An Optimized Editing Approach for Wheat Genes by Improving sgRNA Design and Transformation Strategies
Abstract
1. Introduction
2. Results
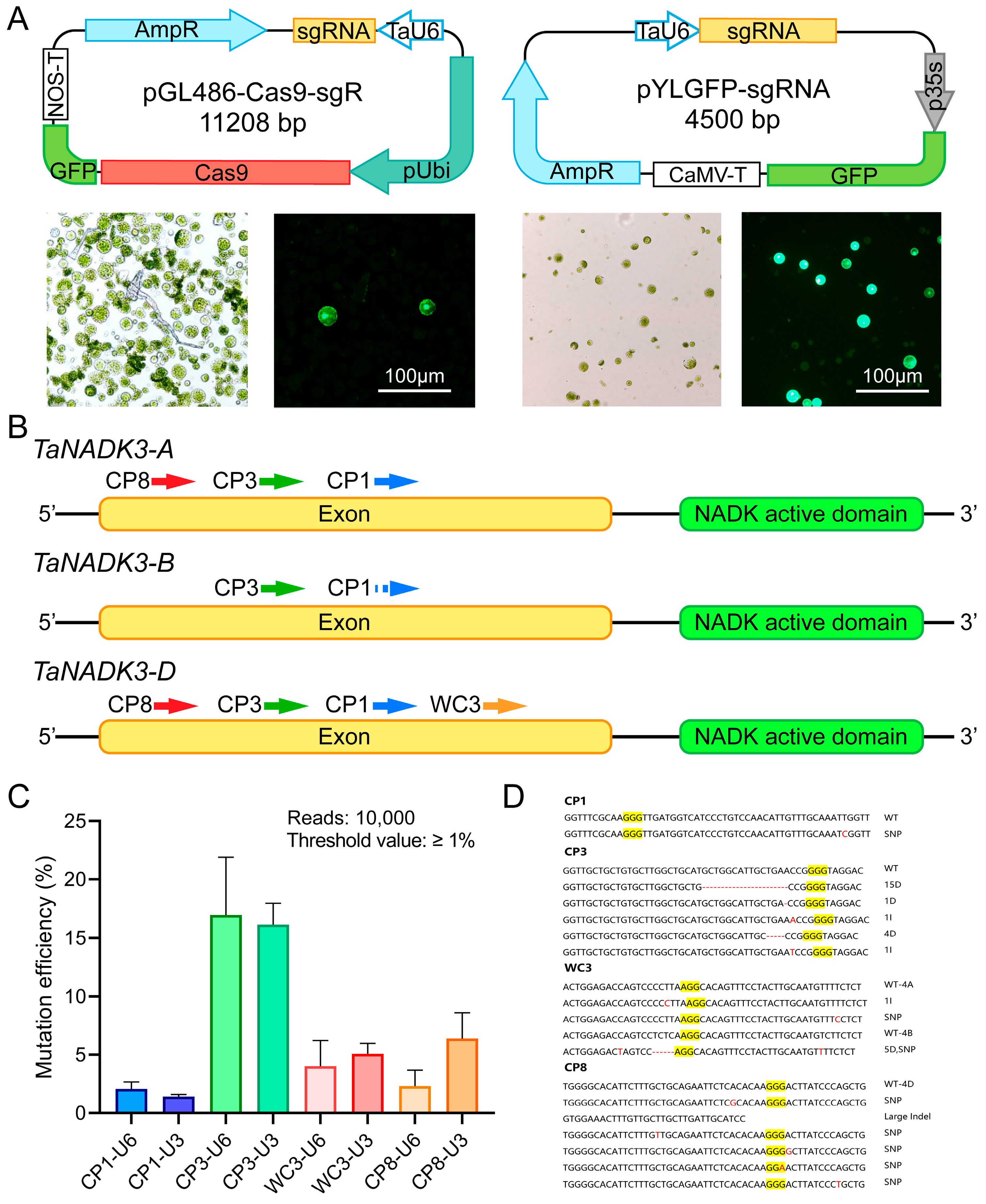
2.1. Evaluation of sgRNA Editing Efficiency in Wheat Protoplasts
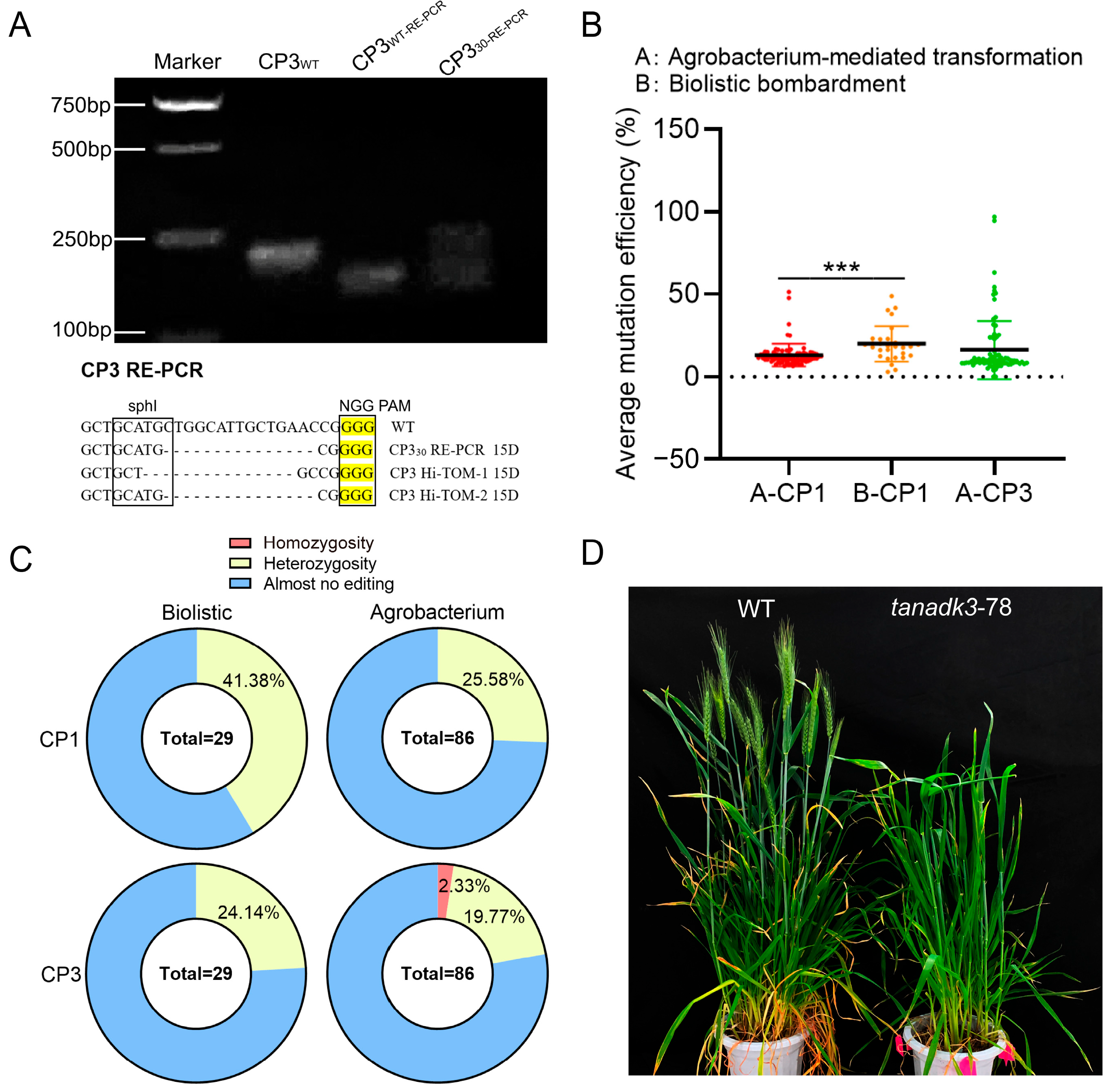
2.2. Comparative Analysis of Transformation Methods for Generating Homozygous Mutants
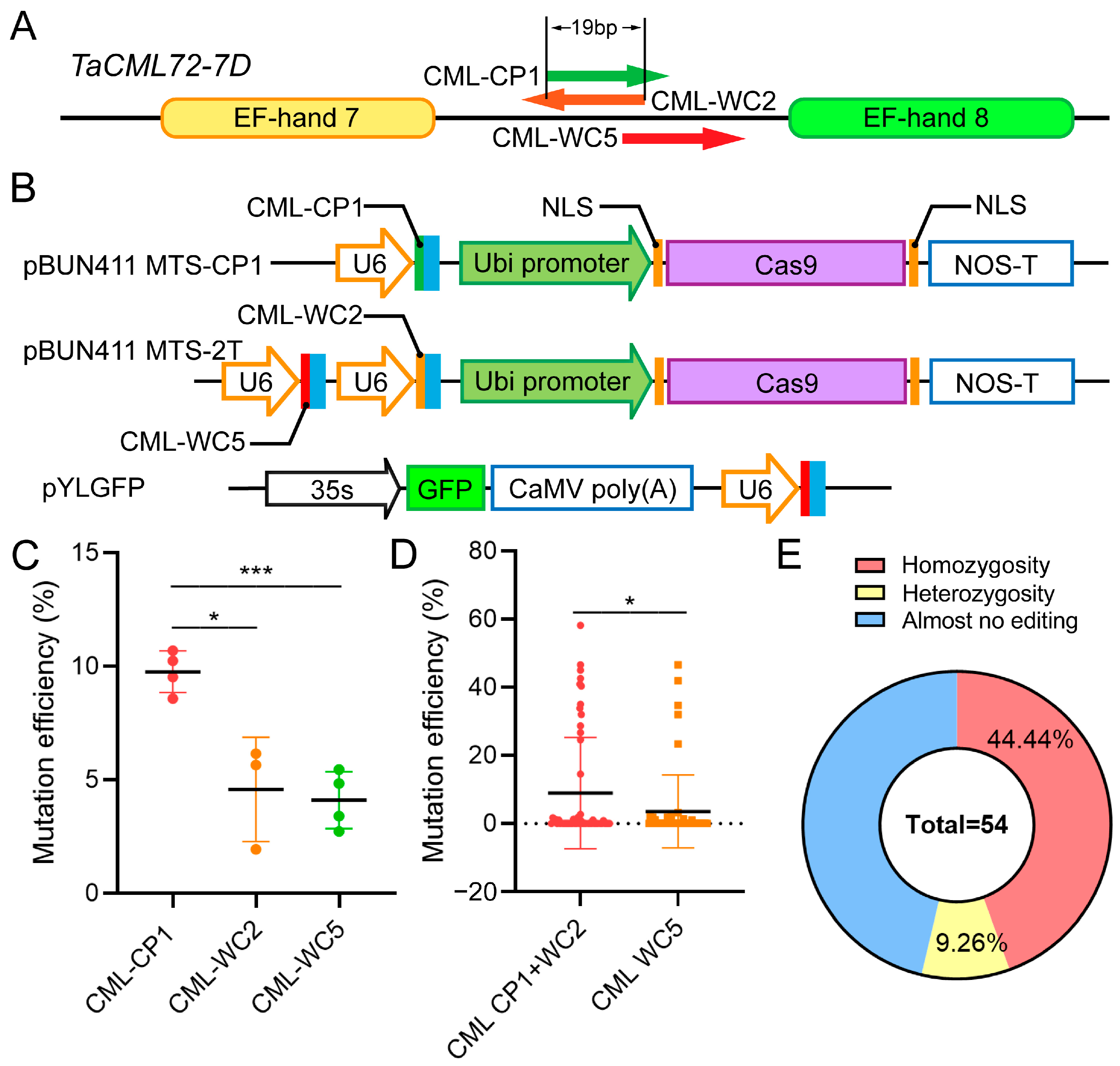
2.3. Impact of sgRNA Target Site Configuration on Gene Editing Efficiency
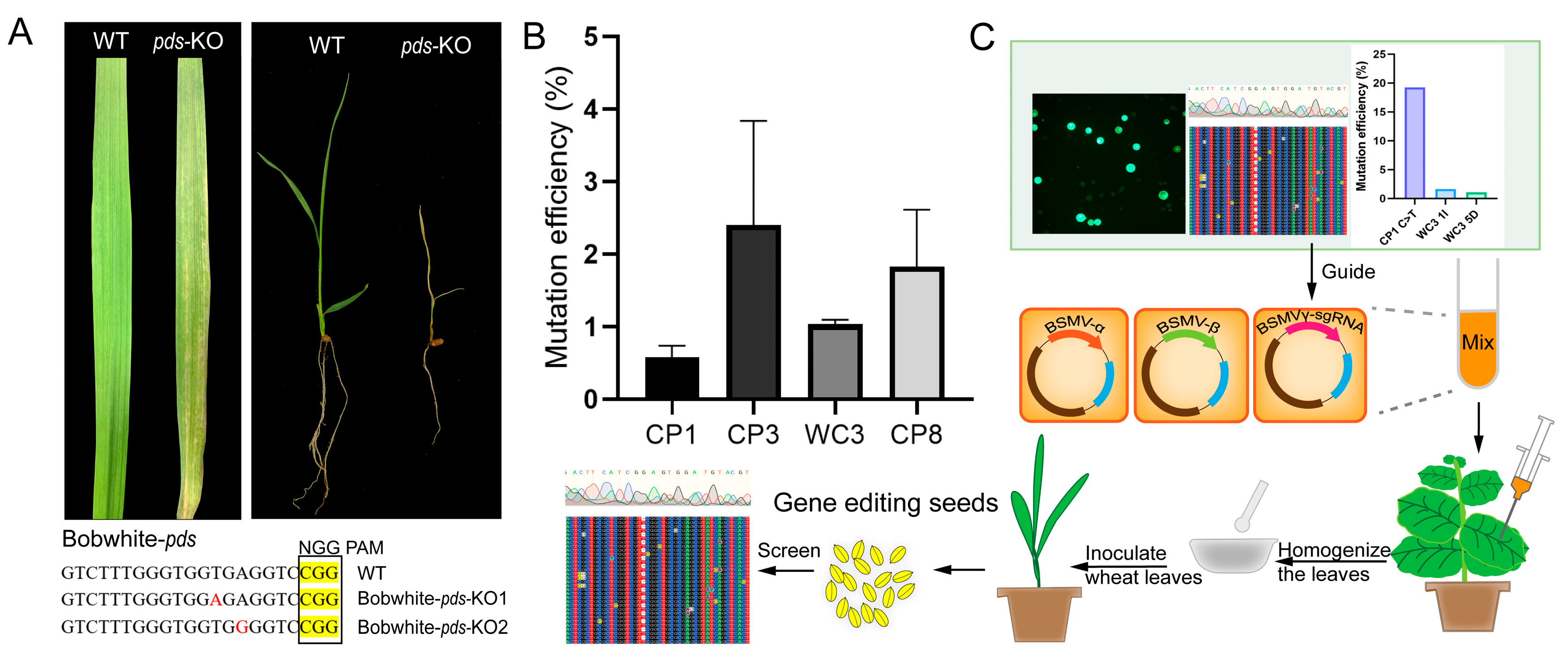
2.4. Development of a BSMV-Mediated Gene Editing System for Wheat
3. Discussion
3.1. Protoplast-Based Evaluation System Enhances sgRNA Design Efficiency
3.2. Multiple Factors Influence sgRNA Editing Efficiency
3.3. Optimized Transformation Strategies for Generating Homozygous Mutants
3.4. BSMV-Mediated Delivery System Offers an Accessible Alternative
3.5. Future Perspectives and Limitations
4. Materials and Methods
4.1. Plant Materials, Plasmids, and Bacteria Strains
4.2. Construction of Plasmids
4.3. Isolation and Transformation of Wheat Protoplasts
4.4. Primers and Guide RNA Design
4.5. Wheat Transformation
4.6. Detection Analysis of Edited Mutations
