High Copy Number Variations Correlate with a Pro-Tumoral Microenvironment and Worse Prognosis in Acral Lentiginous Melanoma
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Characteristics
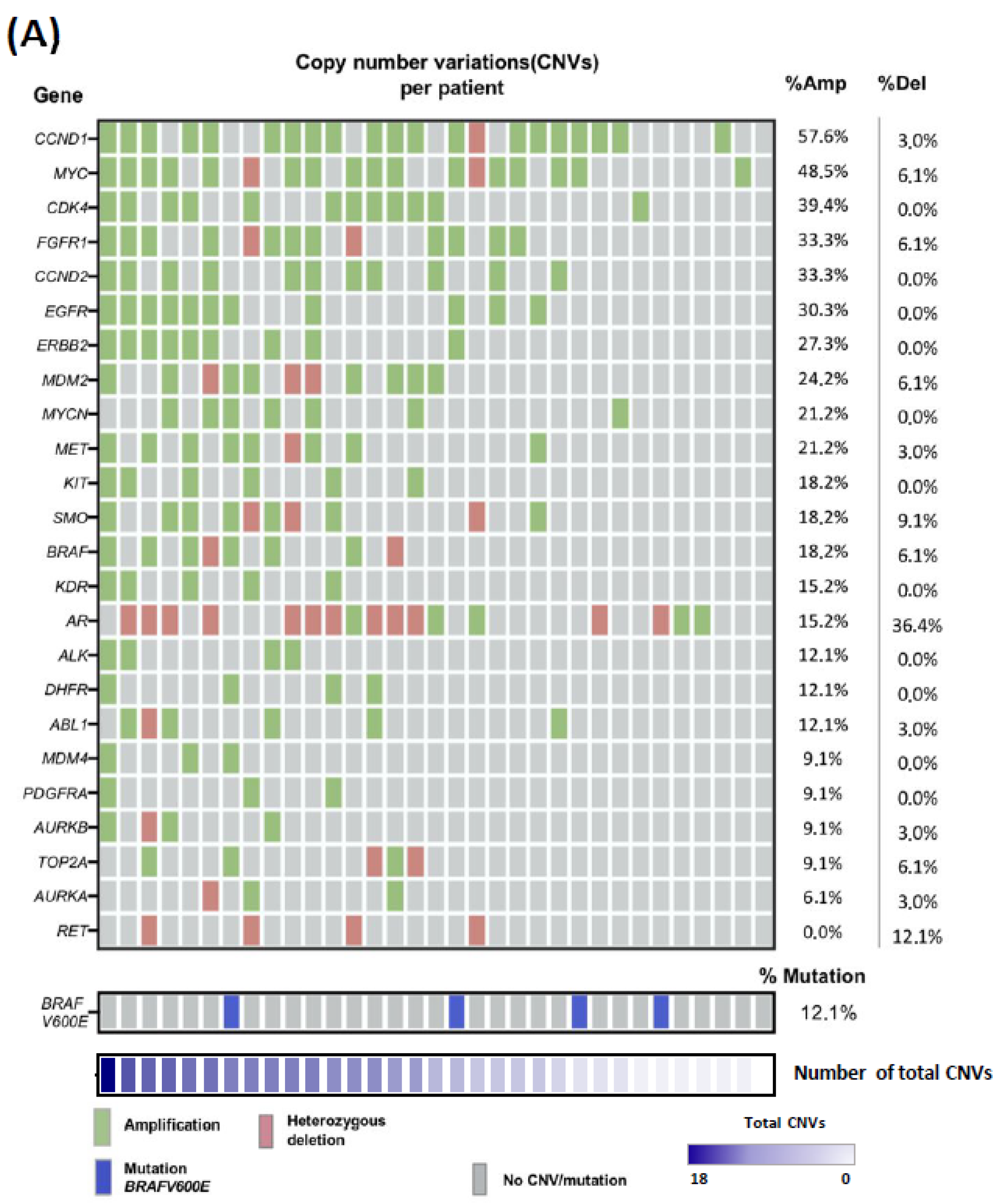
2.2. Copy-Number Variations Landscape in ALM Patient Biopsies
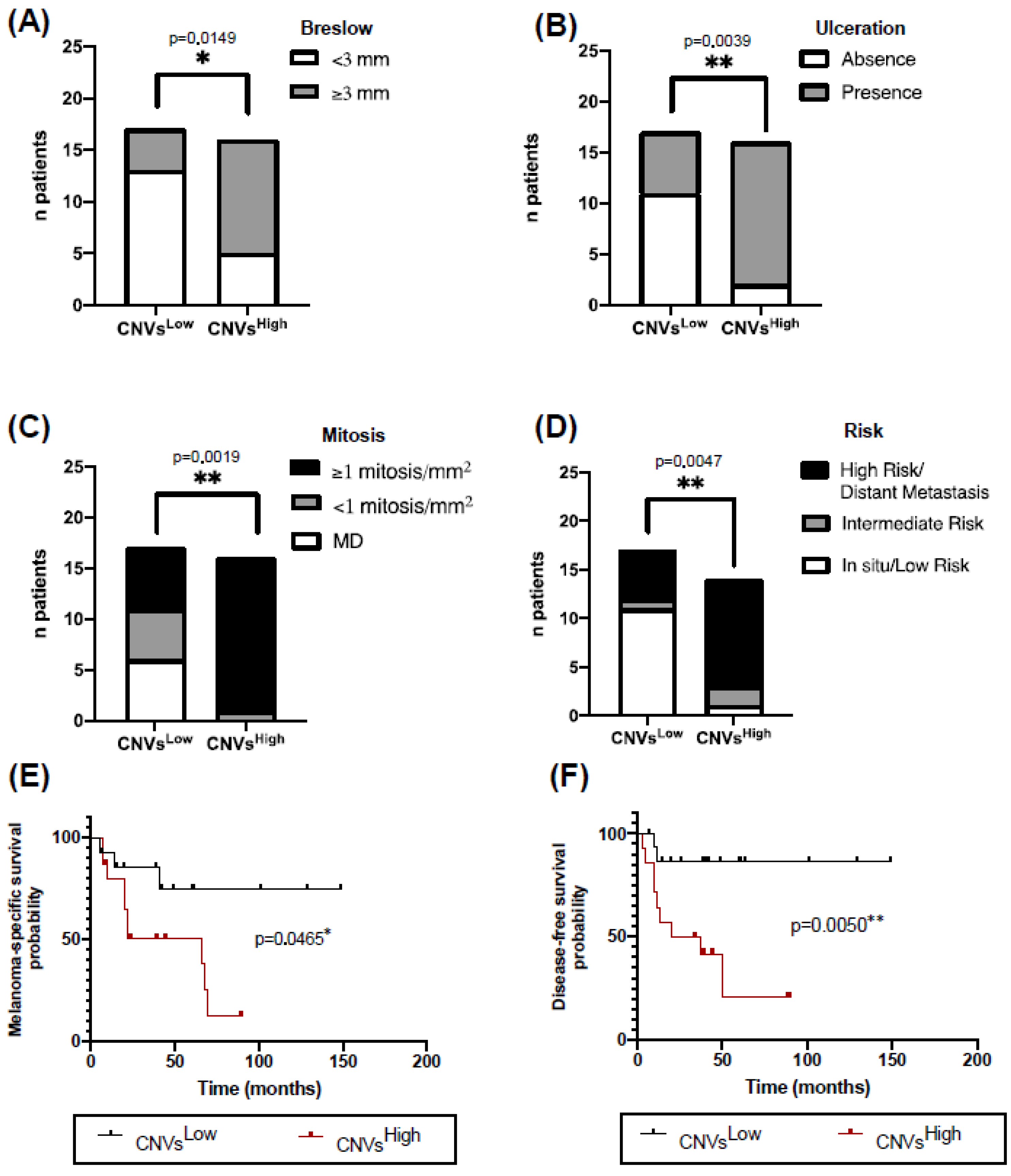
2.3. High CNVs in ALM Biopsies Are Associated with Worse Clinicopathological Characteristics and Prognosis
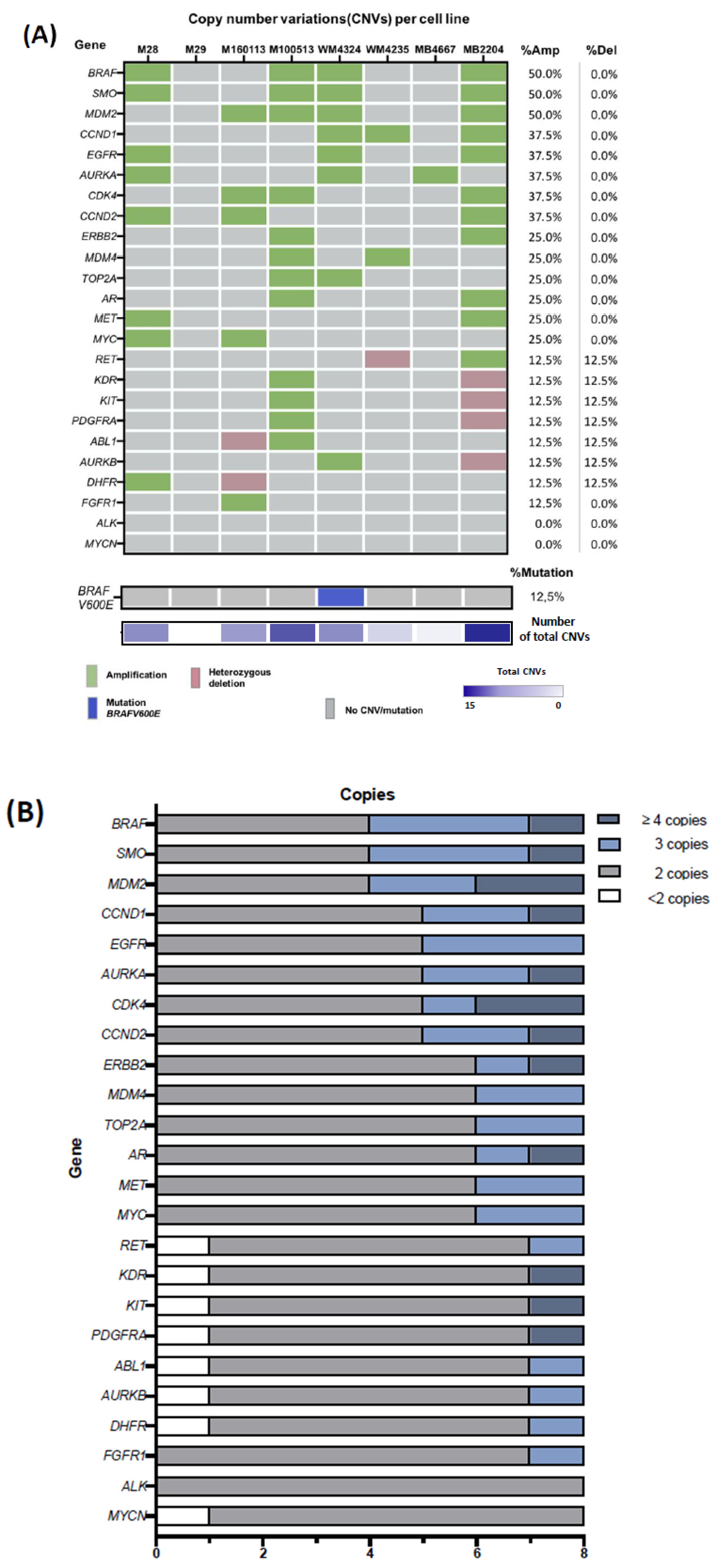
2.4. Characterization of Copy-Number Variations in ALM Cell Lines
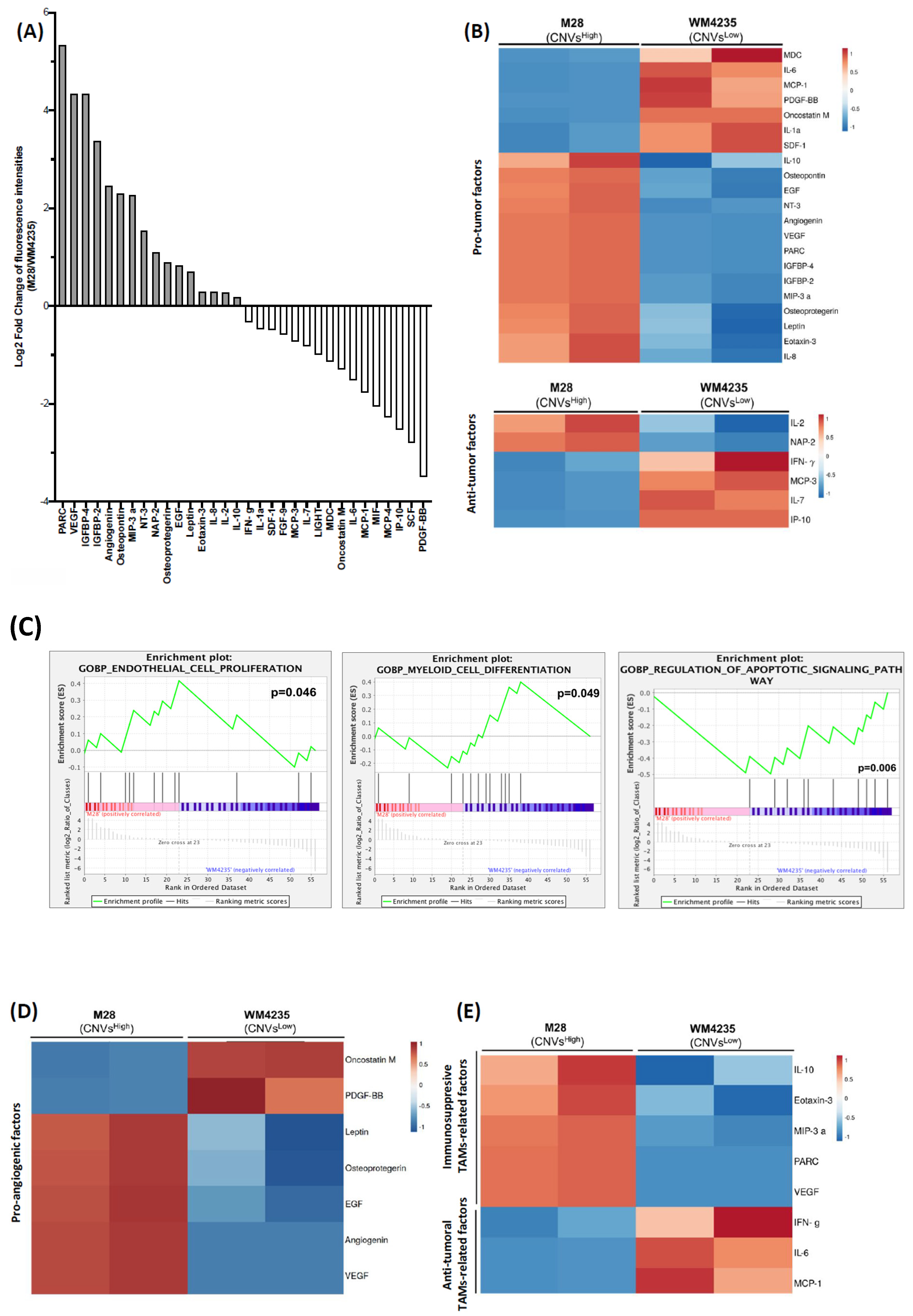
2.5. CNVsHigh Cells Secretome Exhibits an Increased Pro-Tumor Profile
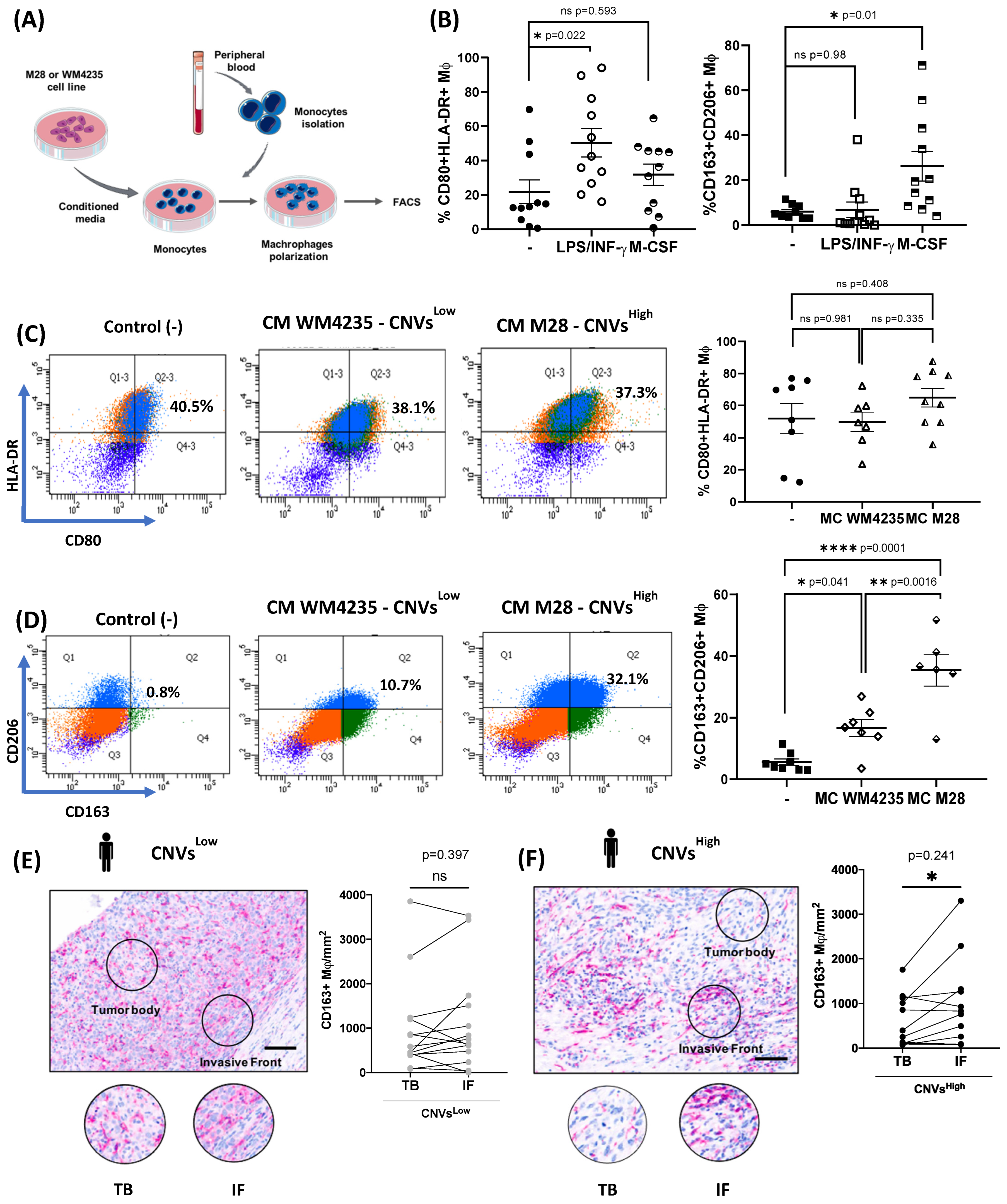
2.6. CNVsHigh Secretome Induce Tumor-Promoting Macrophages Polarization and Enhanced Immunosuppressive TAMs at the Invasive Front of CNVsHigh ALM Biopsies
3. Discussion
4. Materials and Methods
4.1. Cohort Construction and Patient Information
4.2. Patient Biopsies Collection
4.3. ALM Cell Lines
4.4. Conditioned Media (CM) Collection
4.5. DNA Extraction
4.6. Multiplex Ligation-Dependent Probe Amplification (MLPA) Procedure
4.7. Immunohistochemistry (IHC)
4.8. Human Cytokine Array
4.9. Gene Set Enrichment Analysis
4.10. Monocyte Isolation from Human Peripheral Blood
4.11. In Vitro Polarization of Macrophages
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durbec, F.; Martin, L.; Derancourt, C.; Grange, F. Melanoma of the hand and foot: Epidemiological, prognostic and genetic features. A systematic review. Br. J. Dermatol. 2012, 166, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Goydos, J.S.; Shoen, S.L. Acral Lentiginous Melanoma. Cancer Treat. Res. 2016, 167, 321–329. [Google Scholar]
- Newell, F.; Wilmott, J.S.; Johansson, P.A.; Nones, K.; Addala, V.; Mukhopadhyay, P.; Broit, N.; Amato, C.M.; Van Gulick, R.; Kazakoff, S.H.; et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat. Commun. 2020, 11, 5259. [Google Scholar] [CrossRef] [PubMed]
- Rabbie, R.; Ferguson, P.; Molina-Aguilar, C.; Adams, D.J.; Robles-Espinoza, C.D. Melanoma subtypes: Genomic profiles, prognostic molecular markers and therapeutic possibilities. J. Pathol. 2019, 247, 539. [Google Scholar] [CrossRef]
