hTERT and SV40LgT Renal Cell Lines Adjust Their Transcriptional Responses After Copy Number Changes from the Parent Proximal Tubule Cells
Abstract
1. Introduction
2. Results
2.1. RNA-Seq Results
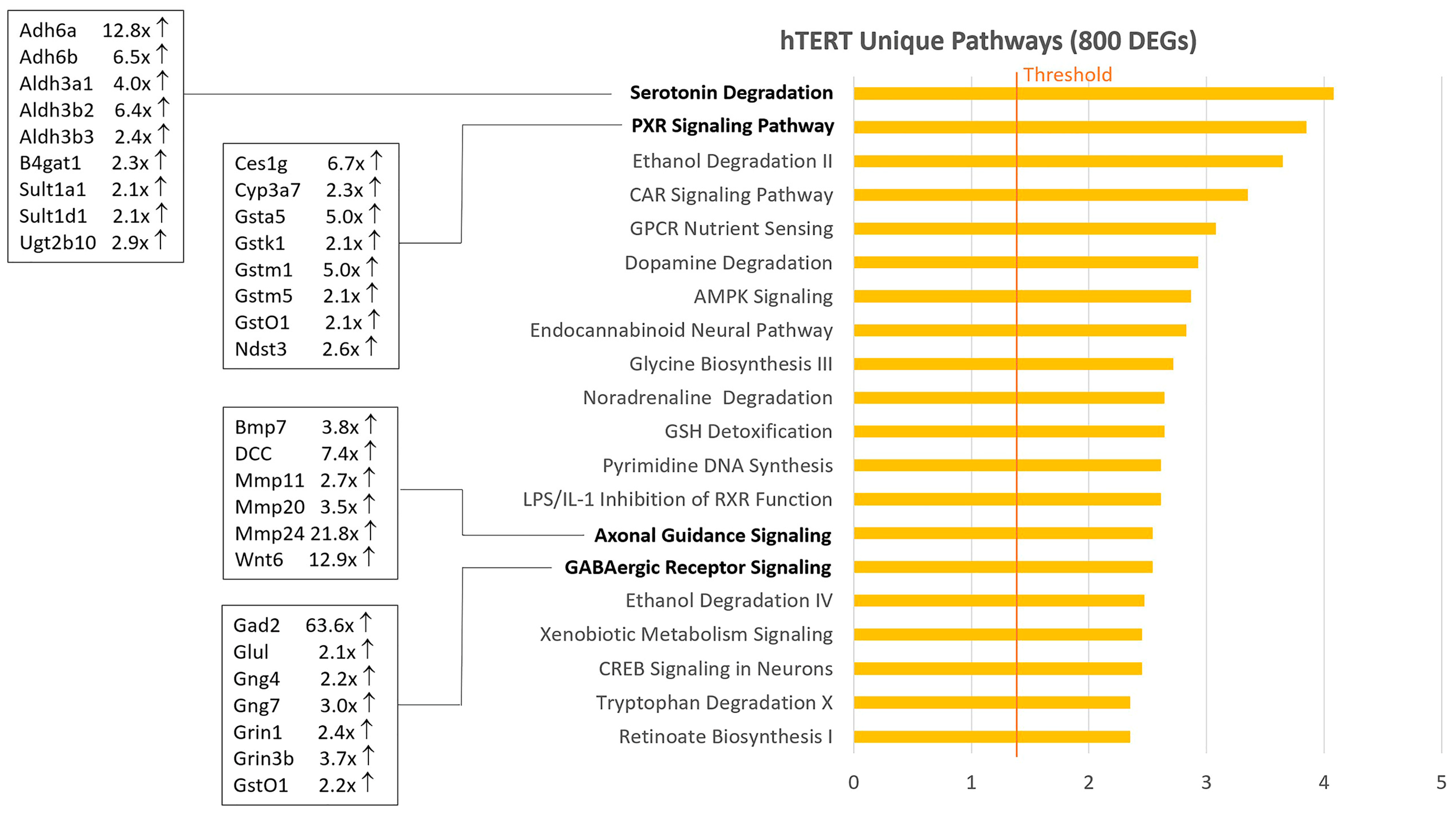
2.2. Venn Diagram and Pathway Analysis
2.3. Differential Expression of Genes in Development, IFN-Response, and Cdk Inhibition
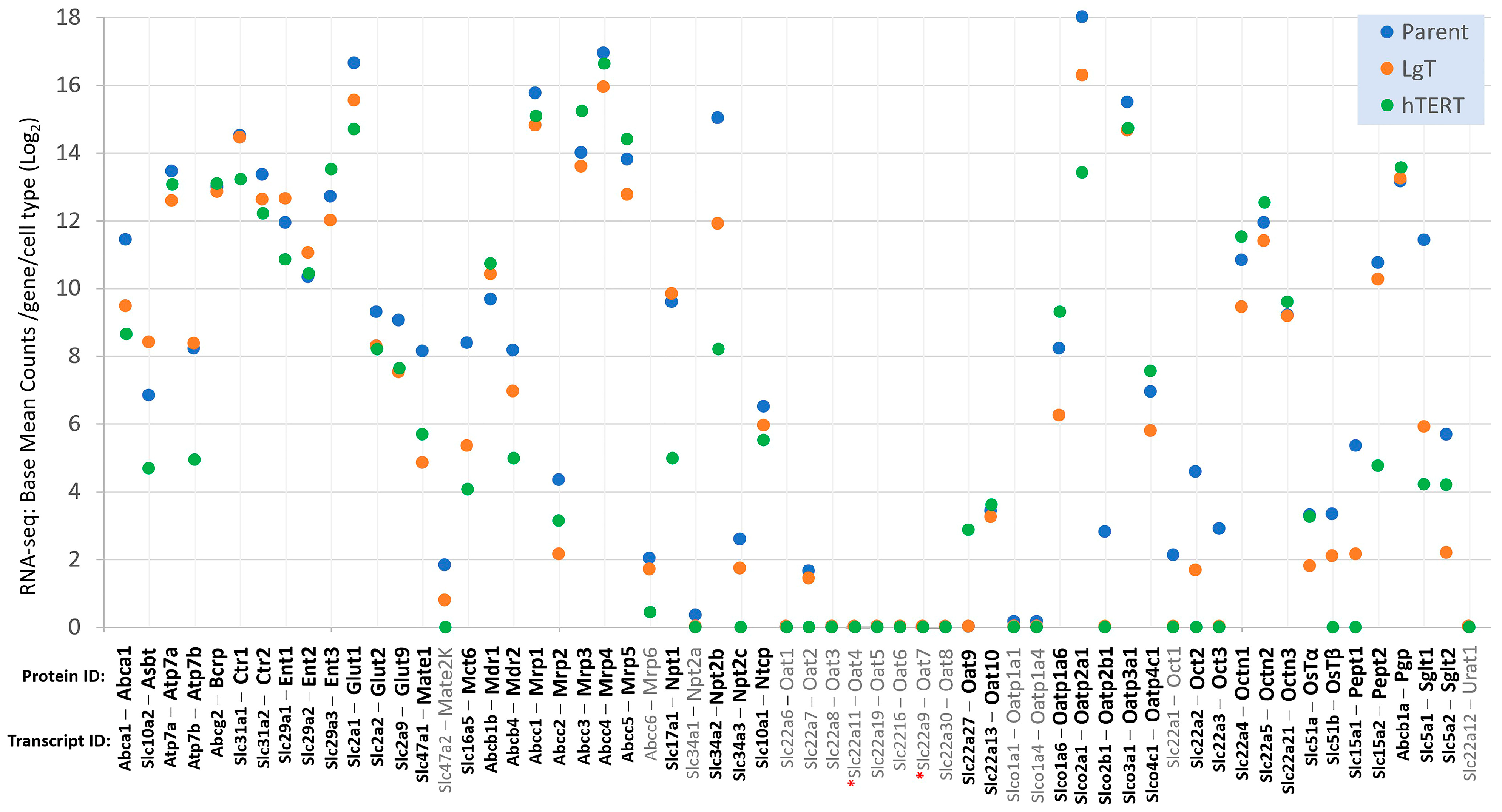
2.4. Differential Expression of Three Gene Classes Underlying Renal Pharmacodynamics
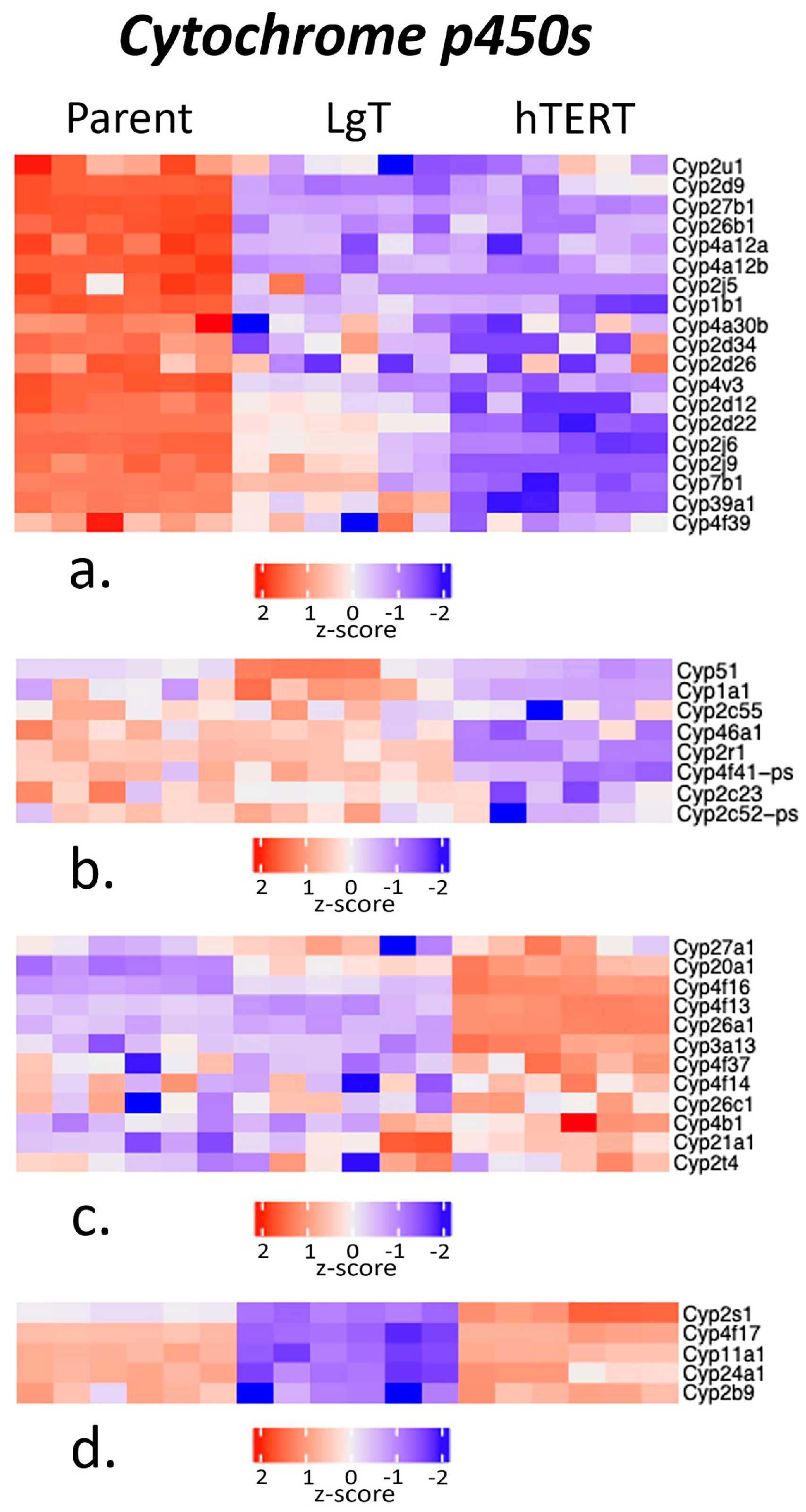
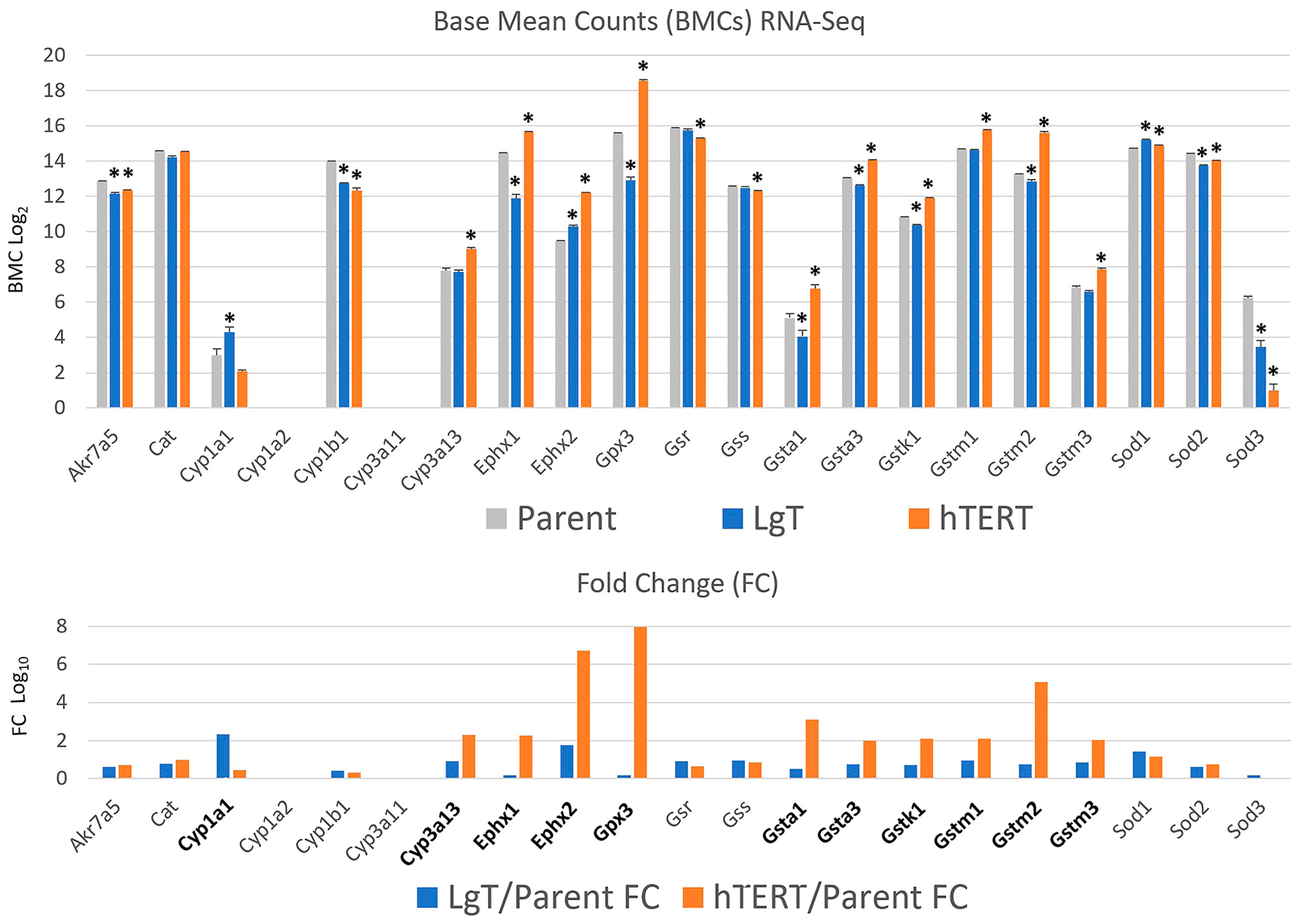
2.5. Cytochrome P450s and Detoxification Enzymes
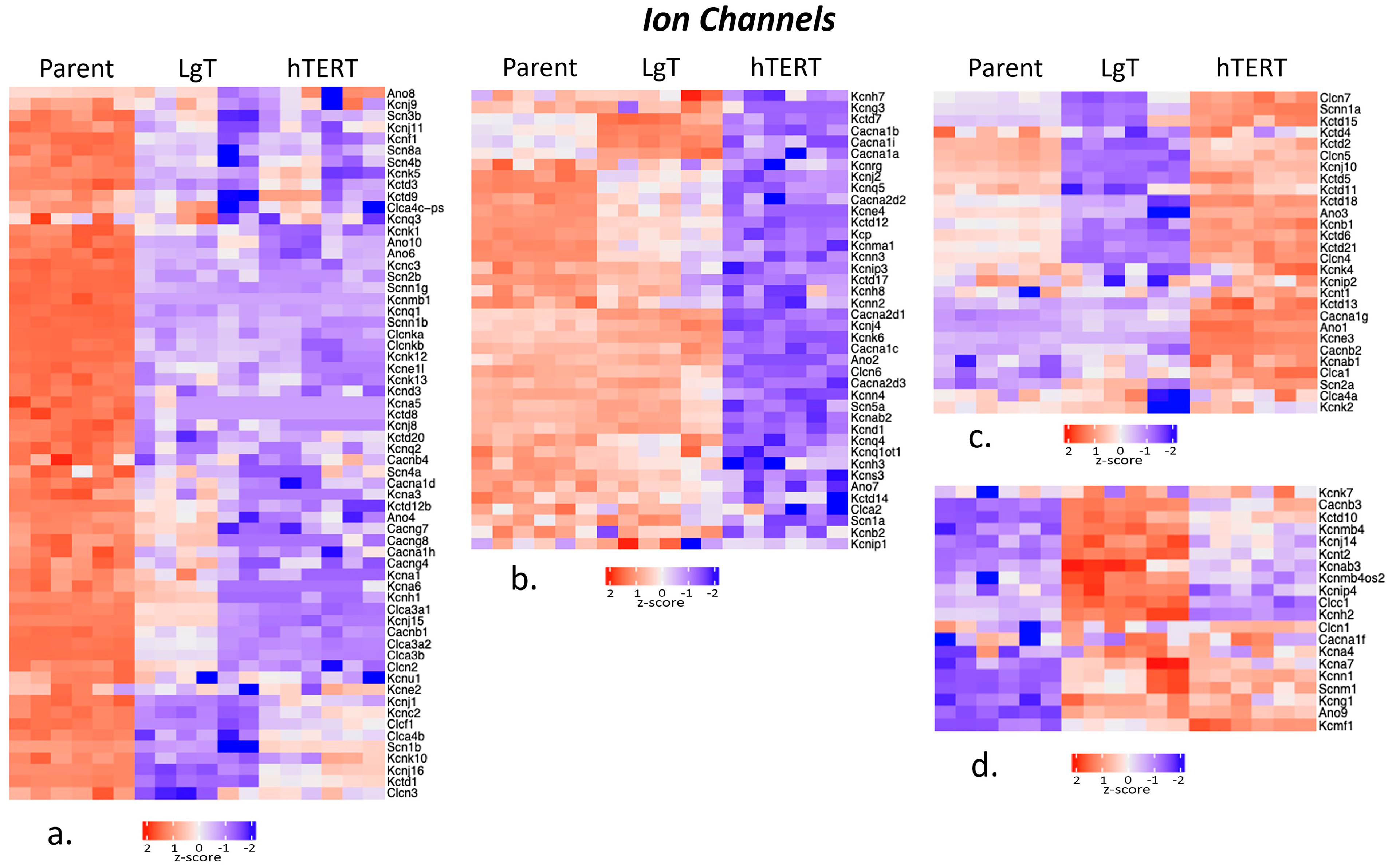
2.6. Ion Channels
2.7. Renal Metabolic Transporters
2.8. Detection of Slc22a11 (mOat4)
2.9. Nanostring Transcript Analysis
2.10. Chemically Inducible Renal Transporters
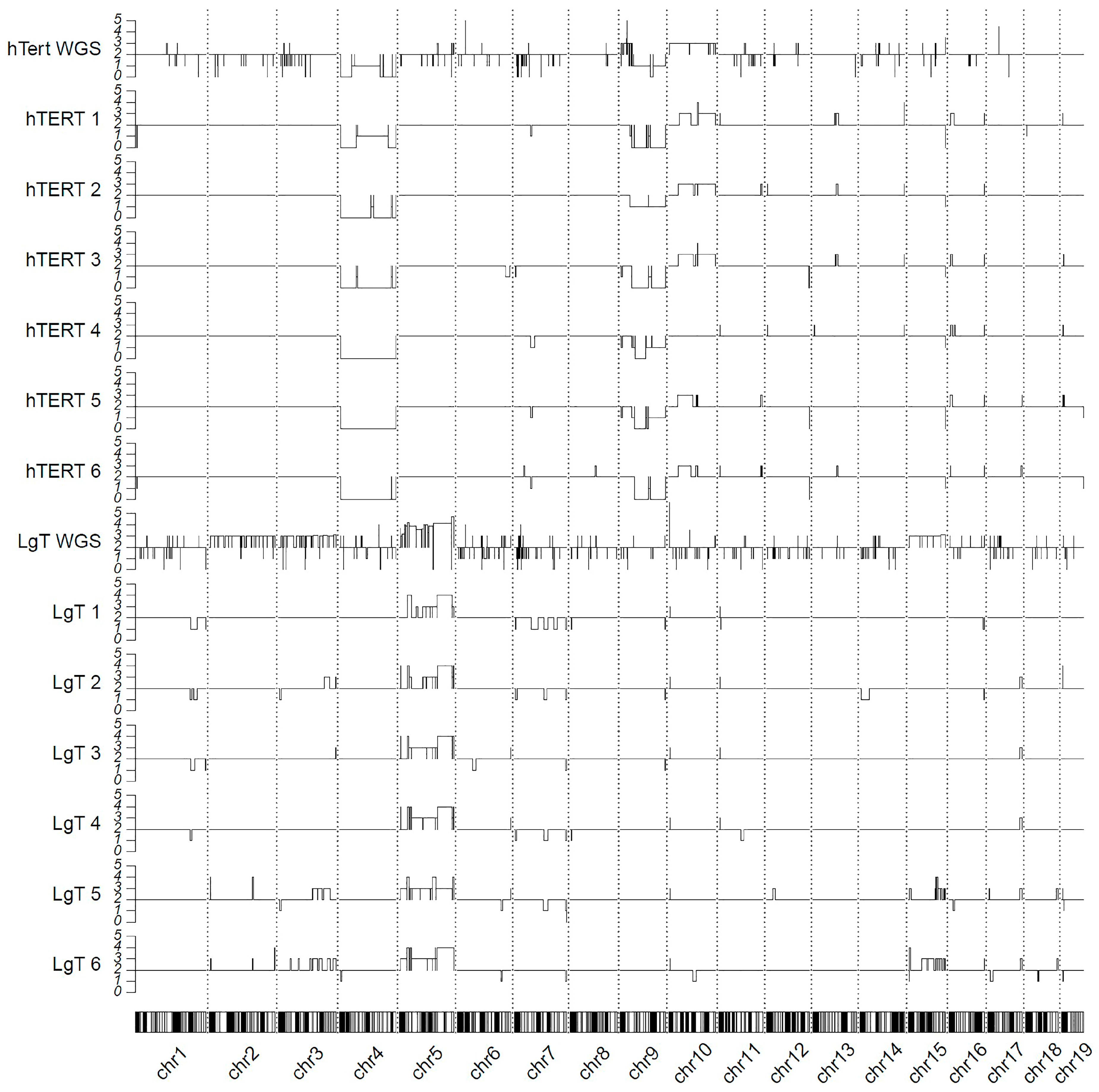
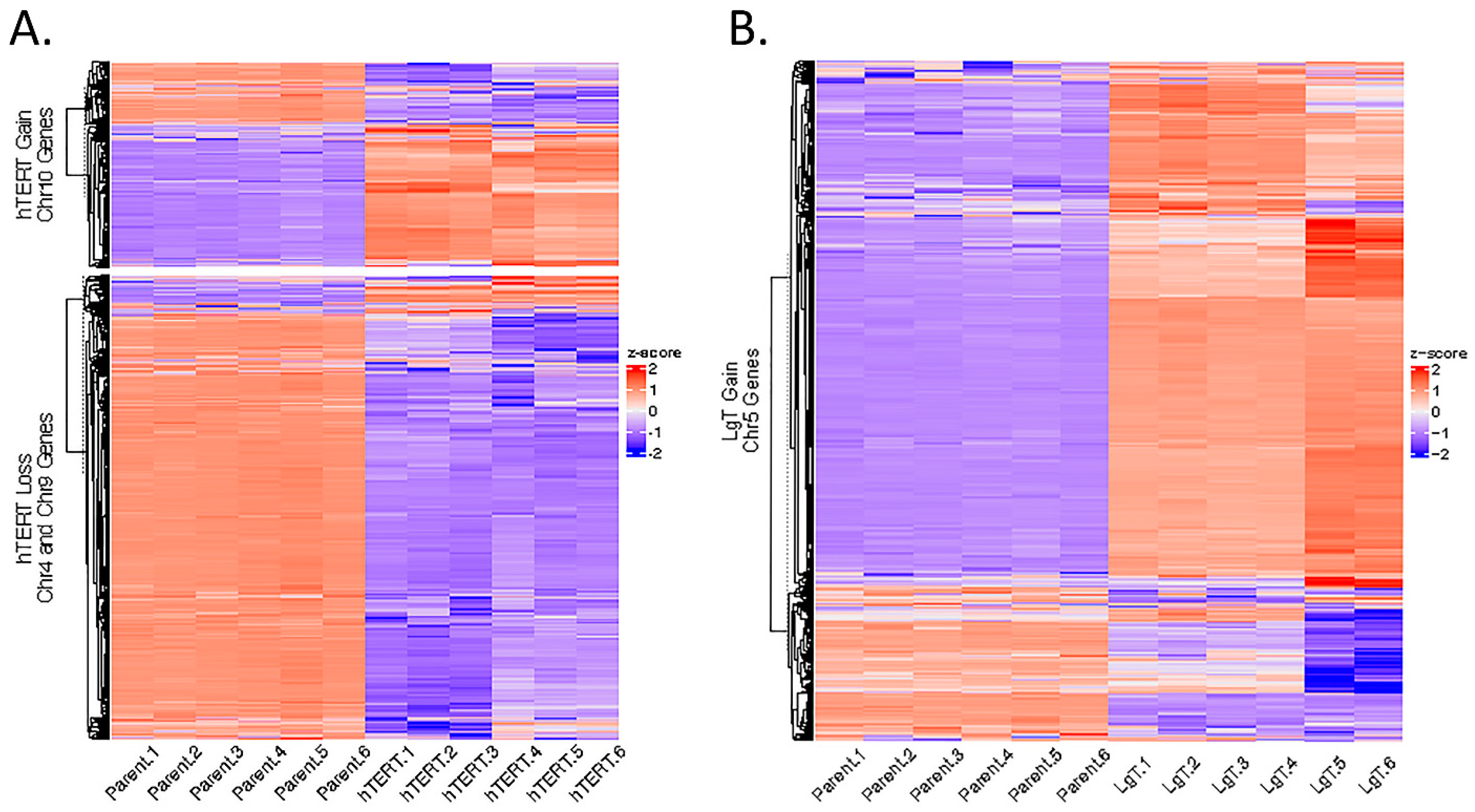
2.11. Genomic Karyotyping and Copy Number
3. Discussion
4. Materials and Methods
4.1. Cell Immortalization and Culture
4.2. RNA and DNA Isolation: RNA-Seq, DNA-Seq
4.3. Genomic Karyotyping: WGS and RNA-Seq
4.4. Differential Expression by RNA-Seq
4.5. Venn Diagram and Pathway Analysis
4.6. Clustering Analysis
4.7. CNVs
4.8. Nanostring Analysis
4.9. ddPCR Analysis for Slc22a11
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abc | ATP-binding cassette |
| Actb | beta-actin |
| AFB1 | aflatoxin B1 |
| Ahr | arylhydrocarbon receptor |
| Ano | anoctamin |
| Bcrp | breast cancer resistance protein |
| BMCs | base mean counts |
| Cacn | voltage-dependent calcium ion channel |
| Cdk | cyclin-dependent kinase |
| CisPt | cisplatin |
| Clca | chloride ion channel |
| CNV | copy number variant |
| Cyp | cytochrome P450 |
| ddPCR | digital droplet PCR |
| DEG | differentially expressed gene |
| Ensembl | European Bioinformatics Institute database |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase |
| Glut | glucose transporter |
| Gst | glutathione S-transferase |
| Hprt | hypoxanthine-guanine phosphoribosyltransferase |
| IPA | Ingenuity Pathway Analysis |
| Kcn | potassium ion channel |
| Mdr | multidrug resistance |
| moRPTECs | mouse renal proximal tubule epithelial cells |
