Genome-Wide Identification of the Dof Gene Family and Functional Analysis of PeSCAP1 in Regulating Guard Cell Maturation in Populus euphratica
Abstract
1. Introduction
2. Results
2.1. Identification and Prediction of the Physicochemical Properties of PeDof Genes
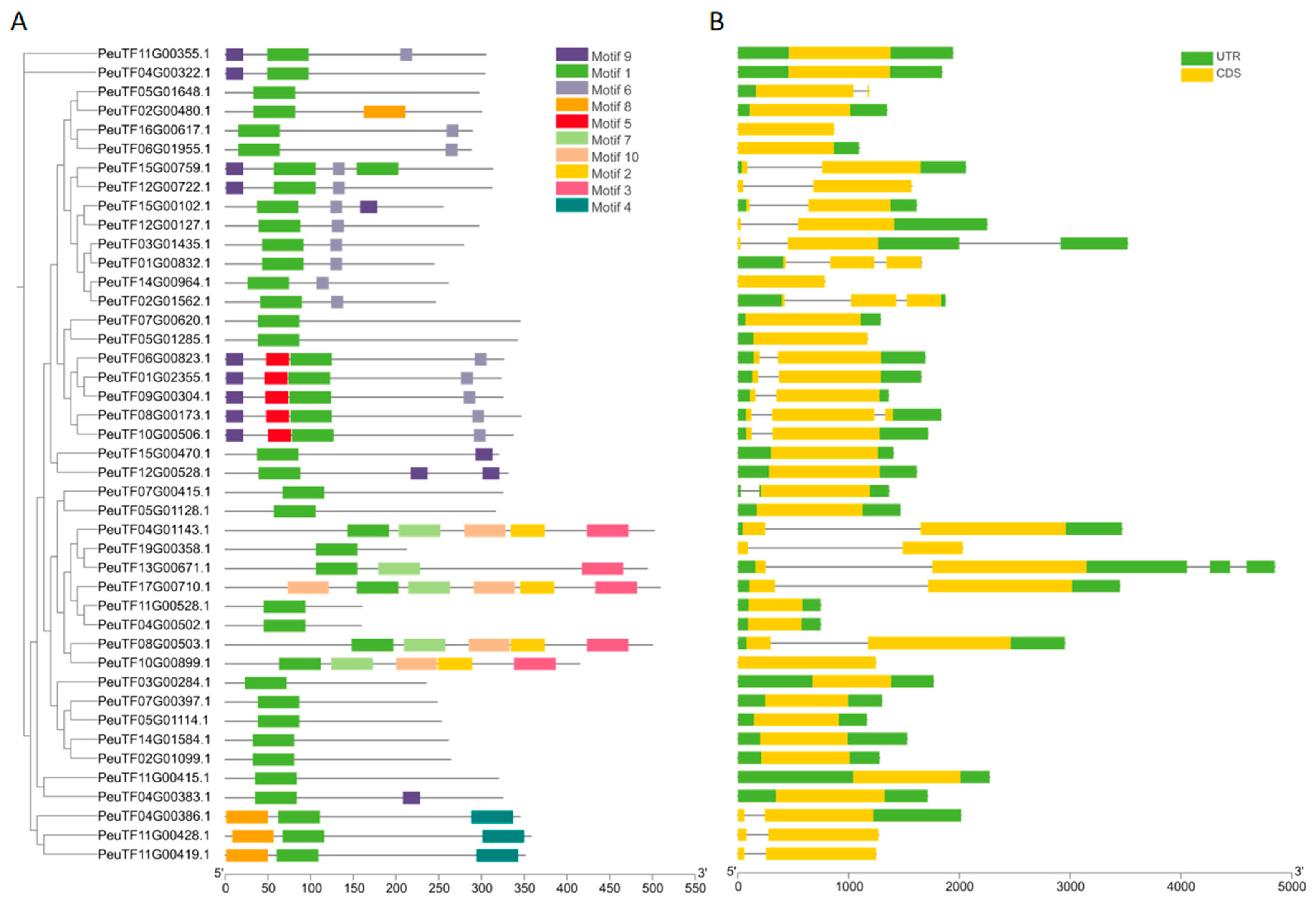
2.2. Gene Structures and Conserved Motifs of PeDofs
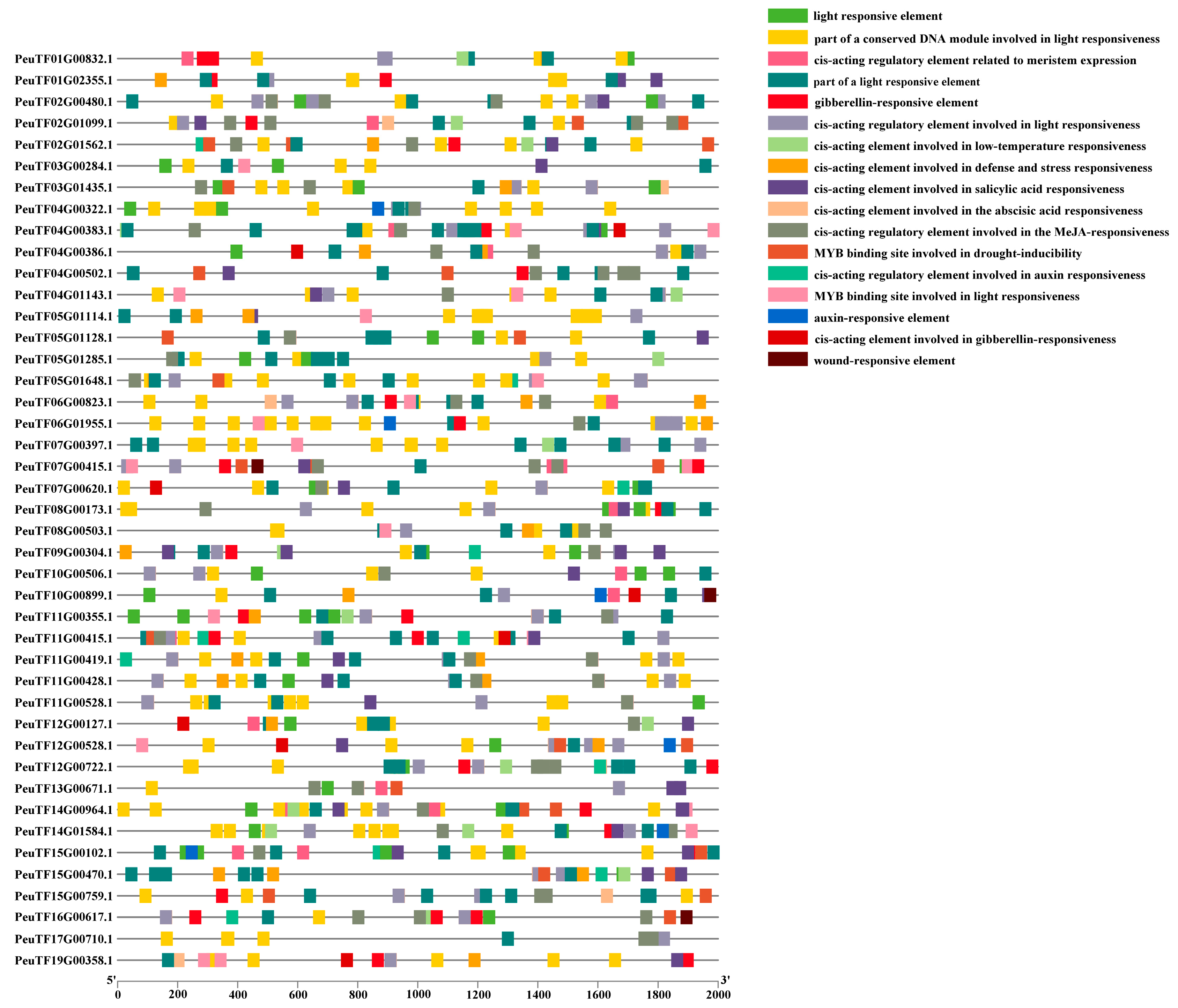
2.3. Prediction of Cis-Acting Elements in PeDofs
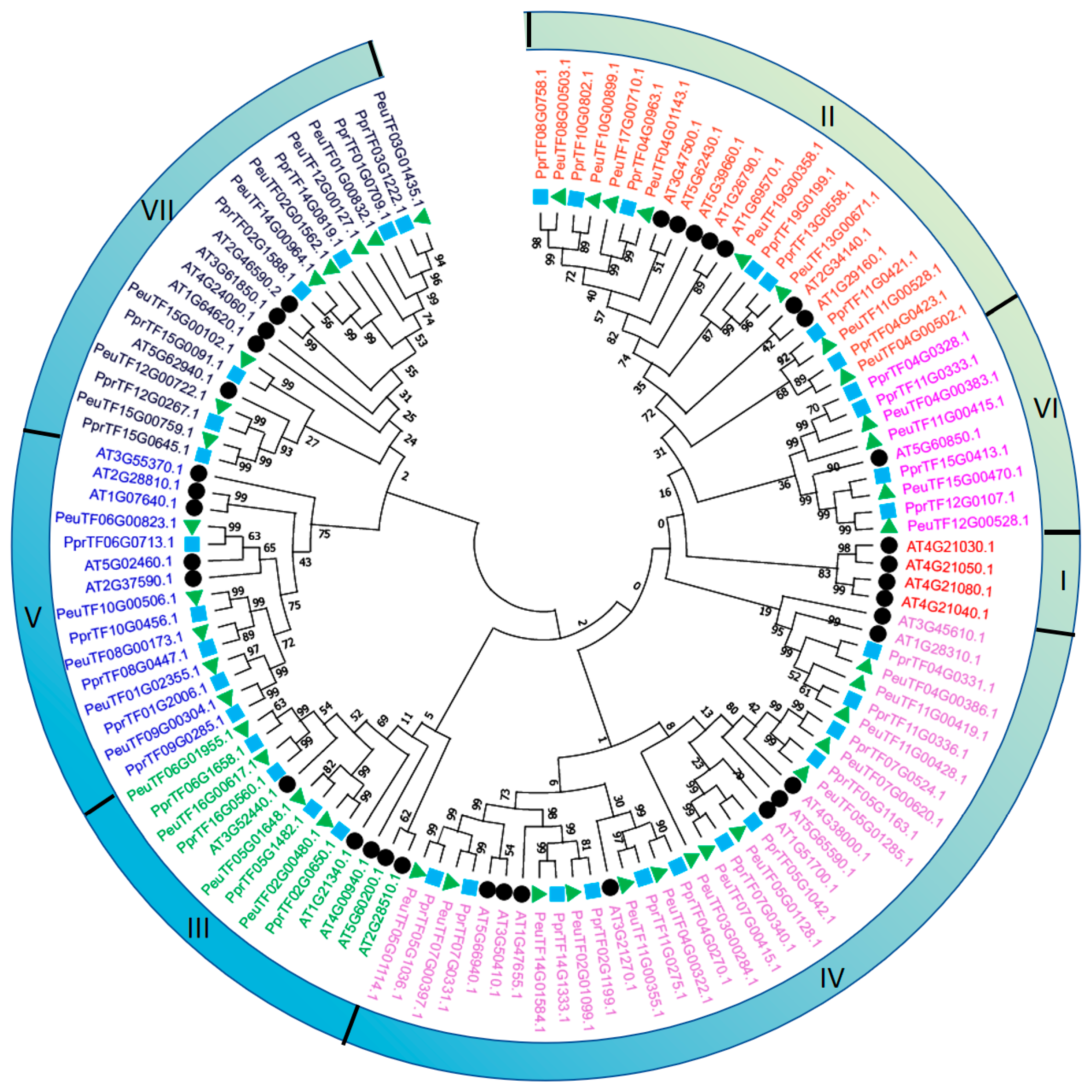
2.4. Collinearity Analysis of Dofs in Multi-Species
2.5. Phylogenetic Tree of PeDofs
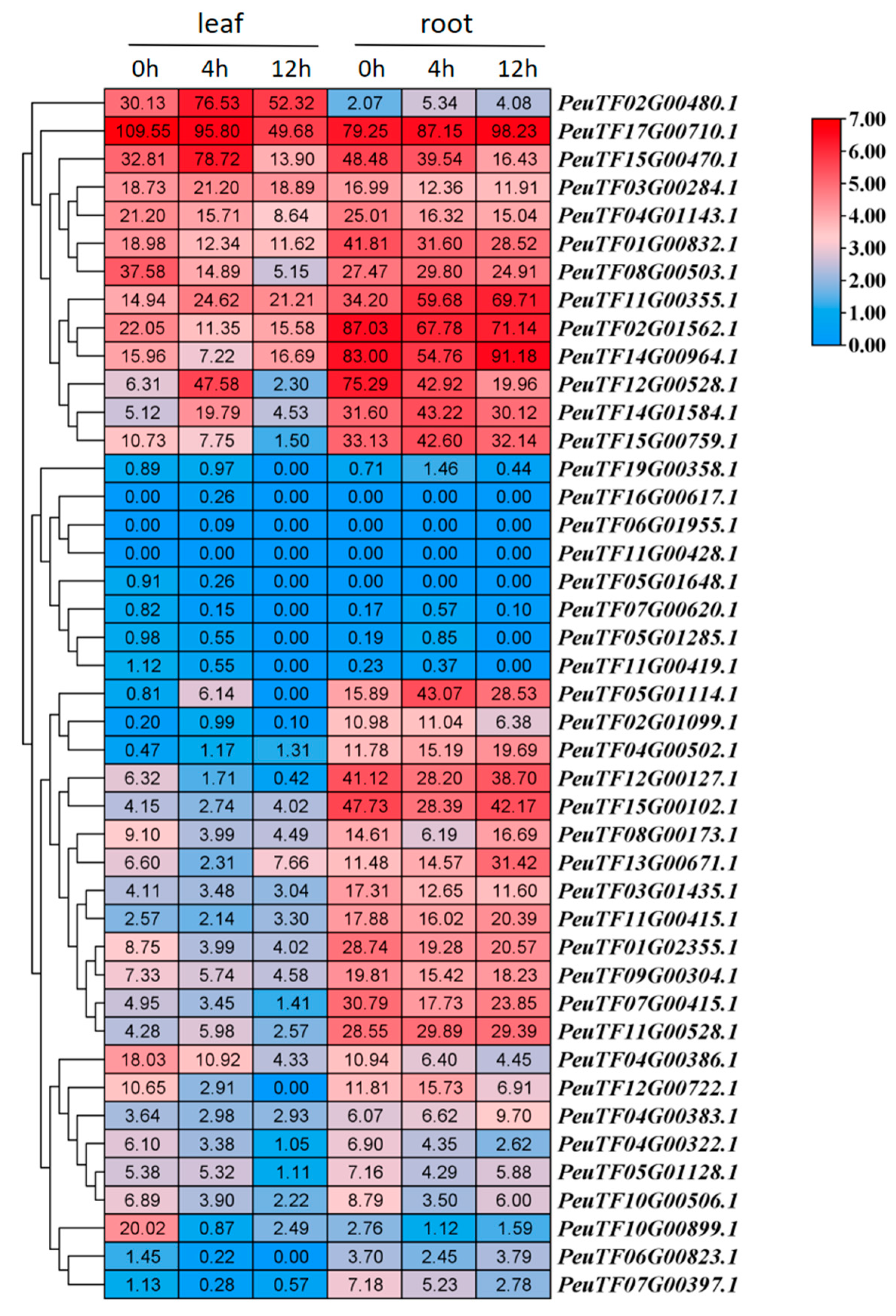
2.6. Expression Patterns of PeDofs in Roots and Leaves Under Drought Treatment
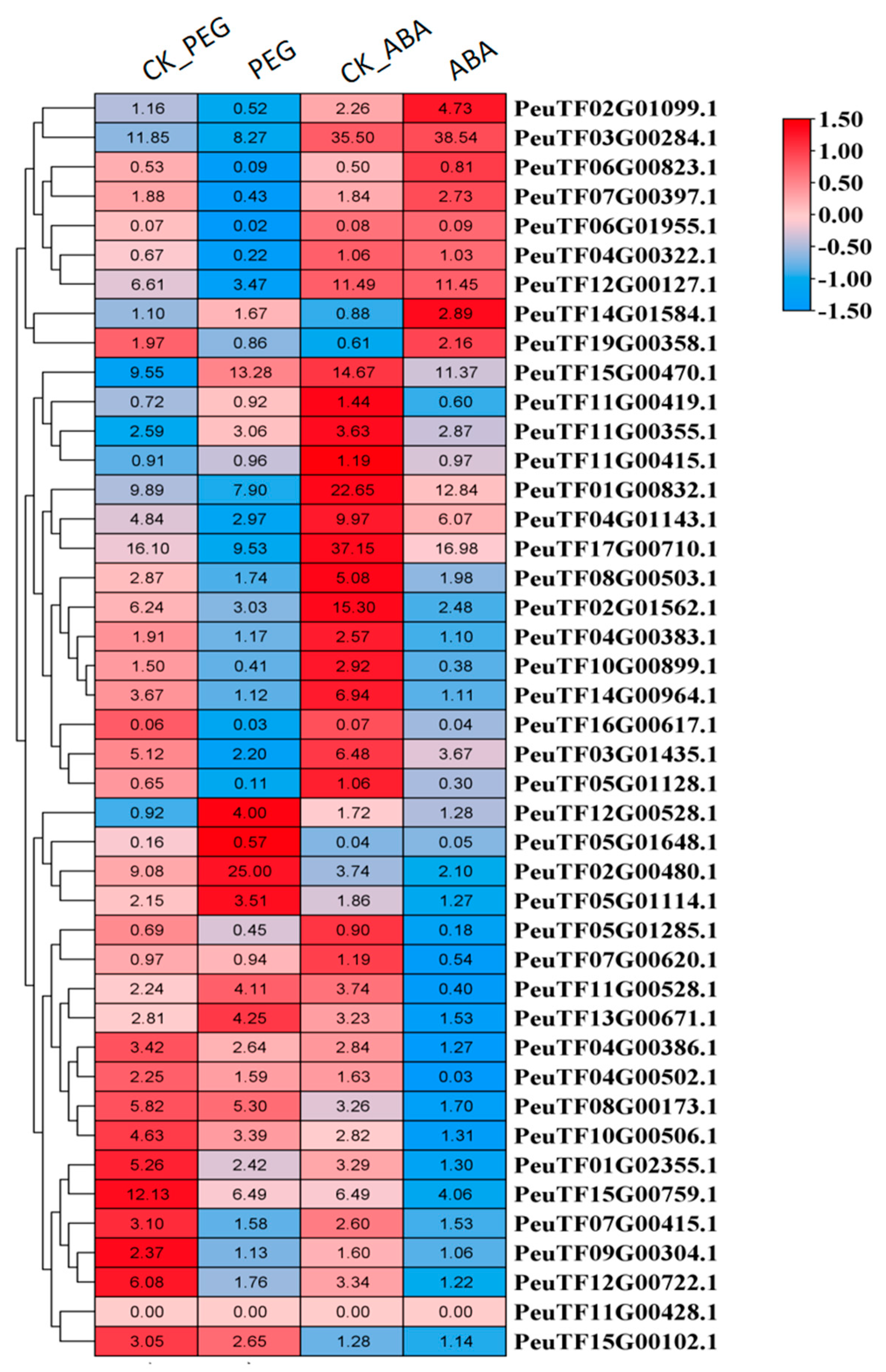
2.7. PeDof Genes Response to Drought or ABA Treatment During Seed Germination
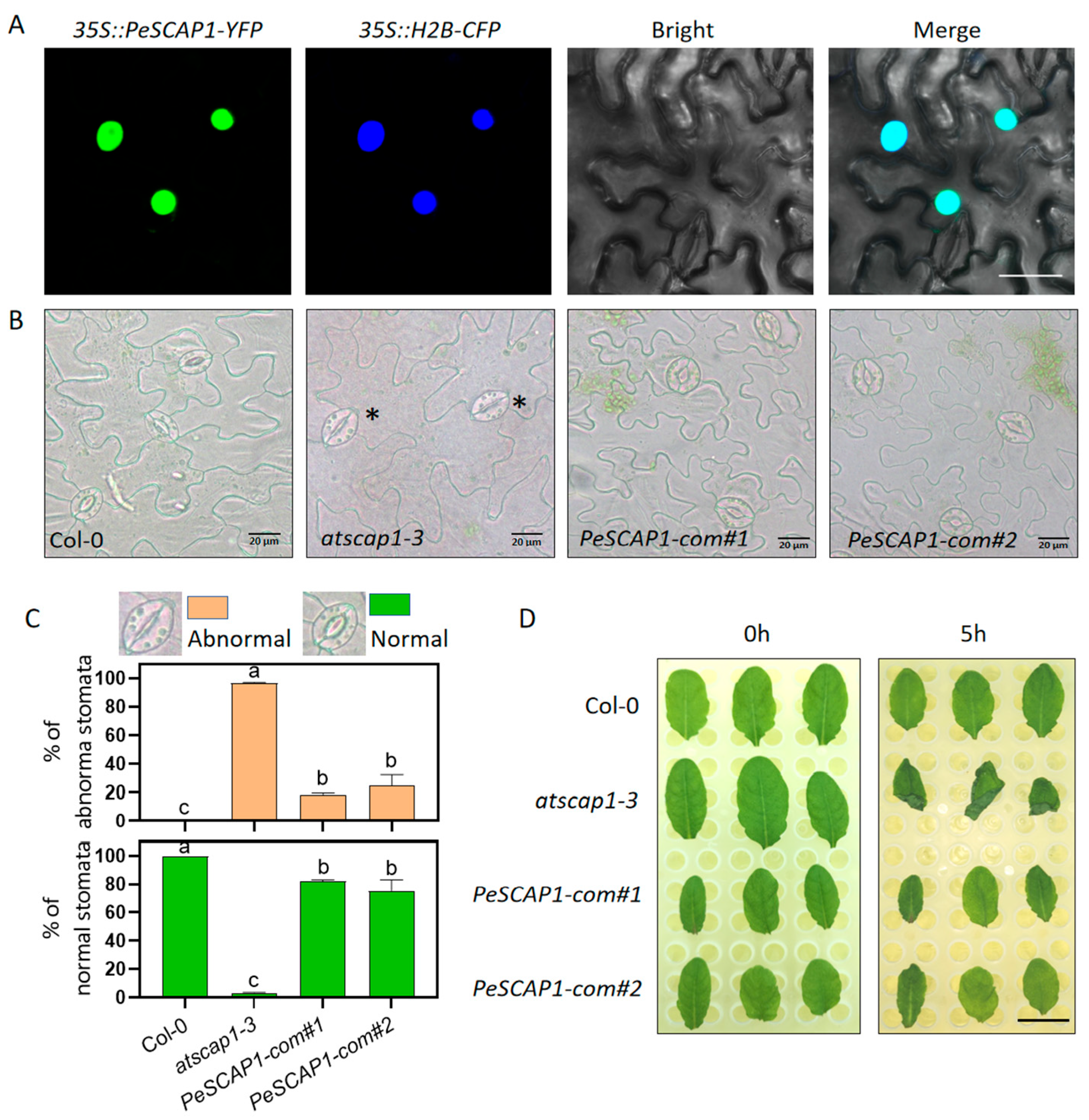
2.8. PeSCAP1 Is Responsible for Maintaining the Integrity of the Morphology and Function of Guard Cells
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification and Physiochemical Predictions of the P. euphratica Dof Gene Family
4.2. Gene Structure and Conserved Motifs Analysis
4.3. Analysis of Cis-Acting Elements in the Promoter Regions of PeDof Genes
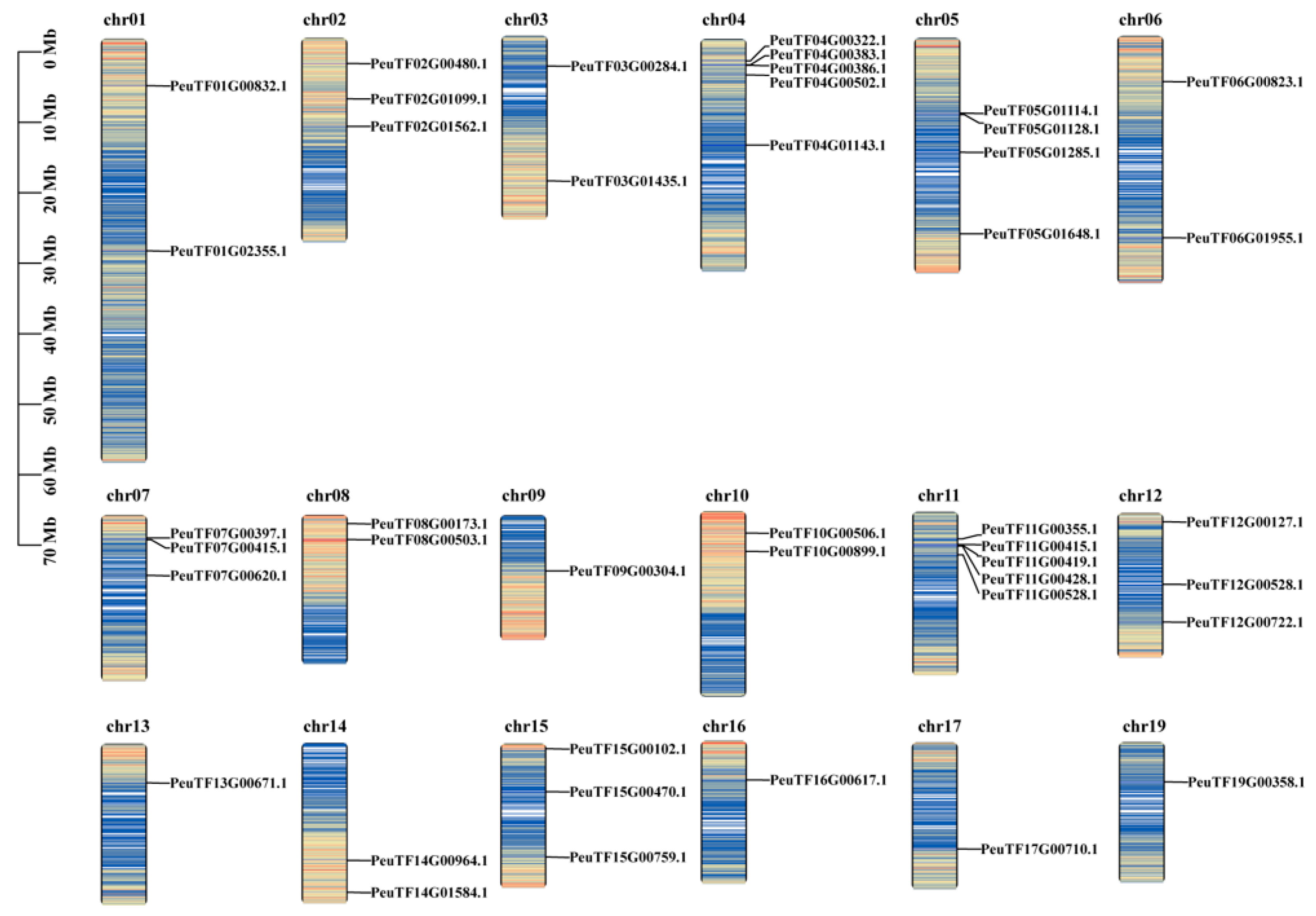
4.4. Multispecies Collinearity and Chromosomal Localization of PeDofs
4.5. Phylogenetic Tree Analysis of PeDofs
4.6. Expression Analysis of PeDofs Under Drought Stress and ABA Treatment
4.7. Subcellular Localization of PeSCAP1
4.8. Plant Materials and Growth Conditions