4.7. Phenotypic Detection of TaNADK3-Edited Mutations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hendelman, A.; Zebell, S.; Rodriguez-Leal, D.; Dukler, N.; Robitaille, G.; Wu, X.; Kostyun, J.; Tal, L.; Wang, P.; Bartlett, M.E.; et al. Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis-regulatory dissection. Cell 2021, 184, 1724–1739.e16. [Google Scholar] [CrossRef] [PubMed]
- CAST. A Paper in the Series on The Need for Agricultural Innovation to Sustainably Feed the World by 2050; CAST: Ames, IA, USA, 2017. [Google Scholar]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, F.; Wang, J.; Wang, K.; Tian, C.; Qi, X.; Lu, F.; Liu, X.; Ye, X.; Jiao, Y. Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nat. Plants 2022, 8, 930–939. [Google Scholar] [CrossRef]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef]
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W.; et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 2022, 185, 2961–2974.e19. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Liu, L.; Kuang, Y.; Yan, F.; Li, S.; Ren, B.; Gosavi, G.; Spetz, C.; Li, X.; Wang, X.; Zhou, X.; et al. Developing a novel artificial rice germplasm for dinitroaniline herbicide resistance by base editing of OsTubA2. Plant Biotechnol. J. 2021, 19, 5–7. [Google Scholar] [CrossRef]
- Tan, J.; Zeng, D.; Zhao, Y.; Wang, Y.; Liu, T.; Li, S.; Xue, Y.; Luo, Y.; Xie, X.; Chen, L.; et al. PhieABEs: A PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 2022, 20, 934–943. [Google Scholar] [CrossRef]