- Broit, N.; Johansson, P.A.; Rodgers, C.B.; Walpole, S.T.; Hayward, N.K.; Pritchard, A.L. Systematic review and meta-analysis of genomic alterations in acral melanoma. Pigment. Cell Melanoma Res. 2022, 35, 369. [Google Scholar] [CrossRef] [PubMed]
- Yeh, I.; Bastian, B.; Yeh, I. Melanoma pathology: New approaches and classification* Funding sources. Br. J. Dermatol. 2021, 185, 282–293. [Google Scholar] [CrossRef]
- Farshidfar, F.; Rhrissorrakrai, K.; Levovitz, C.; Peng, C.; Knight, J.; Bacchiocchi, A.; Su, J.; Yin, M.; Sznol, M.; Ariyan, S.; et al. Integrative molecular and clinical profiling of acral melanoma links focal amplification of 22q11.21 to metastasis. Nat. Commun. 2022, 13, 898. [Google Scholar] [CrossRef]
- Shi, Q.; Liu, L.; Chen, J.; Zhang, W.; Guo, W.; Wang, X.; Wang, H.; Guo, S.; Yue, Q.; Ma, J.; et al. Integrative Genomic Profiling Uncovers Therapeutic Targets of Acral Melanoma in Asian Populations. Clin. Cancer Res. 2022, 28, 2690–2703. [Google Scholar] [CrossRef]
- Asgari, M.M.; Shen, L.; Sokil, M.M.; Yeh, I.; Jorgenson, E. Prognostic factors and survival in acral lentiginous melanoma. Br. J. Dermatol. 2017, 177, 428–435. [Google Scholar] [CrossRef]
- Huang, K.; Fan, J.; Misra, S. Acral Lentiginous Melanoma: Incidence and Survival in the United States, 2006–2015, an Analysis of SEER Registry. J. Surg. Res. 2020, 251, 329–339. [Google Scholar] [CrossRef]
- Egger, M.E.; McMasters, K.M.; Callender, G.G.; Quillo, A.R.; Martin, R.C.; Stromberg, A.J.; Scoggins, C.R. Unique prognostic factors in acral lentiginous melanoma. Am. J. Surg. 2012, 204, 874–880. [Google Scholar] [CrossRef]
- Basurto-Lozada, P.; Molina-Aguilar, C.; Castaneda-Garcia, C.; Vázquez-Cruz, M.E.; Garcia-Salinas, O.I.; Álvarez-Cano, A.; Martínez-Said, H.; Roldán-Marín, R.; Adams, D.J.; Possik, P.A.; et al. Acral lentiginous melanoma: Basic facts, biological characteristics and research perspectives of an understudied disease. Pigment. Cell Melanoma Res. 2021, 34, 59–71. [Google Scholar] [CrossRef]