4.9. Stomatal Phenotype Quantification and Water Loss Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doebley, J.; Lukens, L. Transcriptional regulators and the evolution of plant form. Plant Cell 1998, 10, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Malviya, N.; Kushwaha, H.; Nasim, J.; Bisht, N.C.; Singh, V.K.; Yadav, D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 2015, 241, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Izui, K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 1993, 268, 16028–16036. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Lijavetzky, D.; Carbonero, P.; Vicente-Carbajosa, J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 2003, 3, 17. [Google Scholar] [CrossRef]
- Tabassum, J.; Raza, Q.; Riaz, A.; Ahmad, S.; Rashid, M.A.R.; Javed, M.A.; Ali, Z.; Kang, F.; Khan, I.A.; Atif, R.M.; et al. Exploration of the genomic atlas of Dof transcription factor family across genus Oryza provides novel insights on rice breeding in changing climate. Front. Plant Sci. 2022, 13, 1004359. [Google Scholar] [CrossRef]
- Sun, S.; Wang, B.; Jiang, Q.; Li, Z.; Jia, S.; Wang, Y.; Guo, H. Genome-wide analysis of BpDof genes and the tolerance to drought stress in birch (Betula platyphylla). PeerJ 2021, 9, e11938. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, Y.; Wan, C.; Li, J.; Yang, Y.; Chen, J. Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 2020, 8, e8358. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, B.; Zheng, G.; Zhang, A.; Li, R. Genome-wide characterization of the SiDof gene family in foxtail millet (Setaria italica). Biosystems 2017, 151, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ma, Y.; Lu, Y.; Yue, J.; Ming, R. Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 2021, 11, 14429. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Ludlow, R.A.; Lu, M.; An, H. Genome-Wide Analysis of Dof Genes and Their Response to Abiotic Stress in Rose (Rosa chinensis). Front. Genet. 2021, 12, 538733. [Google Scholar] [CrossRef]
