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. Population genetics software for exact test and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Maruyama, T.; Fuerst, P.A. Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 1985, 111, 675–689. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.L.; Liu, J.X. StructureSelector: A web based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
| ID | Location | N | NA | HO | HE | PHWE | FIS |
|---|---|---|---|---|---|---|---|
| BR | farmed population (Japan, Hiroshima) | 30 | 5.50 | 0.494 | 0.507 | 0.219 | 0.022 |
| GH | Galhwa | 30 | 7.88 | 0.473 | 0.621 | 0.000 *** | 0.242 *** |
| GW | Ganwoldo Island | 30 | 8.50 | 0.437 | 0.604 | 0.000 *** | 0.280 *** |
| PD | Pildo | 30 | 8.44 | 0.455 | 0.667 | 0.000 *** | 0.269 *** |
| Population ID | N | Wilcoxon Signed-Rank Test | Ne | (95% CI) | |||
|---|---|---|---|---|---|---|---|
| PIAM | PTPM | PSMM | Mode-Shift | ||||
| BR | 30 | 0.000 *** | 0.000 *** | 0.011 * | Shifted | 105 | (47–∞) |
| GH | 30 | 0.000 *** | 0.068 | 0.135 | L-shaped | 82 | (48–223) |
| GW | 30 | 0.000 *** | 0.532 | 0.978 | L-shaped | 678 | (104–∞) |
| PD | 30 | 0.000 *** | 0.104 | 0.208 | L-shaped | 403 | (99–∞) |
| BR | GH | GW | PD | |
|---|---|---|---|---|
| BR | - | 0.000 | 0.000 | 0.000 |
| GH | 0.044 * | - | 0.087 | 0.516 |
| GW | 0.040 * | 0.010 | - | 0.184 |
| PD | 0.048 * | 0.004 | 0.009 | - |
| Location Information | Code | Location Name | N | Location |
|---|---|---|---|---|
| Farmed * | BR | farmed | 30 | 34°47′30′′ N 128°25′46′′ E |
| Resource generation population 1 | GH | Galhwa | 30 | 34°53′57′′ N 127°50′06′′ E |
| Resource generation population 1 | PD | Pildo | 30 | 34°51′11′′ N 128°20′39′′ E |
| Wild population | GW | Ganwoldo Island | 30 | 36°36′17′′ N 126°24′37′′ E |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-R.; Park, J.-W.; Park, K.-I.; Lee, H.-J. Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study. Int. J. Mol. Sci. 2025, 26, 4172. https://doi.org/10.3390/ijms26094172
Kim K-R, Park J-W, Park K-I, Lee H-J. Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study. International Journal of Molecular Sciences. 2025; 26(9):4172. https://doi.org/10.3390/ijms26094172
Chicago/Turabian StyleKim, Kang-Rae, Jong-Won Park, Kyung-Il Park, and Hee-Jung Lee. 2025. "Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study" International Journal of Molecular Sciences 26, no. 9: 4172. https://doi.org/10.3390/ijms26094172
APA StyleKim, K.-R., Park, J.-W., Park, K.-I., & Lee, H.-J. (2025). Genetic Diversity and Population Structure in Farmed and Wild Pacific Oysters (Crassostrea gigas): A Comparative Study. International Journal of Molecular Sciences, 26(9), 4172. https://doi.org/10.3390/ijms26094172