- Huang, R.; Shen, G.; Ren, Y.; Zheng, K.; Wang, J.; Shi, Y.; Yin, J.C.; Qin, L.; Zhang, G.; Zhao, M.; et al. Prognostic value of genetic aberrations and tumor immune microenvironment in primary acral melanoma. J. Transl. Med. 2023, 21, 78. [Google Scholar] [CrossRef] [PubMed]
- Teixido, C.; Castillo, P.; Martinez-Vila, C.; Arance, A.; Alos, L. Molecular Markers and Targets in Melanoma. Cells 2021, 10, 2320. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.R.; Choi, Y.D.; Kim, J.M.; Jin, S.; Shin, M.-H.; Shim, H.-J.; Lee, J.-B.; Yun, S.J. Genetic Alterations in Primary Acral Melanoma and Acral Melanocytic Nevus in Korea: Common Mutated Genes Show Distinct Cytomorphological Features. J. Investig. Dermatol. 2018, 138, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Marzagalli, M.; Ebelt, N.D.; Manuel, E.R. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin. Cancer Biol. 2019, 59, 236–250. [Google Scholar] [CrossRef]
- Liu, D.; Yang, X.; Wu, X. Tumor Immune Microenvironment Characterization Identifies Prognosis and Immunotherapy-Related Gene Signatures in Melanoma. Front. Immunol. 2021, 12, 1. [Google Scholar] [CrossRef]
- Li, J.; Smalley, I.; Chen, Z.; Wu, J.-Y.; Phadke, M.S.; Teer, J.K.; Nguyen, T.; Karreth, F.A.; Koomen, J.M.; Sarnaik, A.A.; et al. Single-cell Characterization of the Cellular Landscape of Acral Melanoma Identifies Novel Targets for Immunotherapy. Clin. Cancer Res. 2022, 28, 2131–2146. [Google Scholar] [CrossRef]
- Nakamura, Y.; Zhenjie, Z.; Oya, K.; Tanaka, R.; Ishitsuka, Y.; Okiyama, N.; Watanabe, R.; Fujisawa, Y. Poor Lymphocyte Infiltration to Primary Tumors in Acral Lentiginous Melanoma and Mucosal Melanoma Compared to Cutaneous Melanoma. Front. Oncol. 2020, 10, 524700. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Gao, J.; Liang, Y.; Wang, L. Shaping Polarization of Tumor-Associated Macrophages in Cancer Immunotherapy. Front. Immunol. 2022, 13, 3364. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; Ernst, M. Targeting Macrophages in Cancer: From Bench to Bedside. Front. Oncol. 2018, 8, 1. [Google Scholar] [CrossRef]