| Mrp | multidrug resistance-associated protein |
| NA | non-annotated |
| Npt | sodium-dependent phosphate cotransporter |
| Oat | organic anion transporter |
| Oct | organic cation transporter |
| pAdj | adjusted p-value |
| Pgp | P-glycoprotein |
| Rb | retinoblastoma |
| Rpl32 | ribosomal protein L32 |
| RPTECs | renal proximal tubule epithelial cells |
| Scn | sodium ion channel |
| Sglt | sodium–glucose cotransporter |
| Slc | solute carrier family |
| SV40LgT | LgT |
| Trp53 | transformation-related protein 53 |
| WGS | whole-genome sequencing |
References
- Curthoys, N.P.; Moe, O.W. Proximal tubule function and response to acidosis. Clin. J. Am. Soc. Nephrol. 2014, 9, 1627–1638. [Google Scholar] [CrossRef]
- Bajaj, P.; Chung, G.; Pye, K.; Yukawa, T.; Imanishi, A.; Takai, Y.; Brown, C.; Wagoner, M.P. Freshly isolated primary human proximal tubule cells as an in vitro model for the detection of renal tubular toxicity. Toxicology 2020, 442, 152535. [Google Scholar] [CrossRef]
- Kamiyama, M.; Garner, M.K.; Farragut, K.M.; Kobori, H. The establishment of a primary culture system of proximal tubule segments using specific markers from normal mouse kidneys. Int. J. Mol. Sci. 2012, 13, 5098–5111. [Google Scholar] [CrossRef] [PubMed]
- Vinay, P.; Gougoux, A.; Lemieux, G. Isolation of a pure suspension of rat proximal tubules. Am. J. Physiol. 1981, 241, F403–F411. [Google Scholar] [CrossRef] [PubMed]
- Legouis, D.; Bataille, A.; Hertig, A.; Vandermeersch, S.; Simon, N.; Rondeau, E.; Galichon, P. Ex vivo analysis of renal proximal tubular cells. BMC Cell Biol. 2015, 16, 12. [Google Scholar] [CrossRef][Green Version]
- Sutyagina, O.I.; Beilin, A.K.; Vorotelyak, E.A.; Vasiliev, A.V. Immortalization Reversibility in the Context of Cell Therapy Biosafety. Int. J. Mol. Sci. 2023, 24, 7738. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.K.; Park, N.H. Extension of cell life span using exogenous telomerase. Methods Mol. Biol. 2007, 371, 151–165. [Google Scholar] [CrossRef]
- Ozer, H.L.; Banga, S.S.; Dasgupta, T.; Houghton, J.; Hubbard, K.; Jha, K.K.; Kim, S.H.; Lenahan, M.; Pang, Z.; Pardinas, J.R.; et al. SV40-mediated immortalization of human fibroblasts. Exp. Gerontol. 1996, 31, 303–310. [Google Scholar] [CrossRef]