- Negi, J.; Moriwaki, K.; Konishi, M.; Yokoyama, R.; Nakano, T.; Kusumi, K.; Hashimoto-Sugimoto, M.; Schroeder, J.I.; Nishitani, K.; Yanagisawa, S.; et al. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis. Curr. Biol. 2013, 23, 479–484. [Google Scholar] [CrossRef]
- Martin, G.; Veciana, N.; Boix, M.; Rovira, A.; Henriques, R.; Monte, E. The photoperiodic response of hypocotyl elongation involves regulation of CDF1 and CDF5 activity. Physiol. Plant 2020, 169, 480–490. [Google Scholar] [CrossRef]
- Gao, H.; Song, W.; Severing, E.; Vayssieres, A.; Huettel, B.; Franzen, R.; Richter, R.; Chai, J.; Coupland, G. PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants 2022, 8, 1082–1093. [Google Scholar] [CrossRef]
- Pi, L.; Aichinger, E.; van der Graaff, E.; Llavata-Peris, C.I.; Weijers, D.; Hennig, L.; Groot, E.; Laux, T. Organizer-Derived WOX5 Signal Maintains Root Columella Stem Cells through Chromatin-Mediated Repression of CDF4 Expression. Dev. Cell 2015, 33, 576–588. [Google Scholar] [CrossRef]
- Wei, Z.; Yuan, T.; Tarkowska, D.; Kim, J.; Nam, H.G.; Novak, O.; He, K.; Gou, X.; Li, J. Brassinosteroid Biosynthesis Is Modulated via a Transcription Factor Cascade of COG1, PIF4, and PIF5. Plant Physiol. 2017, 174, 1260–1273. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Papanatsiou, M.; Petersen, J.; Henderson, L.; Wang, Y.; Christie, J.M.; Blatt, M.R. Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 2019, 363, 1456–1459. [Google Scholar] [CrossRef]
- Guo, H.; Xiao, C.; Liu, Q.; Li, R.; Yan, Z.; Yao, X.; Hu, H. Two galacturonosyltransferases function in plant growth, stomatal development, and dynamics. Plant Physiol. 2021, 187, 2820–2836. [Google Scholar] [CrossRef] [PubMed]