- Rodrigues Da Cunha, B.; Domingos, C.; Stefanini, A.C.B.; Henrique, T.; Polachini, G.M.; Castelo-Branco, P.; Tajara, E.H. Cellular Interactions in the Tumor Microenvironment: The Role of Secretome. J. Cancer 2019, 10, 4574–4587. [Google Scholar] [CrossRef]
- Donadelli, M. The cancer secretome and secreted biomarkers. Semin. Cell Dev. Biol. 2018, 78, 1–2. [Google Scholar] [CrossRef]
- Schaaij-Visser, T.B.M.; De Wit, M.; Lam, S.W.; Jiménez, C.R. The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context. Biochim. Biophys. Acta (BBA)-Proteins Proteomics 2013, 1834, 2242–2258. [Google Scholar] [CrossRef]
- Madden, E.C.; Gorman, A.M.; Logue, S.E.; Samali, A. Tumour Cell Secretome in Chemoresistance and Tumour Recurrence. Trends Cancer 2020, 6, 489–505. [Google Scholar] [CrossRef]
- Wei, L.F.; Weng, X.; Huang, X.; Peng, Y.; Guo, H.; Xu, Y. IGFBP2 in cancer: Pathological role and clinical significance (Review). Oncol. Rep. 2021, 45, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Melincovici, C.S.; Boşca, A.B.; Şuşman, S.; Mărginean, M.; Mihu, C.; Istrate, M.; Moldovan, I.M.; Roman, A.L.; Mihu, C.M. Vascular endothelial growth factor (VEGF)-key factor in normal and pathological angiogenesis. Rom. J. Morphol. Embryol. 2018, 59, 455–467. [Google Scholar]
- Cardoso, A.P.; Pinto, M.L.; Castro, F.; Costa, A.M.; Marques-Magalhães, A.; Canha-Borges, A.; Cruz, T.; Velho, S.; Oliveira, M.J. The immunosuppressive and pro-tumor functions of CCL18 at the tumor microenvironment. Cytokine Growth Factor. Rev. 2021, 60, 107–119. [Google Scholar] [CrossRef]
- Olivera, I.; Sanz-Pamplona, R.; Bolaños, E.; Rodriguez, I.; Etxeberria, I.; Cirella, A.; Egea, J.; Garasa, S.; Migueliz, I.; Eguren-Santamaria, I.; et al. A Therapeutically Actionable Protumoral Axis of Cytokines Involving IL-8, TNFα, and IL-1β. Cancer Discov. 2022, 12, 2140–2157. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, U.; Kuroda, E. Regulation of macrophage-derived chemokine (MDC, CCL22) production. Crit. Rev. Immunol. 2002, 22, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Berraondo, P.; Sanmamed, M.F.; Ochoa, M.C.; Etxeberria, I.; Aznar, M.A.; Pérez-Gracia, J.L.; Rodriguez-Ruiz, M.E.; Ponz-Sarvise, M.; Castañón, E.; Melero, I. Cytokines in clinical cancer immunotherapy. Br. J. Cancer 2018, 120, 6–15. [Google Scholar] [CrossRef]
