METTL3 Promotes Cutaneous T-Cell Lymphoma Progression by Regulating ARHGEF12 Expression
Abstract
1. Introduction
2. Results
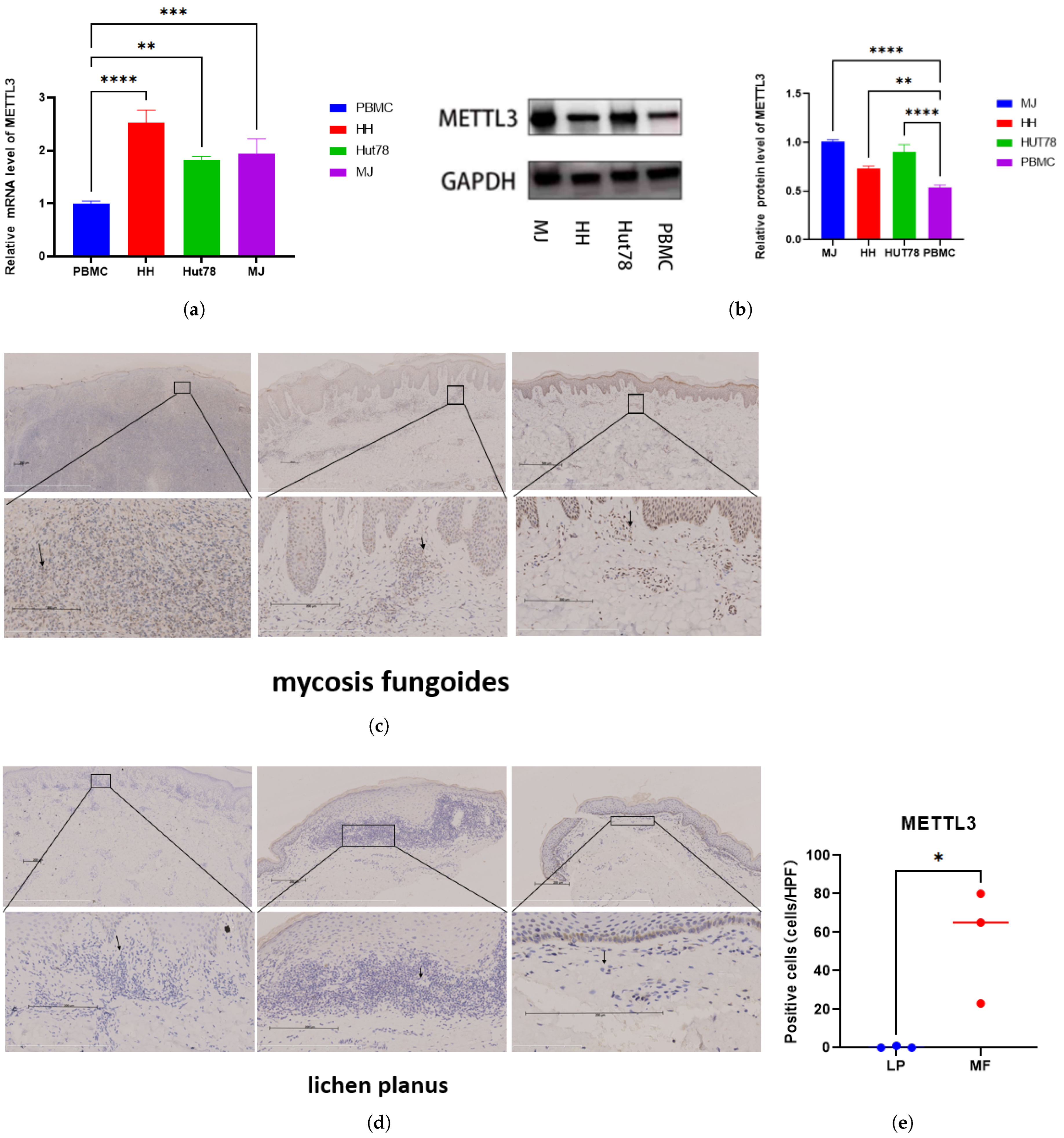
2.1. Elevated METTL3 Expression in MF Tissues and CTCL Cell Lines
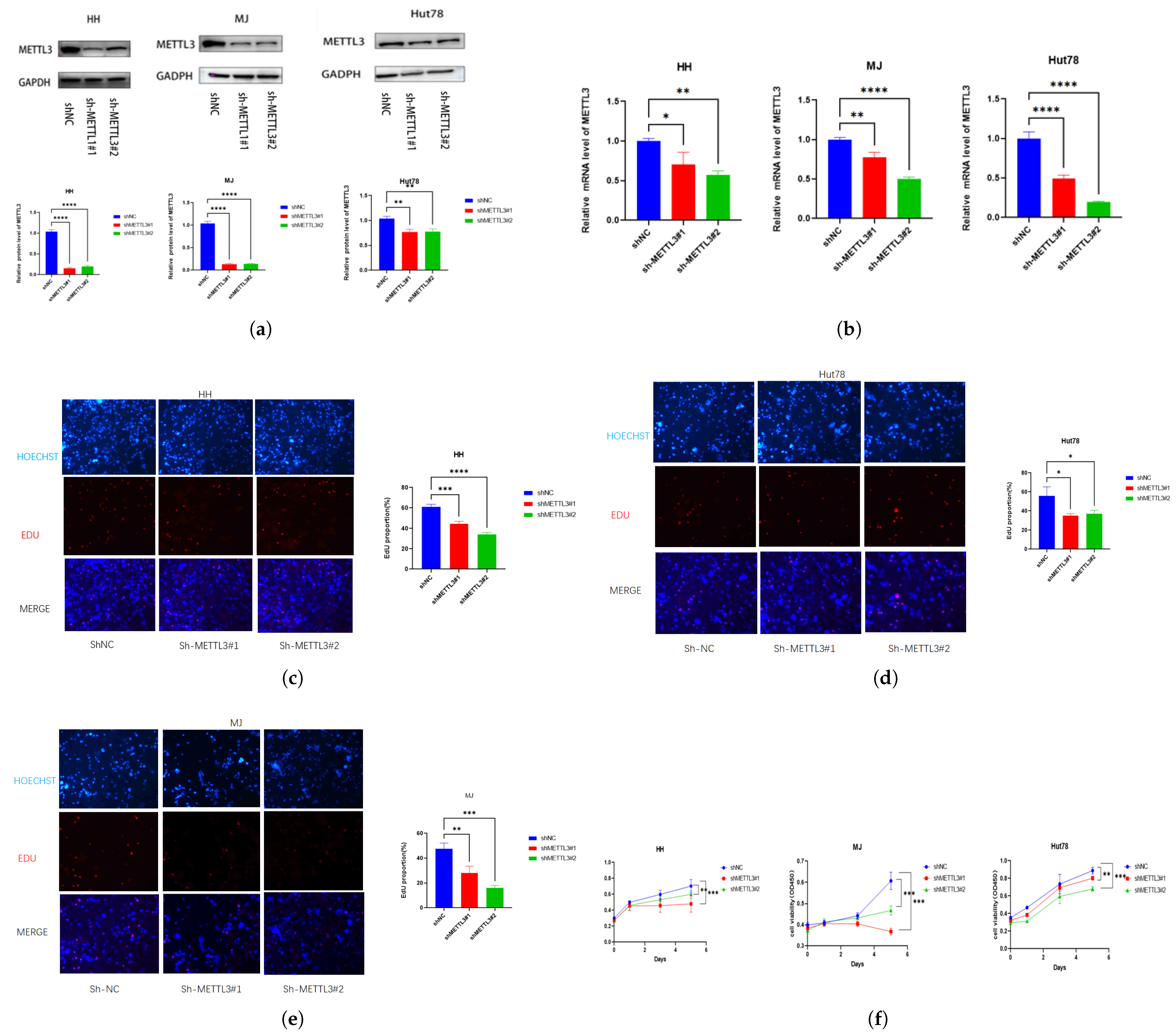
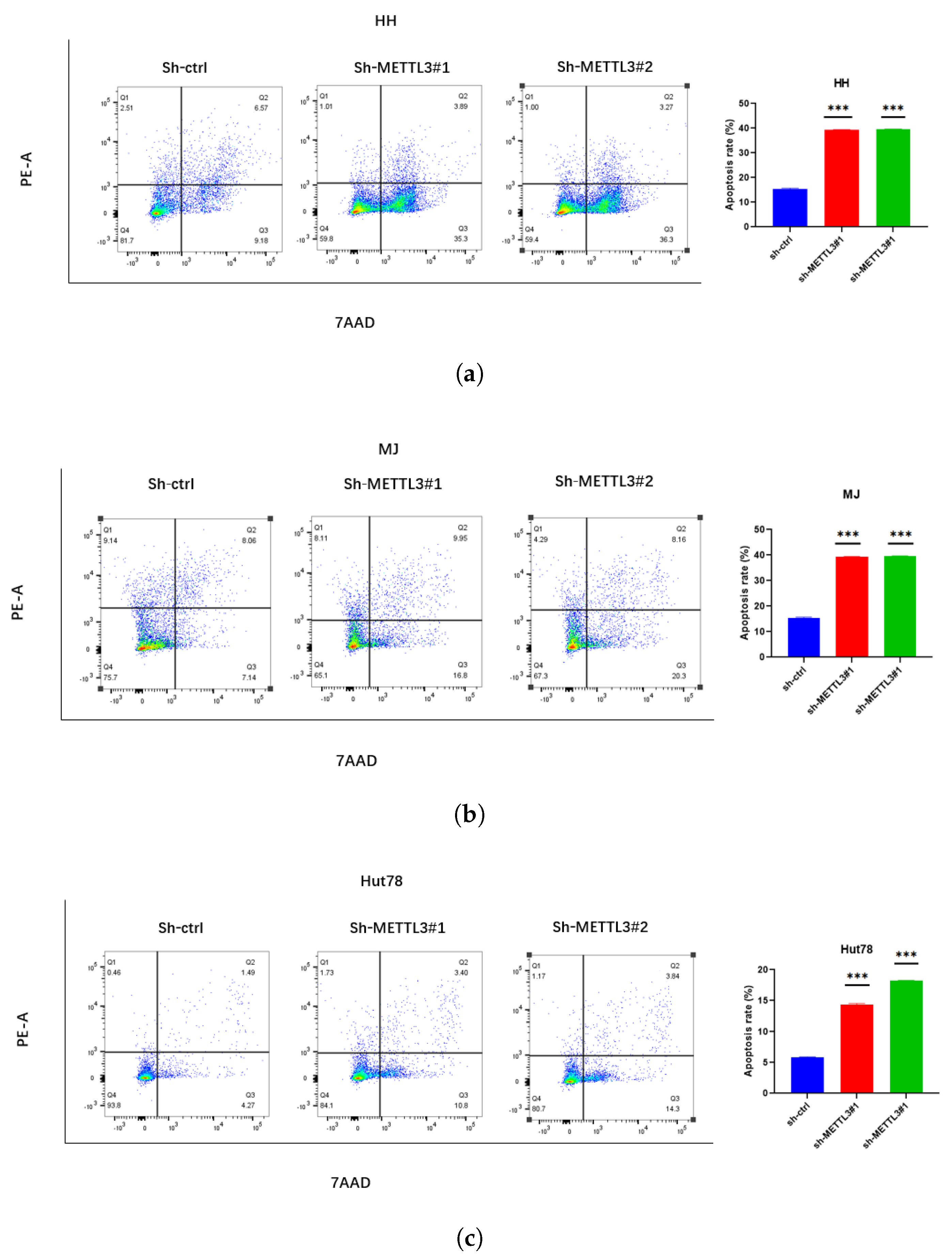
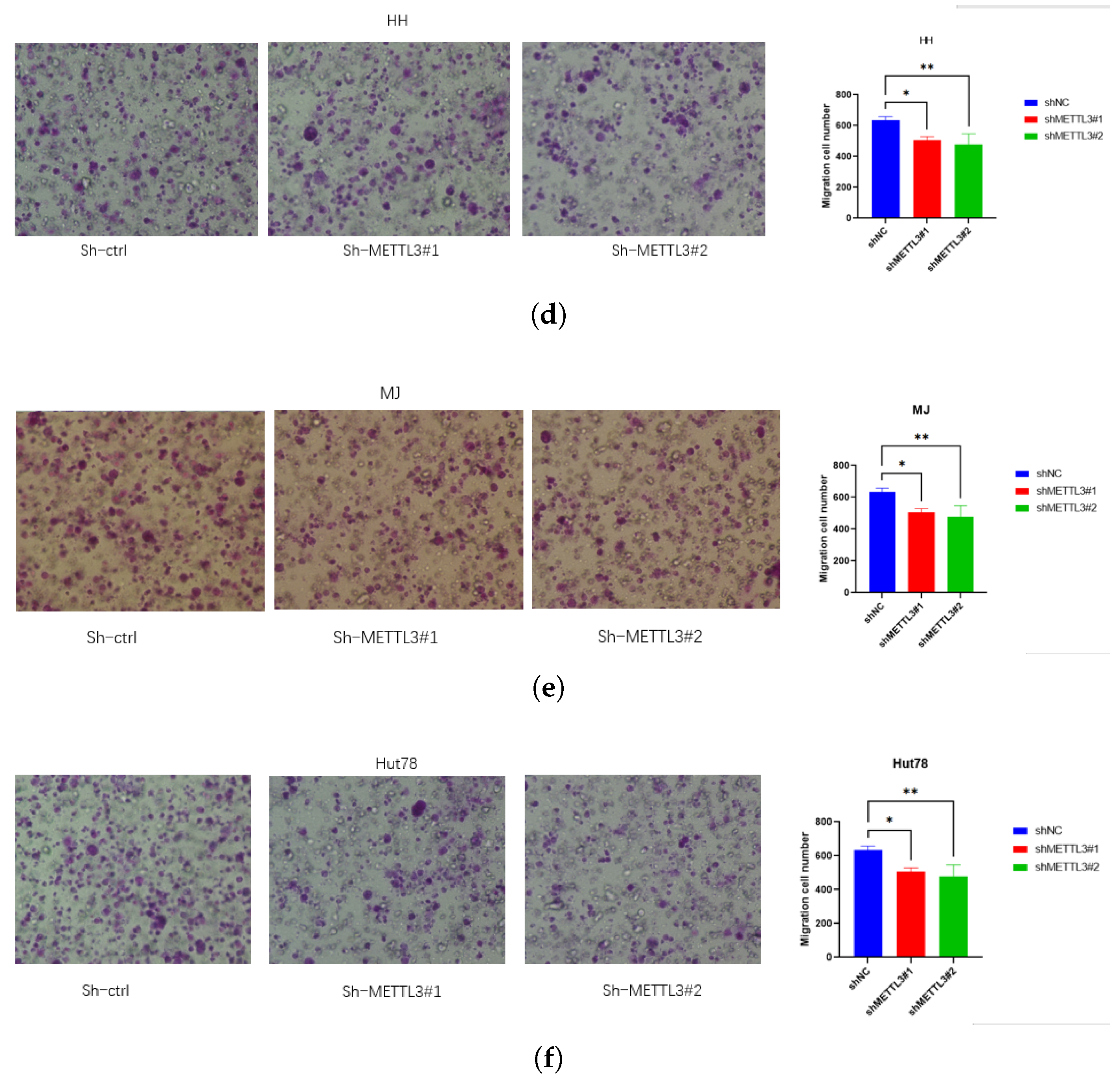
2.2. Knockdown METTL3 Suppresses CTCL Progression
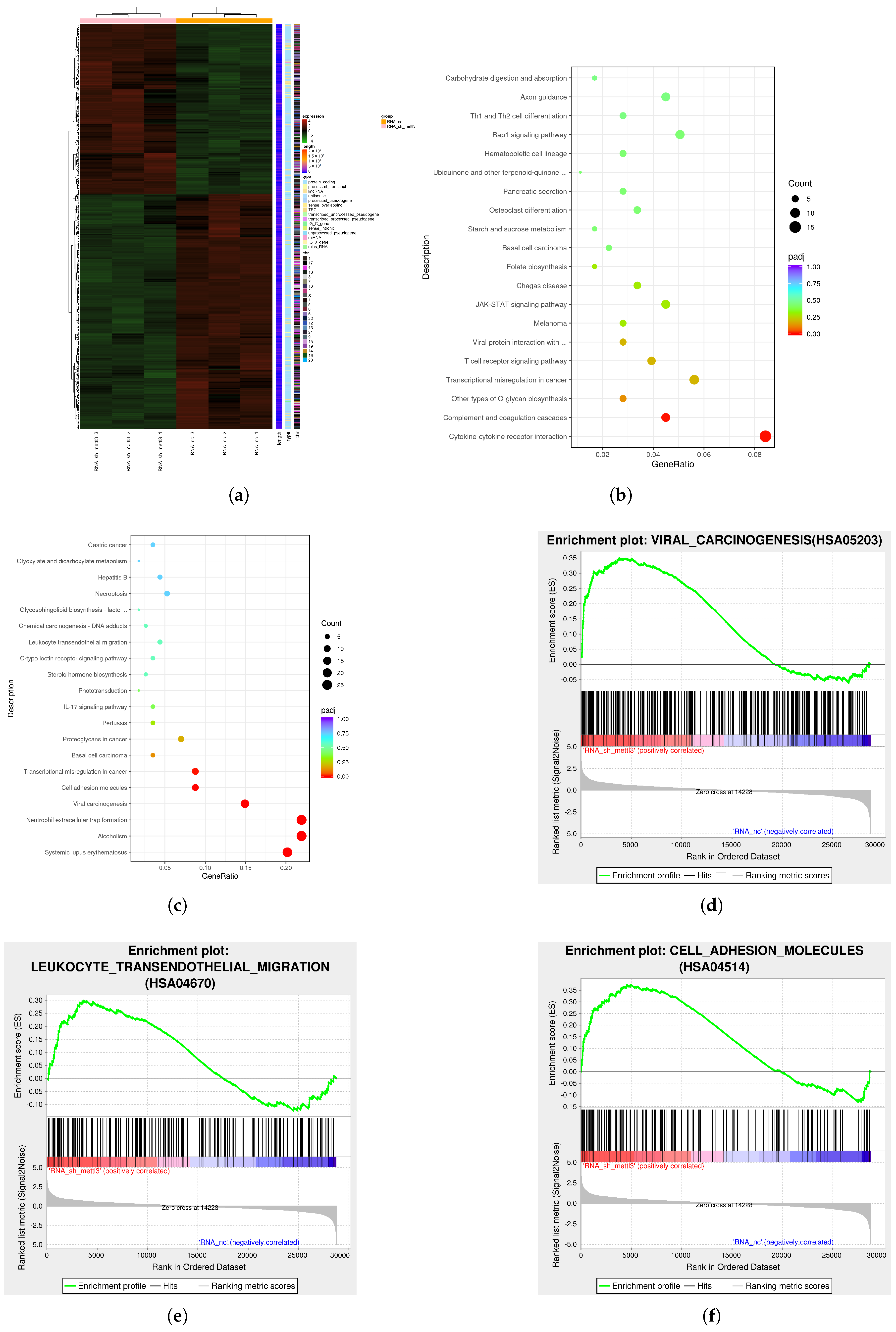
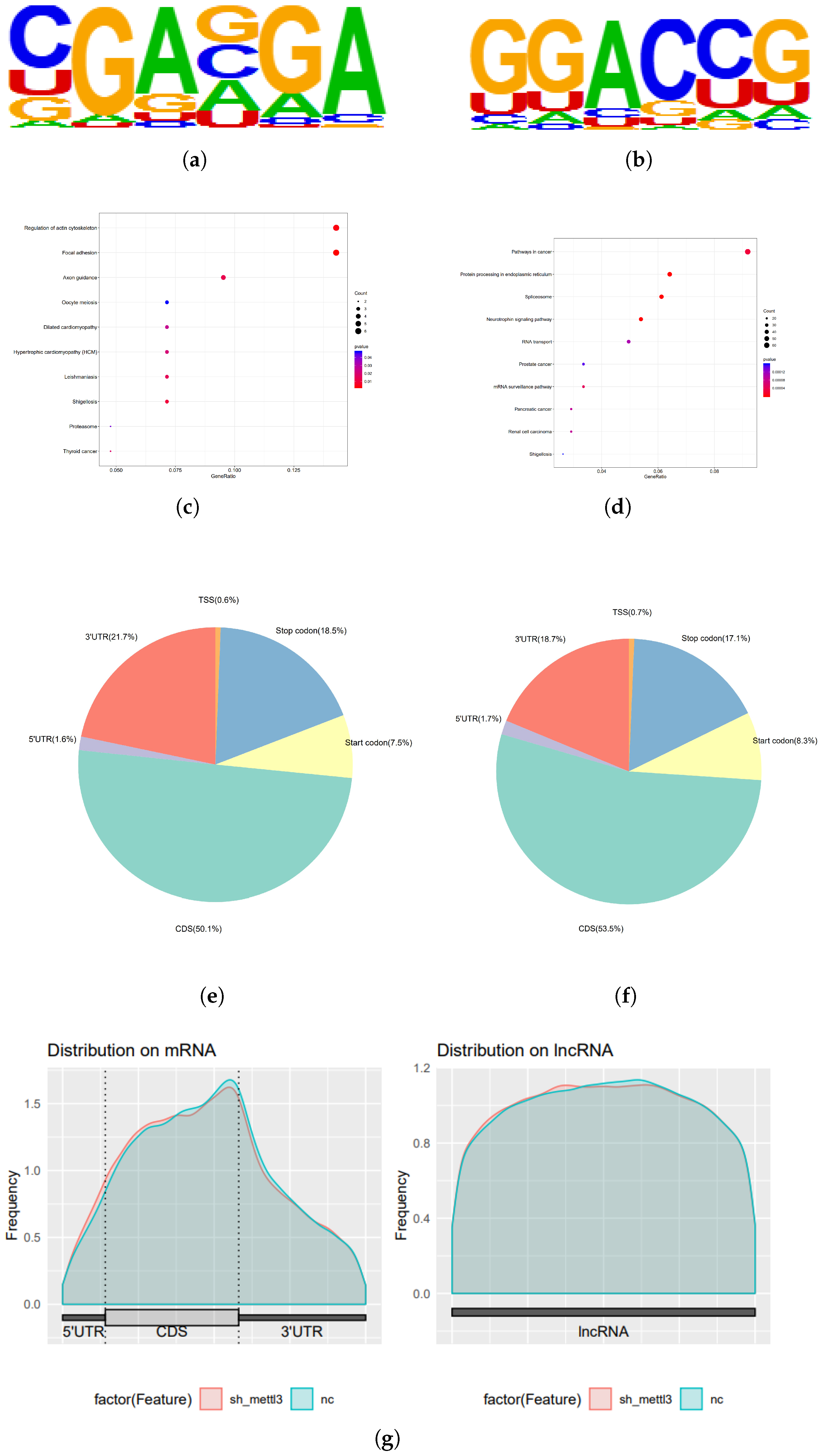
2.3. The Downstream Targets of METTL3 Identified by Multi-Omics Analysis
2.4. METTL3 Reduces ARHGEF12 mRNA Stability
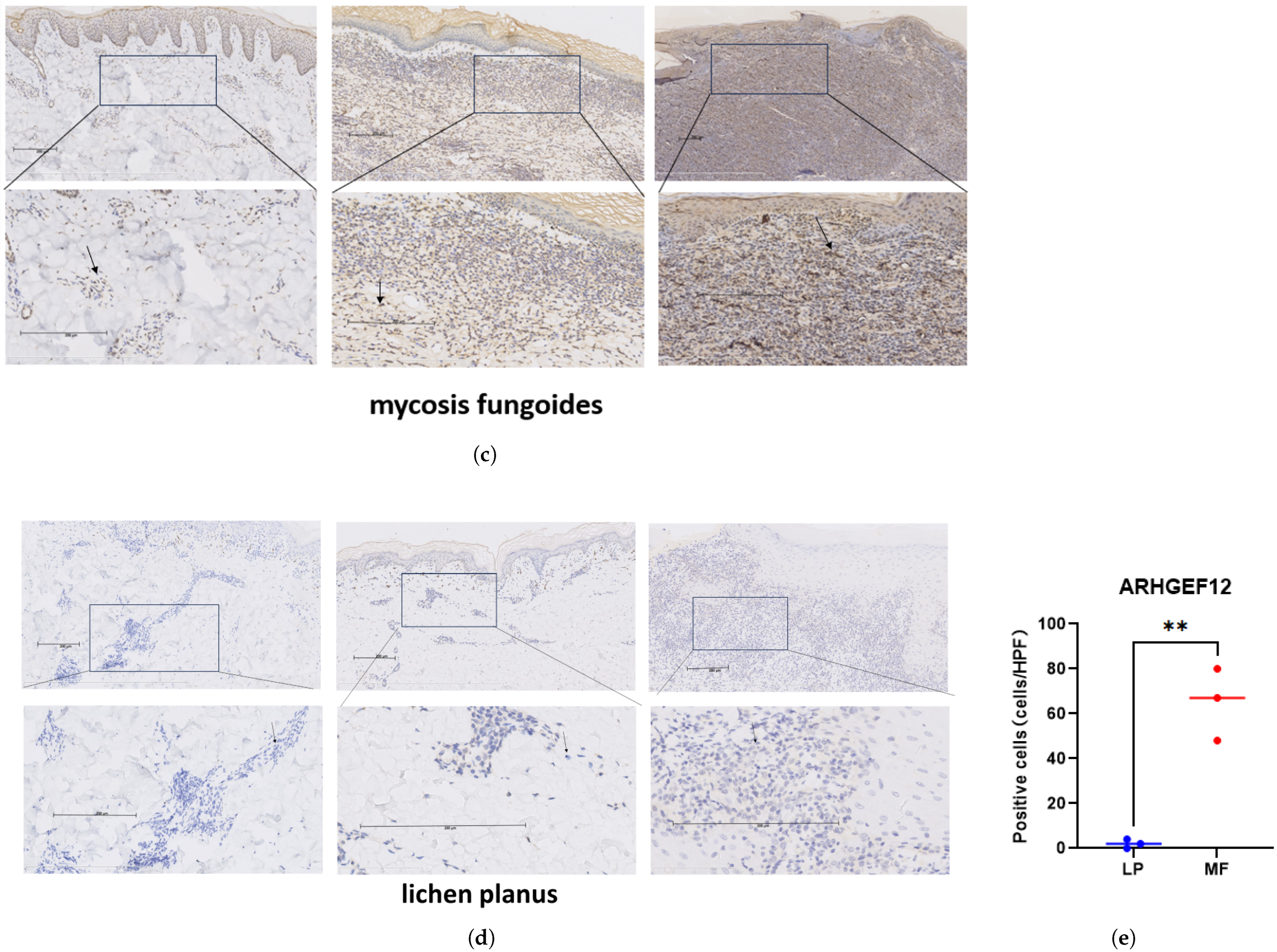
2.5. Elevated ARHGEF12 Expression in CTCL and MF Patients
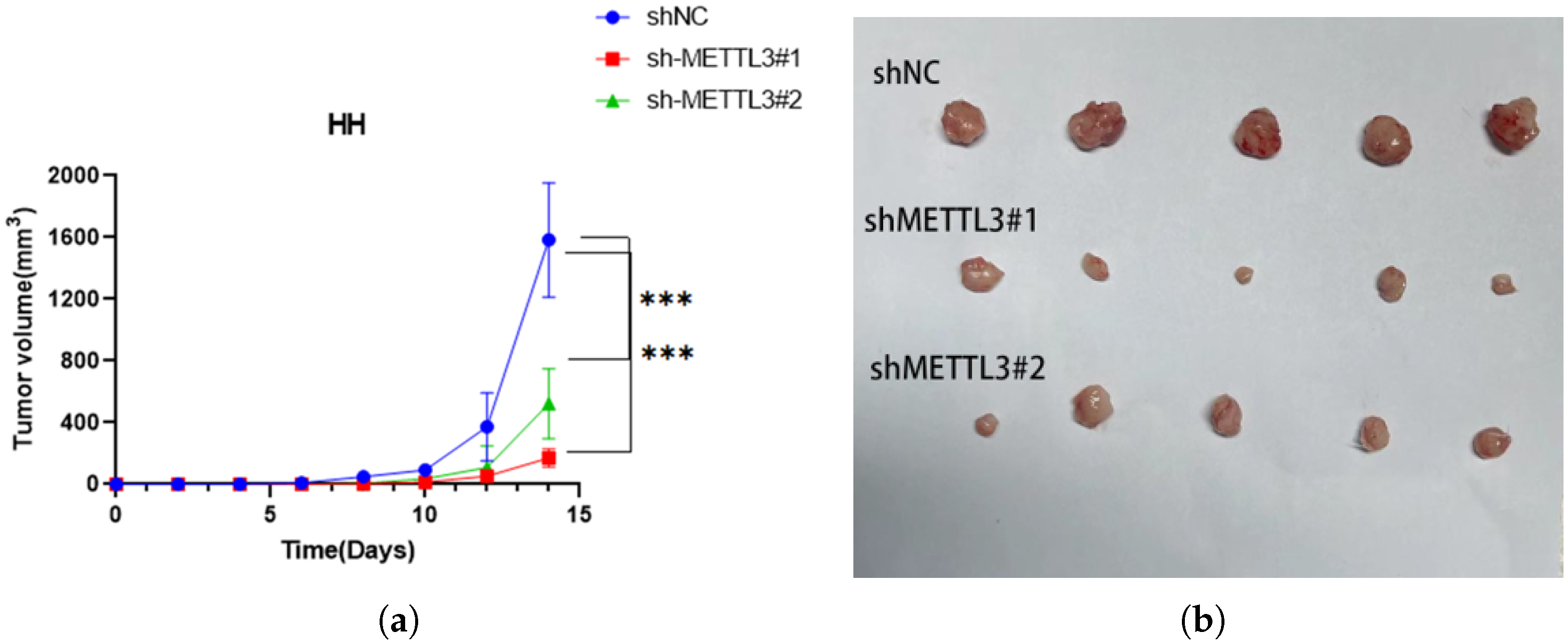
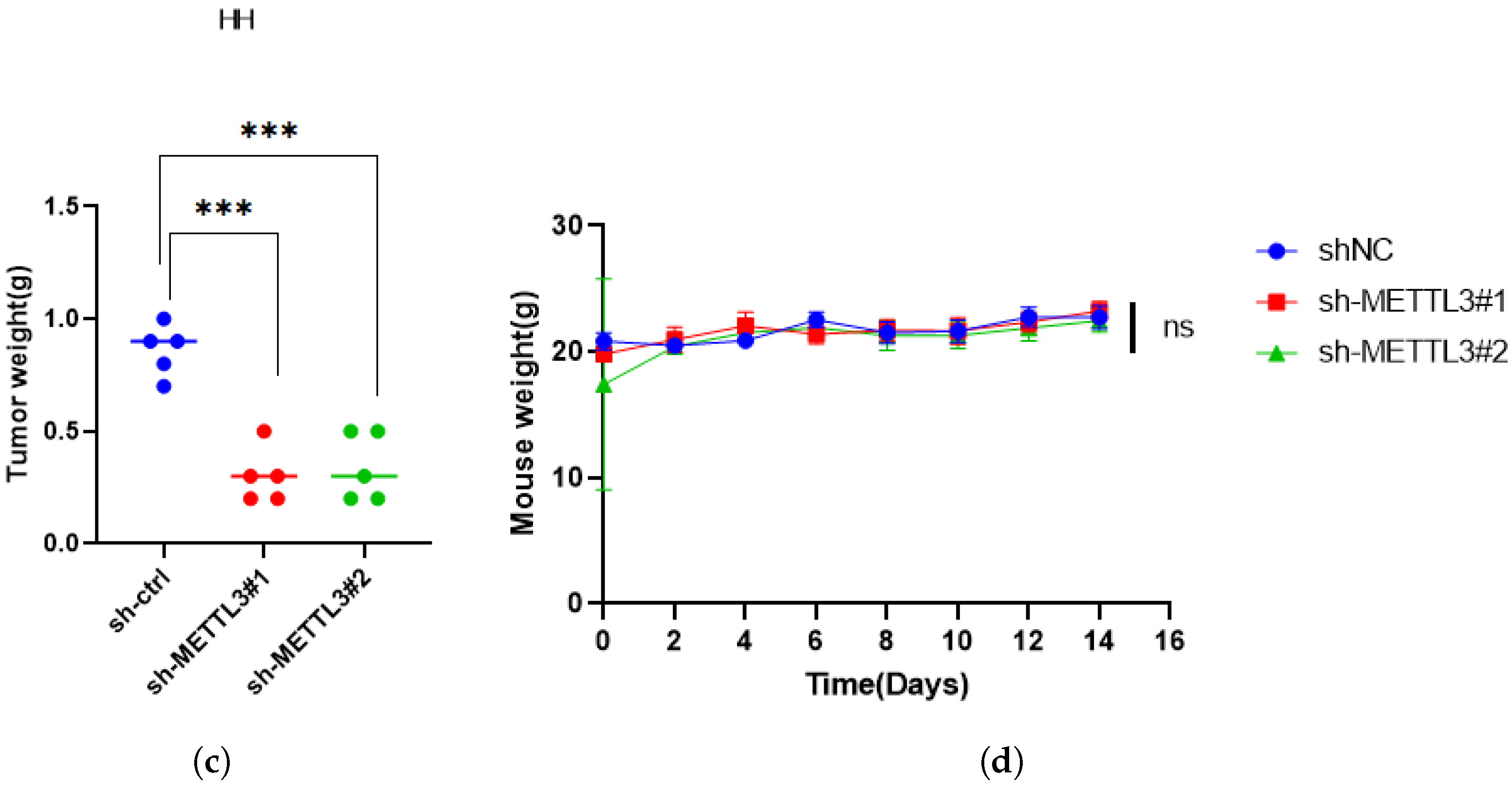
2.6. METTL3 Promotes Tumor Growth In Vivo
3. Discussion
4. Materials and Methods
4.1. Tissue Sample Collection
4.2. Cell Culture
4.3. RNA Isolation and RT-qPCR
4.4. Western Blotting
4.5. Immunohistochemistry (IHC)
4.6. Plasmid Construction and Lentiviral Transfection
4.7. CCK8 Assay
4.8. EdU Cell Proliferation Assay
4.9. Apoptosis Detection
4.10. Transwell Experiment
4.11. RNA High-Throughput Sequencing (RNA-Seq)
4.12. Gene Ontology and KEGG Pathway Analysis
4.13. RNA Stability Assay
4.14. Nude Mouse Xenograft Study
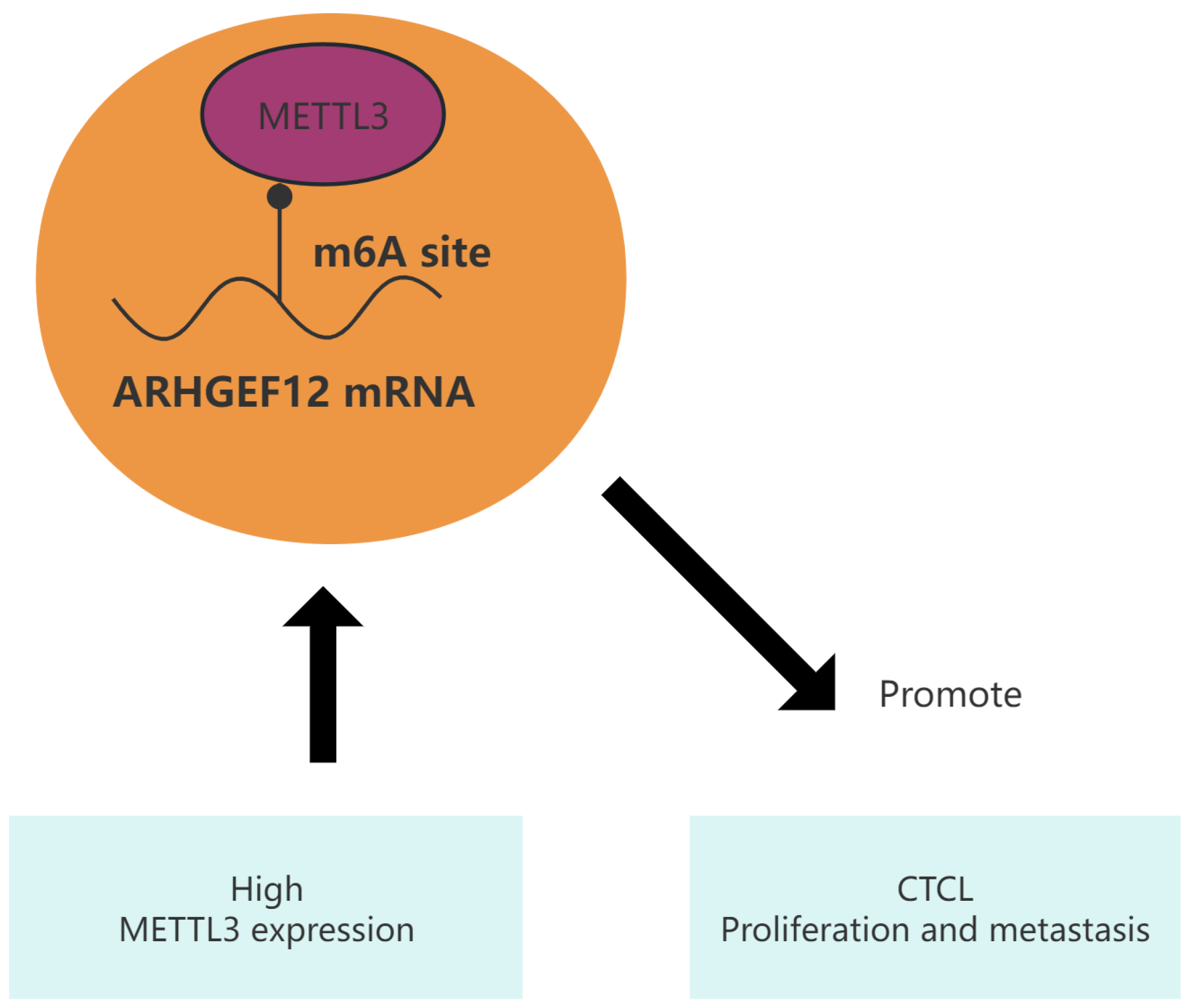