- Zeng, D.; Liu, T.; Tan, J.; Zhang, Y.; Zheng, Z.; Wang, B.; Zhou, D.; Xie, X.; Guo, M.; Liu, Y.G.; et al. PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 2020, 13, 1666–1669. [Google Scholar] [CrossRef]
- Zheng, Z.; Liu, T.; Chai, N.; Zeng, D.; Zhang, R.; Wu, Y.; Hang, J.; Liu, Y.; Deng, Q.; Tan, J.; et al. PhieDBEs: A DBD-containing, PAM-flexible, high-efficiency dual base editor toolbox with wide targeting scope for use in plants. Plant Biotechnol. J. 2024, 22, 3164–3174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.X.; Li, B.B.; Yang, Z.G.; Huang, J.Q.; Sun, W.H.; Bhanbhro, N.; Liu, W.T.; Chen, K.M. Dissecting Plant Gene Functions Using CRISPR Toolsets for Crop Improvement. J. Agric. Food Chem. 2022, 70, 7343–7359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Chai, N.; Liu, T.; Zheng, Z.; Lin, Q.; Xie, X.; Wen, J.; Yang, Z.; Liu, Y.G.; Zhu, Q. The type V effectors for CRISPR/Cas-mediated genome engineering in plants. Biotechnol. Adv. 2024, 74, 108382. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.L.; Chen, Y.H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef]
- Li, S.; Zhang, C.; Li, J.; Yan, L.; Wang, N.; Xia, L. Present and future prospects for wheat improvement through genome editing and advanced technologies. Plant Commun. 2021, 2, 100211. [Google Scholar] [CrossRef]