- Wieser, M.; Stadler, G.; Jennings, P.; Streubel, B.; Pfaller, W.; Ambros, P.; Riedl, C.; Katinger, H.; Grillari, J.; Grillari-Voglauer, R. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am. J. Physiol. Ren. Physiol. 2008, 295, F1365–F1375. [Google Scholar] [CrossRef]
- Toouli, C.D.; Huschtscha, L.I.; Neumann, A.A.; Noble, J.R.; Colgin, L.M.; Hukku, B.; Reddel, R.R. Comparison of human mammary epithelial cells immortalized by simian virus 40 T-Antigen or by the telomerase catalytic subunit. Oncogene 2002, 21, 128–139. [Google Scholar] [CrossRef]
- Liu, S.; Hatton, M.P.; Khandelwal, P.; Sullivan, D.A. Culture, immortalization, and characterization of human meibomian gland epithelial cells. Investig. Ophthalmol. Vis. Sci. 2010, 51, 3993–4005. [Google Scholar] [CrossRef] [PubMed]
- Straszewski-Chavez, S.L.; Abrahams, V.M.; Alvero, A.B.; Aldo, P.B.; Ma, Y.; Guller, S.; Romero, R.; Mor, G. The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 2009, 30, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Yalvac, M.E.; Yilmaz, A.; Mercan, D.; Aydin, S.; Dogan, A.; Arslan, A.; Demir, Z.; Salafutdinov, I.I.; Shafigullina, A.K.; Sahin, F.; et al. Differentiation and neuro-protective properties of immortalized human tooth germ stem cells. Neurochem. Res. 2011, 36, 2227–2235. [Google Scholar] [CrossRef]
- Bernal, A.; Zafon, E.; Dominguez, D.; Bertran, E.; Tusell, L. Generation of Immortalised But Unstable Cells after hTERT Introduction in Telomere-Compromised and p53-Deficient vHMECs. Int. J. Mol. Sci. 2018, 19, 2078. [Google Scholar] [CrossRef]
- Bikkul, M.U.; Faragher, R.G.A.; Worthington, G.; Meinke, P.; Kerr, A.R.W.; Sammy, A.; Riyahi, K.; Horton, D.; Schirmer, E.C.; Hubank, M.; et al. Telomere elongation through hTERT immortalization leads to chromosome repositioning in control cells and genomic instability in Hutchinson-Gilford progeria syndrome fibroblasts, expressing a novel SUN1 isoform. Genes Chromosomes Cancer 2019, 58, 341–356. [Google Scholar] [CrossRef]
- Matsuura, R.; Doi, K.; Rabb, H. Acute kidney injury and distant organ dysfunction-network system analysis. Kidney Int. 2023, 103, 1041–1055. [Google Scholar] [CrossRef]