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CTCL | Cutaneous T-cell lymphoma |
| m6A | N6-methyladenosine |
| MF | Mycosis fungoides |
| pcALCL | Primary cutaneous anaplastic large-cell lymphoma |
| TBST | Tris-buffered saline with Tween 20 |
References
- Mandel, J.; Gleason, L.; Joffe, D.; Bhatti, S.; Nikbakht, N. Immunosequencing applications in cutaneous T-cell lymphoma. Front. Immunol. 2023, 14, 1300061. [Google Scholar] [CrossRef]
- Glinos, G.; Wei, G.; Nosewicz, J.; Abdulla, F.; Chen, P.L.; Chung, C.; Kaffenberger, B.H.; Querfeld, C.; Shinohara, M.M.; Sokol, L.; et al. Characteristics and Outcomes for Hospitalized Patients with Cutaneous T-Cell Lymphoma. JAMA Dermatol. 2023, 159, 192–197. [Google Scholar] [CrossRef]
- Dobos, G.; Pohrt, A.; Ram-Wolff, C.; Lebbé, C.; Bouaziz, J.D.; Battistella, M.; Bagot, M.; de Masson, A. Epidemiology of cutaneous T-cell lymphomas: A systematic review and meta-analysis of 16,953 patients. Cancers 2020, 12, 2921. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Liu, F.; Sun, J.; Gao, S.; Wong, C.C.; Tu, P.; Wang, Y. Abrogation of USP9X Is a Potential Strategy to Decrease PEG10 Levels and Impede Tumor Progression in Cutaneous T-Cell Lymphoma. J. Investig. Dermatol. 2024, 144, 2778–2788.e9. [Google Scholar] [CrossRef]