- Wang, K.; Liu, H.; Du, L.; Ye, X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 2017, 15, 614–623. [Google Scholar] [CrossRef]
- Richardson, T.; Thistleton, J.; Higgins, T.J.; Howitt, C.; Ayliffe, M. Efficient agrobacterium transformation of elite wheat germplasm without selection. Plant Cell Tissue Organ. Cult. 2014, 119, 647–659. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 2021, 19, 427–429. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, R.; Song, G.; Gao, J.; Li, W.; Han, X.; Chen, M.; Li, Y.; Li, G. Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol. 2018, 18, 302. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Li, B.B.; Wang, X.; Tai, L.; Ma, T.T.; Shalmani, A.; Liu, W.T.; Li, W.Q.; Chen, K.M. NAD Kinases: Metabolic Targets Controlling Redox Co-enzymes and Reducing Power Partitioning in Plant Stress and Development. Front. Plant Sci. 2018, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.J.; Yang, B.; Li, W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef]
- Li, J.; Kong, D.; Ke, Y.; Zeng, W.; Miki, D. Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis. BMC Biol. 2024, 22, 6. [Google Scholar] [CrossRef]
- Long, L.; Guo, D.D.; Gao, W.; Yang, W.W.; Hou, L.P.; Ma, X.N.; Miao, Y.C.; Botella, J.R.; Song, C.P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Lv, J.; Huang, H.; Wang, J.; Zhuo, M.; Tan, Z.; Huang, G.; Liu, J.; Liu, Y.; et al. Engineered circular guide RNAs boost CRISPR/Cas12a- and CRISPR/Cas13d-based DNA and RNA editing. Genome Biol. 2023, 24, 145. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-Based Multiplex Genome Editing in Monocot and Dicot Plants. Curr. Protoc. Mol. Biol. 2016, 115, 31.6.1–31.6.21. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Ishida, Y.; Tsunashima, M.; Hiei, Y.; Komari, T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 2015, 1223, 189–198. [Google Scholar]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Sun, T.; Liu, Q.; Chen, X.; Hu, F.; Wang, K. Hi-TOM 2.0: An improved platform for high-throughput mutation detection. Sci. China Life Sci. 2024, 67, 1532–1534. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Foyer, C.H. Oxidative stress and antioxidative systems: Recipes for successful data collection and interpretation. Plant Cell Environ. 2016, 39, 1140–1160. [Google Scholar] [CrossRef]
| Target Gene | Target Site | Off-Target Site Prediction | Off-Target |
|---|---|---|---|
| TaNADK3 | CP1 | TraesCS1B02G376800 | 1.04% |
| TaNADK3 | CP3 | TraesCS1B02G440300 | No |
| TaCML72-7D | CML-CP1 | XM_044587532.1 | 2.09% |
| TaCML72-7D | CML-WC2 | XM_044566133.1 | No |