- Aschauer, L.; Limonciel, A.; Wilmes, A.; Stanzel, S.; Kopp-Schneider, A.; Hewitt, P.; Lukas, A.; Leonard, M.O.; Pfaller, W.; Jennings, P. Application of RPTEC/TERT1 cells for investigation of repeat dose nephrotoxicity: A transcriptomic study. Toxicol. Vitr. 2015, 30 Pt A, 106–116. [Google Scholar] [CrossRef]
- Secker, P.F.; Schlichenmaier, N.; Beilmann, M.; Deschl, U.; Dietrich, D.R. Functional transepithelial transport measurements to detect nephrotoxicity in vitro using the RPTEC/TERT1 cell line. Arch. Toxicol. 2019, 93, 1965–1978. [Google Scholar] [CrossRef] [PubMed]
- Saib, S.; Hodin, S.; Bin, V.; Ollier, E.; Delavenne, X. In Vitro Evaluation of P-gp-Mediated Drug-Drug Interactions Using the RPTEC/TERT1 Human Renal Cell Model. Eur. J. Drug Metab. Pharmacokinet. 2022, 47, 223–233. [Google Scholar] [CrossRef]
- Limonciel, A.; Ates, G.; Carta, G.; Wilmes, A.; Watzele, M.; Shepard, P.J.; VanSteenhouse, H.C.; Seligmann, B.; Yeakley, J.M.; van de Water, B.; et al. Comparison of base-line and chemical-induced transcriptomic responses in HepaRG and RPTEC/TERT1 cells using TempO-Seq. Arch. Toxicol. 2018, 92, 2517–2531. [Google Scholar] [CrossRef]
- Poolsri, W.; Noitem, R.; Jutabha, P.; Raveesunthornkiat, M.; Danova, A.; Chavasiri, W.; Muanprasat, C. Discovery of a chalcone derivative as an anti-fibrotic agent targeting transforming growth factor-beta1 signaling: Potential therapy of renal fibrosis. Biomed. Pharmacother. 2023, 165, 115098. [Google Scholar] [CrossRef] [PubMed]
- van der Stel, W.; Carta, G.; Eakins, J.; Darici, S.; Delp, J.; Forsby, A.; Bennekou, S.H.; Gardner, I.; Leist, M.; Danen, E.H.J.; et al. Multiparametric assessment of mitochondrial respiratory inhibition in HepG2 and RPTEC/TERT1 cells using a panel of mitochondrial targeting agrochemicals. Arch. Toxicol. 2020, 94, 2707–2729. [Google Scholar] [CrossRef] [PubMed]
- Sakolish, C.; Tsai, H.D.; Lin, H.C.; Bajaj, P.; Villenave, R.; Ferguson, S.S.; Stanko, J.P.; Becker, R.A.; Hewitt, P.; Chiu, W.A.; et al. Comparative Analysis of Proximal Tubule Cell Sources for In Vitro Studies of Renal Proximal Tubule Toxicity. Biomedicines 2025, 13, 563. [Google Scholar] [CrossRef]