- Stuver, R.; Geller, S. Advances in the treatment of mycoses fungoides and Sézary syndrome: A narrative update in skin-directed therapies and immune-based treatments. Front. Immunol. 2023, 14, 1284045. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jia, B.; Xu, W.; Li, W.; Liu, T.; Liu, P.; Zhao, W.; Zhang, H.; Sun, X.; Yang, H.; et al. Chidamide in relapsed or refractory peripheral T cell lymphoma: A multicenter real-world study in China. J. Hematol. Oncol. 2017, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Muniesa, C.; Gallardo, F.; García-Doval, I.; Estrach, M.T.; Combalia, A.; Morillo-Andújar, M.; De Cruz-Vicente, F.; Machan, S.; Moya-Martínez, C.; Rovira, R.; et al. Brentuximab vedotin in the treatment of cutaneous T-cell lymphomas: Data from the Spanish Primary Cutaneous Lymphoma Registry. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 57–64. [Google Scholar] [CrossRef]
- Ishitsuka, K.; Yasukawa, T.; Tsuji, Y. Safety and effectiveness of mogamulizumab in relapsed or refractory CC chemokine receptor 4-positive peripheral T-cell lymphoma and relapsed or refractory cutaneous T-cell lymphoma: A post-marketing surveillance in Japan. Hematol. Oncol. 2024, 42, e3292. [Google Scholar] [CrossRef]
- Kohnken, R.; Mishra, A. MicroRNAs in cutaneous T-cell lymphoma: The future of therapy. J. Investig. Dermatol. 2019, 139, 528–534. [Google Scholar] [CrossRef]