- Garo, L.P.; Gopal, M. Role of cytokines in tumor immunity and immune tolerance to cancer. In Cancer Immunology: A Translational Medicine Context, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 205–233. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Simińska, D.; Bohatyrewicz, R.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of the Ligands of Receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci. 2020, 21, 8412. [Google Scholar] [CrossRef]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Cantaluppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Mediators Inflamm. 2017, 2017, 4049098. [Google Scholar] [CrossRef]
- Kadomoto, S.; Izumi, K.; Mizokami, A. The CCL20-CCR6 Axis in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 5186. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, W.; Wang, P.; Deng, Y.; Dong, Y.-T.; Liu, F.-F.; Huang, R.; Zhang, P.; Duan, Y.-Q.; Liu, X.-D.; et al. CCL7 recruits cDC1 to promote antitumor immunity and facilitate checkpoint immunotherapy to non-small cell lung cancer. Nat. Commun. 2020, 11, 6119. [Google Scholar] [CrossRef] [PubMed]
- Keyser, J.; Schultz, J.; Ladell, K.; Elzaouk, L.; Heinzerling, L.; Pavlovic, J.; Moelling, K. IP-10-encoding plasmid DNA therapy exhibits anti-tumor and anti-metastatic efficiency. Exp. Dermatol. 2004, 13, 380–390. [Google Scholar] [CrossRef]
- Struyf, S.; Schutyser, E.; Gouwy, M.; Gijsbers, K.; Proost, P.; Benoit, Y.; Opdenakker, G.; Van Damme, J.; Laureys, G. PARC/CCL18 is a plasma CC chemokine with increased levels in childhood acute lymphoblastic leukemia. Am. J. Pathol. 2003, 163, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Gajanin, V.; Krivokuća, Z.; Kostić, K.; Gajanin, R.; Sladojević, I. Significance of vascular endothelial growth factor expression in skin melanoma. Vojnosanit. Pregl. 2010, 67, 747–754. [Google Scholar] [CrossRef]