- Adelfio, M.; Bonzanni, M.; Levin, M.; Kaplan, D.L. Impact of Membrane Voltage on Formation and Stability of Human Renal Proximal Tubules in Vitro. ACS Biomater. Sci. Eng. 2022, 8, 1239–1246. [Google Scholar] [CrossRef]
- Adelfio, M.; Szymkowiak, S.; Kaplan, D.L. Matrigel-Free Laminin-Entactin Matrix to Induce Human Renal Proximal Tubule Structure Formation In Vitro. ACS Biomater. Sci. Eng. 2020, 6, 6618–6625. [Google Scholar] [CrossRef] [PubMed]
- Piossek, F.; Beneke, S.; Schlichenmaier, N.; Mucic, G.; Drewitz, S.; Dietrich, D.R. Physiological oxygen and co-culture with human fibroblasts facilitate in vivo-like properties in human renal proximal tubular epithelial cells. Chem. Biol. Interact. 2022, 361, 109959. [Google Scholar] [CrossRef]
- Sakolish, C.; Moyer, H.L.; Tsai, H.D.; Ford, L.C.; Dickey, A.N.; Bajaj, P.; Villenave, R.; Hewitt, P.; Ferguson, S.S.; Stanko, J.; et al. Comparative analysis of the physiological and transport functions of various sources of renal proximal tubule cells under static and fluidic conditions in PhysioMimix T12 platform. Drug Metab. Dispos. 2025, 53, 100001. [Google Scholar] [CrossRef]
- Dasgupta, P.; Kulkarni, P.; Majid, S.; Hashimoto, Y.; Shiina, M.; Shahryari, V.; Bhat, N.S.; Tabatabai, L.; Yamamura, S.; Saini, S.; et al. LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis. 2020, 11, 660. [Google Scholar] [CrossRef]
- Pletcher, J.P.; Bhattacharjee, S.; Doan, J.P.; Wynn, R.; Sindhwani, P.; Nadiminty, N.; Petros, F.G. The Emerging Role of Poly (ADP-Ribose) Polymerase Inhibitors as Effective Therapeutic Agents in Renal Cell Carcinoma. Front. Oncol. 2021, 11, 681441. [Google Scholar] [CrossRef]
- Poplawski, P.; Zarychta-Wisniewska, W.; Burdzinska, A.; Boguslawska, J.; Adamiok-Ostrowska, A.; Hanusek, K.; Rybicka, B.; Bialas, A.; Kossowska, H.; Iwanicka-Rokicka, R.; et al. Renal cancer secretome induces migration of mesenchymal stromal cells. Stem Cell Res. Ther. 2023, 14, 200. [Google Scholar] [CrossRef]
- Arakawa, H.; Matsushita, K.; Ishiguro, N. Advanced in vitro evaluation of drug-induced kidney injury using microphysiological systems in drug discovery and development. Drug Metab. Pharmacokinet. 2025, 61, 101056. [Google Scholar] [CrossRef] [PubMed]