- Dong, Z.; Zhu, X.; Li, Y.; Gan, L.; Chen, H.; Zhang, W.; Sun, J. Oncogenomic analysis identifies novel biomarkers for tumor stage mycosis fungoides. Medicine 2018, 97, e10871. [Google Scholar] [CrossRef]
- Gan, L.; Shi, H.; Zhang, Y.; Sun, J.; Chen, H. Proteomic screening and verification of biomarkers in different stages of mycosis fungoides: A pilot study. Front. Cell Dev. Biol. 2021, 9, 747017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Zhang, J.; Zhu, J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer 2019, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, Q.; Zhou, Q.; Fang, F.; Lei, K.; Liu, Z.; Zheng, G.; Zhu, L.; Huo, J.; Li, X.; et al. METTL3-m6A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023, 559, 216122. [Google Scholar] [CrossRef]
- Xie, H.; Yao, J.; Wang, Y.; Ni, B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022, 29, 1257–1271. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Dong, W.; Zhang, C.; Hu, M.; Ma, W.; Jiang, X.; Li, H.; Yang, P.; Xiang, D. N6-methyladenosine–mediated up-regulation of FZD10 regulates liver cancer stem cells’ properties and lenvatinib resistance through WNT/β-catenin and hippo signaling pathways. Gastroenterology 2023, 164, 990–1005. [Google Scholar] [CrossRef]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO 3-mediated autophagy. EMBO J. 2020, 39, e103181. [Google Scholar] [CrossRef] [PubMed]