- Piliang, M.P. Acral Lentiginous Melanoma. Surg. Pathol. Clin. 2009, 2, 535–541. [Google Scholar] [CrossRef]
- Nakamura, Y.; Fujisawa, Y. Diagnosis and Management of Acral Lentiginous Melanoma. Curr. Treat. Options Oncol. 2018, 19, 42. [Google Scholar] [CrossRef] [PubMed]
- Yeh, I.; Jorgenson, E.; Shen, L.; Xu, M.; North, J.P.; Shain, A.H.; Reuss, D.; Wu, H.; Robinson, W.A.; Olshen, A.; et al. Targeted Genomic Profiling of Acral Melanoma. J. Natl. Cancer Inst. 2019, 111, 1068–1077. [Google Scholar] [CrossRef]
- Wang, M.; Banik, I.; Shain, A.H.; Yeh, I.; Bastian, B.C. Integrated genomic analyses of acral and mucosal melanomas nominate novel driver genes. Genome Med. 2022, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Fukushima, S.; Sheen, Y.-S.; Ramelyte, E.; Cruz-Pacheco, N.; Shi, C.; Liu, S.; Banik, I.; Aquino, J.D.; Acosta, M.S.; et al. The genetic evolution of acral melanoma. bioRxiv 2023. [Google Scholar] [CrossRef]
- Grist, E.; Friedrich, S.; Brawley, C.; Mendes, L.; Parry, M.; Ali, A.; Haran, A.; Hoyle, A.; Gilson, C.; Lall, S.; et al. Accumulation of copy number alterations and clinical progression across advanced prostate cancer. Genome Med. 2022, 14, 102. [Google Scholar] [CrossRef]
- Liang, L.; Fang, J.Y.; Xu, J. Gastric cancer and gene copy number variation: Emerging cancer drivers for targeted therapy. Oncogene 2016, 35, 1475–1482. [Google Scholar] [PubMed]
- Ashida, A.; Takata, M.; Murata, H.; Kido, K.; Saida, T. Pathological activation of KIT In metastatic tumors of acral and mucosal melanomas. Int. J. Cancer 2009, 124, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Furney, S.J.; Turajlic, S.; Fenwick, K.; Lambros, M.B.; MacKay, A.; Ricken, G.; Mitsopoulos, C.; Kozarewa, I.; Hakas, J.; Zvelebil, M.; et al. Genomic characterisation of acral melanoma cell lines. Pigment. Cell Melanoma Res. 2012, 25, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Sheng, X.; Wu, X.; Yan, J.; Ma, M.; Yu, J.; Si, L.; Chi, Z.; Cui, C.; Dai, J.; et al. Frequent genetic aberrations in the CDK4 pathway in acral melanoma indicate the potential for CDK4/6 inhibitors in targeted therapy. Clin. Cancer Res. 2017, 23, 6946–6957. [Google Scholar] [CrossRef]