- Amsbury, S.; Hunt, L.; Elhaddad, N.; Baillie, A.; Lundgren, M.; Verhertbruggen, Y.; Scheller, H.V.; Knox, J.P.; Fleming, A.J.; Gray, J.E. Stomatal Function Requires Pectin De-methyl-esterification of the Guard Cell Wall. Curr. Biol. 2016, 26, 2899–2906. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, W.; Turner, J.A.; Anderson, C.T. PECTATE LYASE LIKE12 patterns the guard cell wall to coordinate turgor pressure and wall mechanics for proper stomatal function in Arabidopsis. Plant Cell 2021, 33, 3134–3150. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Liang, Y.; Chen, S.; Yuan, Y.; Chen, Y.; Hu, H. Bna.EPF2 Enhances Drought Tolerance by Regulating Stomatal Development and Stomatal Size in Brassica napus. Int. J. Mol. Sci. 2023, 24, 8007. [Google Scholar] [CrossRef]
- Mohammed, U.; Caine, R.S.; Atkinson, J.A.; Harrison, E.L.; Wells, D.; Chater, C.C.; Gray, J.E.; Swarup, R.; Murchie, E.H. Rice plants overexpressing OsEPF1 show reduced stomatal density and increased root cortical aerenchyma formation. Sci. Rep. 2019, 9, 5584. [Google Scholar] [CrossRef]
- Wang, C.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.; Xia, X.; Yin, W. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 2016, 14, 849–860. [Google Scholar] [CrossRef]
- Hepworth, C.; Doheny-Adams, T.; Hunt, L.; Cameron, D.D.; Gray, J.E. Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol. 2015, 208, 336–341. [Google Scholar] [CrossRef]
- Hughes, J.; Hepworth, C.; Dutton, C.; Dunn, J.A.; Hunt, L.; Stephens, J.; Waugh, R.; Cameron, D.D.; Gray, J.E. Reducing Stomatal Density in Barley Improves Drought Tolerance without Impacting on Yield. Plant Physiol. 2017, 174, 776–787. [Google Scholar] [CrossRef]