- Pomaville, M.; Chennakesavalu, M.; Wang, P.; Jiang, Z.; Sun, H.L.; Ren, P.; Borchert, R.; Gupta, V.; Ye, C.; Ge, R.; et al. Small-molecule inhibition of the METTL3/METTL14 complex suppresses neuroblastoma tumor growth and promotes differentiation. Cell Rep. 2024, 43, 114165. [Google Scholar] [CrossRef]
- Wang, X.; Hu, M.; Yu, L.; Wang, X.; Jiang, X.; Zhang, G.; Ding, K. The “m6A writer” METTL3 and the “m6A reader” IGF2BP2 regulate cutaneous T-cell lymphomas progression via CDKN2A. Hematol. Oncol. 2022, 40, 567–576. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, S.; Cai, J.; Liang, J.; Zhang, Y.; Xie, Y.; Luo, F.; Tang, J.; Gao, Y.; Shen, S.; et al. Targeting ARHGEF12 promotes neuroblastoma differentiation, MYCN degradation, and reduces tumorigenicity. Cell. Oncol. 2023, 46, 133–143. [Google Scholar] [CrossRef]
- Schafernak, K.T.; Williams, J.A.; Clyde, B.I.; Marcus, C.; Decker, B.; Toydemir, R.M. Identification of KMT2A-ARHGEF12 fusion in a child with a high-grade B-cell lymphoma. Cancer Genet. 2021, 258, 23–26. [Google Scholar] [CrossRef]