- Zúñiga-Castillo, M.; Pereira, N.V.; Sotto, M.N. High density of M2-macrophages in acral lentiginous melanoma compared to superficial spreading melanoma. Histopathology 2018, 72, 1189–1198. [Google Scholar] [CrossRef]
- Georgouli, M.; Herraiz, C.; Crosas-Molist, E.; Fanshawe, B.; Maiques, O.; Perdrix, A.; Pandya, P.; Rodriguez-Hernandez, I.; Ilieva, K.M.; Cantelli, G.; et al. Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment. Cell 2019, 176, 757. [Google Scholar] [CrossRef]
- Liu, H.; Gao, J.; Feng, M.; Cheng, J.; Tang, Y.; Cao, Q.; Zhao, Z.; Meng, Z.; Zhang, J.; Zhang, G.; et al. Integrative molecular and spatial analysis reveals evolutionary dynamics and tumor-immune interplay of in situ and invasive acral melanoma. Cancer Cell 2024, 42, 1067–1085.e11. [Google Scholar] [CrossRef]
- Maiques, O.; Macià, A.; Moreno, S.; Barceló, C.; Santacana, M.; Vea, A.; Herreros, J.; Gatius, S.; Ortega, E.; Valls, J.; et al. Immunohistochemical analysis of T-type calcium channels in acquired melanocytic naevi and melanoma. Br. J. Dermatol. 2017, 176, 1247–1258. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Barceló, C.; Sisó, P.; de la Rosa, I.; Megino-Luque, C.; Navaridas, R.; Maiques, O.; Urdanibia, I.; Eritja, N.; Soria, X.; Potrony, M.; et al. M-CSF as a therapeutic target in BRAFV600E melanoma resistant to BRAF inhibitors. Br. J. Cancer 2022, 127, 1142–1152. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Moral, L.; Paul, T.; Martori, C.; Font-Díaz, J.; Sanjurjo, L.; Aran, G.; Téllez, É.; Blanco, J.; Carrillo, J.; Ito, M.; et al. Macrophage CD5L is a target for cancer immunotherapy. EBioMedicine 2023, 91, 104555. [Google Scholar] [CrossRef] [PubMed]

| Gender | |
| Male | 15 (45.5) |
| Female | 18 (54.5) |
| Age (years) | |
| Mean (±SD) | 70.2 (±15.9) |
| Median (IQR) | 75 (54–83) |
| Primary tumor location | |
| Hands n (%) | 8 (24.2) |
| Subungual | 6 (18.2) |
| Finger | 2 (6.1) |
| Feet n (%) | 25 (75.8) |
| Subungual | 8 (24.2) |
| Sole | 11 (33.3) |