- Jones-Isaac, K.A.; Lidberg, K.A.; Yeung, C.K.; Yang, J.; Bain, J.; Ruiz, M.; Koenig, G.; Koenig, P.; Countryman, S.; Himmelfarb, J.; et al. Development of a kidney microphysiological system hardware platform for microgravity studies. NPJ Microgravity 2024, 10, 54. [Google Scholar] [CrossRef]
- Nieskens, T.T.G.; Persson, M.; Kelly, E.J.; Sjogren, A.K. A Multicompartment Human Kidney Proximal Tubule-on-a-Chip Replicates Cell Polarization-Dependent Cisplatin Toxicity. Drug Metab. Dispos. 2020, 48, 1303–1311. [Google Scholar] [CrossRef]
- Rusyn, I.; Sakolish, C.; Kato, Y.; Stephan, C.; Vergara, L.; Hewitt, P.; Bhaskaran, V.; Davis, M.; Hardwick, R.N.; Ferguson, S.S.; et al. Microphysiological Systems Evaluation: Experience of TEX-VAL Tissue Chip Testing Consortium. Toxicol. Sci. 2022, 188, 143–152. [Google Scholar] [CrossRef]
- Marx, U.; Beken, S.; Chen, Z.; Dehne, E.M.; Doherty, A.; Ewart, L.; Fitzpatrick, S.C.; Griffith, L.G.; Gu, Z.; Hartung, T.; et al. Biology-inspired dynamic microphysiological system approaches to revolutionize basic research, healthcare and animal welfare. ALTEX—Altern. Anim. Exp. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Giacomini, K.M.; Yee, S.W.; Koleske, M.L.; Zou, L.; Matsson, P.; Chen, E.C.; Kroetz, D.L.; Miller, M.A.; Gozalpour, E.; Chu, X. New and Emerging Research on Solute Carrier and ATP Binding Cassette Transporters in Drug Discovery and Development: Outlook from the International Transporter Consortium. Clin. Pharmacol. Ther. 2022, 112, 540–561. [Google Scholar] [CrossRef] [PubMed]
- Chalumeau, C.; Lamblin, D.; Bourgeois, S.; Borensztein, P.; Chambrey, R.; Bruneval, P.; Huyen, J.P.; Froissart, M.; Biber, J.; Paillard, M.; et al. Kidney cortex cells derived from SV40 transgenic mice retain intrinsic properties of polarized proximal tubule cells. Kidney Int. 1999, 56, 559–570. [Google Scholar] [CrossRef]
- Cartier, N.; Lacave, R.; Vallet, V.; Hagege, J.; Hellio, R.; Robine, S.; Pringault, E.; Cluzeaud, F.; Briand, P.; Kahn, A.; et al. Establishment of renal proximal tubule cell lines by targeted oncogenesis in transgenic mice using the L-pyruvate kinase-SV40 (T) antigen hybrid gene. J. Cell Sci. 1993, 104 Pt 3, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Riccaldi, D.; Robic, D.; Bens, M.; Cluzeaud, F.; Wu, M.S.; Bourbouze, R.; Vandewalle, A. Cultured proximal cells derived from transgenic mouse provide a model to study drug toxicity. Kidney Int. 1995, 48, 722–730. [Google Scholar] [CrossRef]