- Klos, C.; Roynard, P.; Berthon, C.; Soenen, V.; Marceau, A.; Fournier, E.; Fenwarth, L.; Daudignon, A.; Roche, C.; Guermouche, H. Unravelling a KMT2A::ARHGEF12 fusion within chromoanagenesis in acute myeloid leukemia using Optical Genome Mapping. Ann. Hematol. 2024, 103, 4793–4795. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.C.; Tien, Y.C.; Shi, Y.H.; Chen, S.; Zhu, Y.Q.; Huang, X.T.; Huang, C.S.; Zhao, W.; Yin, X.Y. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene 2022, 41, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Z.; Xiong, J.S.; Gan, L.; Zhang, Y.; Zhang, C.C.; Kong, Y.Q.; Miao, Q.J.; Tian, C.C.; Li, R.; Liu, J.Q.; et al. N6-methyladenosine reader YTHDF3 regulates melanoma metastasis via its ‘executor’ LOXL3. Clin. Transl. Med. 2022, 12, e1075. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Saleh, J.S.; Subtil, A.; Hristov, A.C. Primary cutaneous T-cell lymphoma: A review of the most common entities with focus on recent updates. Hum. Pathol. 2023, 138, 76–102. [Google Scholar] [CrossRef]
- Dummer, R.; Vermeer, M.H.; Scarisbrick, J.J.; Kim, Y.H.; Stonesifer, C.; Tensen, C.P.; Geskin, L.J.; Quaglino, P.; Ramelyte, E. Cutaneous T cell lymphoma. Nat. Rev. Dis. Prim. 2021, 7, 61. [Google Scholar] [CrossRef]
- Song, X.; Chang, S.; Seminario-Vidal, L.; de Mingo Pulido, A.; Tordesillas, L.; Song, X.; Reed, R.A.; Harkins, A.; Whiddon, S.; Nguyen, J.V.; et al. Genomic and single-cell landscape reveals novel drivers and therapeutic vulnerabilities of transformed cutaneous T-cell lymphoma. Cancer Discov. 2022, 12, 1294–1313. [Google Scholar] [CrossRef]