| Toe | 3 (9.1) |
| Lateral | 2 (6.1) |
| N/A | 1 (3.0) |
| Breslow thickness (mm) | |
| Mean (±SD) | 4.2 (±3.9) |
| Median (IQR) | 2.7 (0.89–7.9) |
| Ulceration n (%) | |
| Yes | 20 (60.6) |
| No | 13 (39.4) |
| Mitotic index n (%) | |
| <1/mm2 | 6 (18.2) |
| ≥1/mm2 | 21 (63.6) |
| N/A | 6 (18.2) |
| Live status n (%) | |
| Alive | 16 (48.4) |
| Melanoma death | 13 (39.4) |
| Other causes | 4 (12.1) |
| Relapse (metastasis) n (%) | |
| Yes | 12 (36.3) |
| No | 18 (54.5) |
| Distant metastasis at diagnosis | 3 (9.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de la Rosa, I.; Sisó, P.; Ríos, C.; Gracia, J.; Cuevas, D.; Maiques, O.; Eritja, N.; Soria, X.; Angel-Baldó, J.; Gatius, S.; et al. High Copy Number Variations Correlate with a Pro-Tumoral Microenvironment and Worse Prognosis in Acral Lentiginous Melanoma. Int. J. Mol. Sci. 2025, 26, 4097. https://doi.org/10.3390/ijms26094097
de la Rosa I, Sisó P, Ríos C, Gracia J, Cuevas D, Maiques O, Eritja N, Soria X, Angel-Baldó J, Gatius S, et al. High Copy Number Variations Correlate with a Pro-Tumoral Microenvironment and Worse Prognosis in Acral Lentiginous Melanoma. International Journal of Molecular Sciences. 2025; 26(9):4097. https://doi.org/10.3390/ijms26094097
Chicago/Turabian Stylede la Rosa, Inés, Pol Sisó, Christopher Ríos, Judith Gracia, Dolors Cuevas, Oscar Maiques, Núria Eritja, Xavier Soria, Joan Angel-Baldó, Sonia Gatius, and et al. 2025. "High Copy Number Variations Correlate with a Pro-Tumoral Microenvironment and Worse Prognosis in Acral Lentiginous Melanoma" International Journal of Molecular Sciences 26, no. 9: 4097. https://doi.org/10.3390/ijms26094097
APA Stylede la Rosa, I., Sisó, P., Ríos, C., Gracia, J., Cuevas, D., Maiques, O., Eritja, N., Soria, X., Angel-Baldó, J., Gatius, S., Sanchez-Moral, L., Sarrias, M.-R., Matias-Guiu, X., Martí, R. M., & Macià, A. (2025). High Copy Number Variations Correlate with a Pro-Tumoral Microenvironment and Worse Prognosis in Acral Lentiginous Melanoma. International Journal of Molecular Sciences, 26(9), 4097. https://doi.org/10.3390/ijms26094097