- Brunner, A.M.; Busov, V.B.; Strauss, S.H. Poplar genome sequence: Functional genomics in an ecologically dominant plant species. Trends Plant Sci. 2004, 9, 49–56. [Google Scholar] [CrossRef]
- Qiu, Q.; Ma, T.; Hu, Q.; Liu, B.; Wu, Y.; Zhou, H.; Wang, Q.; Wang, J.; Liu, J. Genome-scale transcriptome analysis of the desert poplar, Populus euphratica. Tree Physiol. 2011, 31, 452–461. [Google Scholar] [CrossRef]
- Li, B.; Qin, Y.; Duan, H.; Yin, W.; Xia, X. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 2011, 62, 3765–3779. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Wu, Z.; Wang, X.; Jiang, Z.; Wang, Y.; Liu, H.; Qin, R.; Li, Z. Short-term transcriptomic responses of Populus euphratica roots and leaves to drought stress. J. For. Res. 2021, 32, 841–853. [Google Scholar] [CrossRef]
- Han, X.L.; Qiu, C.; Sun, J.H.; Xu, J.D.; Zhang, X.; Zhai, J.T.; Zhang, S.H.; Wu, Z.H.; Li, Z.J. Identification of AP2/ERF gene family of Salicaceae and their response to salt stress, abscisic acid, and gibberellic acid in Populus euphratica seeds. Biol. Plant. 2023, 67, 88–99. [Google Scholar] [CrossRef]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef]
- Waqas, M.; Shahid, L.; Shoukat, K.; Aslam, U.; Azeem, F.; Atif, R.M. Chapter 1—Role of DNA-binding with one finger (Dof) transcription factors for abiotic stress tolerance in plants. In Transcription Factors for Abiotic Stress Tolerance in Plants; Wani, S.H., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 1–14. [Google Scholar]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef]
- He, L.; Su, C.; Wang, Y.; Wei, Z. ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 2015, 110, 17–24. [Google Scholar] [CrossRef]