- Ernest, S.; Bello-Reuss, E. Expression and function of P-glycoprotein in a mouse kidney cell line. Am. J. Physiol. 1995, 269 Pt 1, C323–C333. [Google Scholar] [CrossRef]
- Loghman-Adham, M.; Nauli, S.M.; Soto, C.E.; Kariuki, B.; Zhou, J. Immortalized epithelial cells from human autosomal dominant polycystic kidney cysts. Am. J. Physiol. Renal Physiol. 2003, 285, F397–F412. [Google Scholar] [CrossRef]
- Villegas, G.; Lange-Sperandio, B.; Tufro, A. Autocrine and paracrine functions of vascular endothelial growth factor (VEGF) in renal tubular epithelial cells. Kidney Int. 2005, 67, 449–457. [Google Scholar] [CrossRef]
- Takaya, K.; Koya, D.; Isono, M.; Sugimoto, T.; Sugaya, T.; Kashiwagi, A.; Haneda, M. Involvement of ERK pathway in albumin-induced MCP-1 expression in mouse proximal tubular cells. Am. J. Physiol. Renal Physiol. 2003, 284, F1037–F1045. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.A.; Martin, N.P.; Brooks, A.M.; Foley, J.F.; Dunlap, P.E.; Ramaiahgari, S.; Fannin, R.D.; Gerrish, K.E. Insights into Repeated Renal Injury Using RNA-Seq with Two New RPTEC Cell Lines. Int. J. Med. Sci. 2023, 24, 14228. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Cellular and molecular pathways of renal repair after acute kidney injury. Kidney Int. 2018, 93, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, V.; Zhou, Y.; Ciorba, M.A.; Lingle, C.J. The LRRC family of BK channel regulatory subunits: Potential roles in health and disease. J. Physiol. 2022, 600, 1357–1371. [Google Scholar] [CrossRef]
- Kefaloyianni, E.; Keerthi Raja, M.R.; Schumacher, J.; Muthu, M.L.; Krishnadoss, V.; Waikar, S.S.; Herrlich, A. Proximal Tubule-Derived Amphiregulin Amplifies and Integrates Profibrotic EGF Receptor Signals in Kidney Fibrosis. J. Am. Soc. Nephrol. 2019, 30, 2370–2383. [Google Scholar] [CrossRef]
- Sang, Y.; Tsuji, K.; Nakanoh, H.; Fukushima, K.; Kitamura, S.; Wada, J. Role of Semaphorin 3A in Kidney Development and Diseases. Diagnostics 2023, 13, 3038. [Google Scholar] [CrossRef]