| TaCML72-7D | CML-WC5 | TraesCS1D02G312500 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.-X.; Zhang, Y.-F.; Yang, H.; Zhang, X.-D.; Yang, Z.-G.; Li, B.-B.; Sun, W.-H.; Yang, Z.; Liu, W.-T.; Chen, K.-M. An Optimized Editing Approach for Wheat Genes by Improving sgRNA Design and Transformation Strategies. Int. J. Mol. Sci. 2025, 26, 3796. https://doi.org/10.3390/ijms26083796
Zhang R-X, Zhang Y-F, Yang H, Zhang X-D, Yang Z-G, Li B-B, Sun W-H, Yang Z, Liu W-T, Chen K-M. An Optimized Editing Approach for Wheat Genes by Improving sgRNA Design and Transformation Strategies. International Journal of Molecular Sciences. 2025; 26(8):3796. https://doi.org/10.3390/ijms26083796
Chicago/Turabian StyleZhang, Rui-Xiang, Yun-Fei Zhang, Hao Yang, Xiao-Dong Zhang, Zheng-Guang Yang, Bin-Bin Li, Wei-Hang Sun, Zi Yang, Wen-Ting Liu, and Kun-Ming Chen. 2025. "An Optimized Editing Approach for Wheat Genes by Improving sgRNA Design and Transformation Strategies" International Journal of Molecular Sciences 26, no. 8: 3796. https://doi.org/10.3390/ijms26083796
APA StyleZhang, R.-X., Zhang, Y.-F., Yang, H., Zhang, X.-D., Yang, Z.-G., Li, B.-B., Sun, W.-H., Yang, Z., Liu, W.-T., & Chen, K.-M. (2025). An Optimized Editing Approach for Wheat Genes by Improving sgRNA Design and Transformation Strategies. International Journal of Molecular Sciences, 26(8), 3796. https://doi.org/10.3390/ijms26083796