- Morgenroth, S.; Roggo, A.; Pawlik, L.; Dummer, R.; Ramelyte, E. What is New in cutaneous T cell lymphoma? Curr. Oncol. Rep. 2023, 25, 1397–1408. [Google Scholar] [CrossRef]
- Wu, S.; Wang, H.; Yang, Q.; Liu, Z.; Du, J.; Wang, L.; Chen, S.; Lu, Q.; Yang, D.H. METTL3 regulates M6A methylation-modified EBV-pri-miR-BART3-3p to promote NK/T cell lymphoma growth. Cancer Lett. 2024, 597, 217058. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, T.; Ding, M.; Cai, Y.; Yu, Z.; Zhou, X.; Wang, X. Targeting YTHDF2 inhibits tumorigenesis of diffuse large B-cell lymphoma through ACER2-mediated ceramide catabolism. J. Adv. Res. 2024, 63, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Chen, D.; Wang, X.; Li, Y.; Li, Z.; Li, B.; Zhang, Y.; Li, W.; Zhang, J.; Ye, J.; et al. Inhibition of the m6A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia 2022, 36, 2180–2188. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ji, M.; Xia, Y.; Han, X.; Li, M.; Li, W.; Sun, T.; Zhang, J.; Lu, F.; Sun, Y.; et al. Silencing of IRF8 Mediated by m6A Modification Promotes the Progression of T-Cell Acute Lymphoblastic Leukemia. Adv. Sci. 2023, 10, 2201724. [Google Scholar] [CrossRef]