- Dong, C.; Hu, H.; Xie, J. Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 2016, 59, 1085–1100. [Google Scholar] [CrossRef]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Y.; Zhang, J.; Ma, X.; Li, Y.; Li, M.; Wang, D.; Kang, M.; Wu, H.; Yang, Y.; et al. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica). Mol. Ecol. Resour. 2020, 20, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Wu, H.; Chen, Y.; Li, X.; Hou, J.; Lu, J.; Wei, S.; Dai, X.; Olson, M.S.; Liu, J.; et al. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat. Commun. 2020, 11, 5893. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Xia, Y.Z.; Li, K.; Li, J.J.; Wang, T.Q.; Gu, L.C.; Xun, L.Y. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. Nucleic Acids Res. 2019, 47, e15. [Google Scholar] [CrossRef]

| Gene ID | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|
| PeuTF01G00832.1 | 244 | 27,021.21 | 8.17 | 52.46 | 63.11 | −0.671 |
| PeuTF01G02355.1 | 323 | 34,585.48 | 9.48 | 63.19 | 59.16 | −0.583 |
| PeuTF02G00480.1 | 300 | 34,034.88 | 4.77 | 53.38 | 63.40 | −0.675 |
| PeuTF02G01099.1 | 264 | 27565.24 | 6.30 | 45.47 | 46.97 | −0.541 |
| PeuTF02G01562.1 | 246 | 27,015.15 | 9.28 | 46.80 | 53.17 | −0.748 |
| PeuTF03G00284.1 | 235 | 25,200.50 | 8.79 | 46.44 | 43.62 | −0.930 |
| PeuTF03G01435.1 | 279 | 30,697.23 | 8.63 | 52.31 | 62.58 | −0.641 |
| PeuTF04G00322.1 | 304 | 33,730.41 | 8.73 | 53.13 | 61.88 | −0.791 |
| PeuTF04G00383.1 | 325 | 35,635.15 | 9.3 | 61.05 | 52.80 | −0.916 |
| PeuTF04G00386.1 | 345 | 38,108.04 | 8.42 | 53.79 | 54.23 | −0.757 |
| PeuTF04G00502.1 | 159 | 17,653.88 | 8.99 | 49.20 | 49.62 | −0.826 |
| PeuTF04G01143.1 | 502 | 54,941.55 | 5.85 | 54.11 | 50.54 | −0.894 |