- Ao, K.; Yin, M.; Lyu, X.; Xiao, Y.; Chen, X.; Zhong, S.; Wen, X.; Yuan, J.; Ye, M.; Zhang, J. METTL3-mediated HSPA9 m6A modification promotes malignant transformation and inhibits cellular senescence by regulating exosomal mortalin protein in cervical cancer. Cancer Lett. 2024, 587, 216658. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, L.; Kong, Y.; Shi, H.; Zhang, C.; Tian, C.; Chen, H. METTL3 Promotes Cutaneous T-Cell Lymphoma Progression by Regulating ARHGEF12 Expression. Int. J. Mol. Sci. 2025, 26, 3640. https://doi.org/10.3390/ijms26083640
Gan L, Kong Y, Shi H, Zhang C, Tian C, Chen H. METTL3 Promotes Cutaneous T-Cell Lymphoma Progression by Regulating ARHGEF12 Expression. International Journal of Molecular Sciences. 2025; 26(8):3640. https://doi.org/10.3390/ijms26083640
Chicago/Turabian StyleGan, Lu, Yingqi Kong, Haoze Shi, Congcong Zhang, Cuicui Tian, and Hao Chen. 2025. "METTL3 Promotes Cutaneous T-Cell Lymphoma Progression by Regulating ARHGEF12 Expression" International Journal of Molecular Sciences 26, no. 8: 3640. https://doi.org/10.3390/ijms26083640
APA StyleGan, L., Kong, Y., Shi, H., Zhang, C., Tian, C., & Chen, H. (2025). METTL3 Promotes Cutaneous T-Cell Lymphoma Progression by Regulating ARHGEF12 Expression. International Journal of Molecular Sciences, 26(8), 3640. https://doi.org/10.3390/ijms26083640