| PeuTF05G01114.1 | 253 | 26,039.95 | 8.44 | 41.24 | 56.72 | −0.378 |
| PeuTF05G01128.1 | 316 | 33,705.48 | 9.49 | 45.06 | 63.96 | −0.516 |
| PeuTF05G01285.1 | 342 | 37,143.24 | 8.92 | 44.88 | 59.68 | −0.608 |
| PeuTF05G01648.1 | 297 | 33,543.19 | 4.61 | 43.52 | 61.08 | −0.695 |
| PeuTF06G00823.1 | 326 | 34,715.73 | 9.10 | 54.35 | 50.95 | −0.542 |
| PeuTF06G01955.1 | 288 | 31,953.02 | 5.98 | 62.88 | 50.80 | −0.826 |
| PeuTF07G00397.1 | 248 | 25,508.40 | 8.57 | 41.51 | 60.24 | −0.344 |
| PeuTF07G00415.1 | 325 | 34,483.32 | 8.96 | 49.11 | 63.97 | −0.452 |
| PeuTF07G00620.1 | 345 | 37,195.00 | 8.28 | 44.45 | 56.35 | −0.599 |
| PeuTF08G00173.1 | 346 | 36,955.26 | 9.18 | 51.97 | 63.99 | −0.545 |
| PeuTF08G00503.1 | 500 | 54,216.45 | 6.51 | 56.44 | 48.60 | −0.833 |
| PeuTF09G00304.1 | 325 | 34,474.38 | 9.35 | 62.58 | 51.32 | −0.524 |
| PeuTF10G00506.1 | 337 | 35,634.61 | 9.30 | 50.81 | 58.72 | −0.569 |
| PeuTF10G00899.1 | 415 | 45,414.71 | 9.33 | 60.22 | 50.72 | −0.782 |
| PeuTF11G00355.1 | 305 | 33,930.65 | 7.57 | 58.18 | 61.05 | −0.775 |
| PeuTF11G00415.1 | 320 | 35,125.52 | 9.26 | 68.67 | 50.91 | −0.912 |
| PeuTF11G00419.1 | 351 | 38,445.75 | 8.73 | 49.97 | 58.06 | −0.646 |
| PeuTF11G00428.1 | 358 | 39,314.79 | 8.57 | 50.56 | 59.11 | −0.642 |
| PeuTF11G00528.1 | 160 | 17,804.13 | 9.28 | 56.11 | 54.19 | −0.778 |
| PeuTF12G00127.1 | 297 | 32,814.02 | 7.14 | 48.17 | 46.40 | −0.844 |
| PeuTF12G00528.1 | 331 | 36,602.88 | 6.30 | 49.99 | 53.90 | −0.911 |
| PeuTF12G00722.1 | 312 | 34,251.20 | 6.64 | 43.32 | 57.47 | −0.628 |
| PeuTF13G00671.1 | 494 | 53,843.54 | 6.57 | 46.71 | 60.61 | −0.580 |
| PeuTF14G00964.1 | 261 | 28,561.58 | 9.14 | 58.13 | 49.35 | −0.808 |
| PeuTF14G01584.1 | 261 | 27,480.08 | 5.84 | 51.41 | 46.36 | −0.572 |
| PeuTF15G00102.1 | 255 | 28,187.27 | 8.24 | 50.45 | 47.49 | −0.770 |
| PeuTF15G00470.1 | 320 | 35,176.36 | 7.70 | 49.66 | 54.25 | −0.838 |
| PeuTF15G00759.1 | 313 | 34,601.65 | 6.19 | 49.33 | 57.57 | −0.650 |
| PeuTF16G00617.1 | 289 | 32,307.57 | 5.77 | 57.43 | 50.93 | −0.764 |
| PeuTF17G00710.1 | 509 | 55,583.03 | 5.49 | 51.25 | 49.86 | −0.913 |
| PeuTF19G00358.1 | 212 | 23,388.24 | 7.60 | 36.75 | 57.08 | −0.787 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yuan, Y.; Jia, M.; Yang, H.; Jiao, P.; Guo, H. Genome-Wide Identification of the Dof Gene Family and Functional Analysis of PeSCAP1 in Regulating Guard Cell Maturation in Populus euphratica. Int. J. Mol. Sci. 2025, 26, 3798. https://doi.org/10.3390/ijms26083798
Chen Y, Yuan Y, Jia M, Yang H, Jiao P, Guo H. Genome-Wide Identification of the Dof Gene Family and Functional Analysis of PeSCAP1 in Regulating Guard Cell Maturation in Populus euphratica. International Journal of Molecular Sciences. 2025; 26(8):3798. https://doi.org/10.3390/ijms26083798
Chicago/Turabian StyleChen, Yongqiang, Yang Yuan, Mingyu Jia, Huiyun Yang, Peipei Jiao, and Huimin Guo. 2025. "Genome-Wide Identification of the Dof Gene Family and Functional Analysis of PeSCAP1 in Regulating Guard Cell Maturation in Populus euphratica" International Journal of Molecular Sciences 26, no. 8: 3798. https://doi.org/10.3390/ijms26083798
APA StyleChen, Y., Yuan, Y., Jia, M., Yang, H., Jiao, P., & Guo, H. (2025). Genome-Wide Identification of the Dof Gene Family and Functional Analysis of PeSCAP1 in Regulating Guard Cell Maturation in Populus euphratica. International Journal of Molecular Sciences, 26(8), 3798. https://doi.org/10.3390/ijms26083798