- Dwyer, K.M.; Kishore, B.K.; Robson, S.C. Conversion of extracellular ATP into adenosine: A master switch in renal health and disease. Nat. Rev. Nephrol. 2020, 16, 509–524. [Google Scholar] [CrossRef]
- Brooker, A.S.; Berkowitz, K.M. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol. Biol. 2014, 1170, 229–266. [Google Scholar] [CrossRef]
- Johnson, D.G.; Walker, C.L. Cyclins and cell cycle checkpoints. Annu. Rev. Pharmacol. Toxicol. 1999, 39, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Gorodetska, I.; Kozeretska, I.; Dubrovska, A. BRCA Genes: The Role in Genome Stability, Cancer Stemness and Therapy Resistance. J. Cancer 2019, 10, 2109–2127. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Zou, L. Hallmarks of DNA replication stress. Mol. Cell 2022, 82, 2298–2314. [Google Scholar] [CrossRef] [PubMed]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef]
- Miller-Hodges, E.; Hohenstein, P. WT1 in disease: Shifting the epithelial-mesenchymal balance. J. Pathol. 2012, 226, 229–240. [Google Scholar] [CrossRef]
- Padariya, M.; Sznarkowska, A.; Kote, S.; Gomez-Herranz, M.; Mikac, S.; Pilch, M.; Alfaro, J.; Fahraeus, R.; Hupp, T.; Kalathiya, U. Functional Interfaces, Biological Pathways, and Regulations of Interferon-Related DNA Damage Resistance Signature (IRDS) Genes. Biomolecules 2021, 11, 622. [Google Scholar] [CrossRef]
- Goel, B.; Tripathi, N.; Bhardwaj, N.; Jain, S.K. Small Molecule CDK Inhibitors for the Therapeutic Management of Cancer. Curr. Top. Med. Chem. 2020, 20, 1535–1563. [Google Scholar] [CrossRef]
- Nijjar, T.; Wigington, D.; Garbe, J.C.; Waha, A.; Stampfer, M.R.; Yaswen, P. p57KIP2 expression and loss of heterozygosity during immortal conversion of cultured human mammary epithelial cells. Cancer Res. 1999, 59, 5112–5118. [Google Scholar]
- Deng, J.; Zhao, L.; Zhang, N.Y.; Karrow, N.A.; Krumm, C.S.; Qi, D.S.; Sun, L.H. Aflatoxin B(1) metabolism: Regulation by phase I and II metabolizing enzymes and chemoprotective agents. Mutat. Res. Rev. Mutat. Res. 2018, 778, 79–89. [Google Scholar] [CrossRef]
- Hammoudeh, N.; Soukkarieh, C.; Murphy, D.J.; Hanano, A. Involvement of hepatic lipid droplets and their associated proteins in the detoxification of aflatoxin B(1) in aflatoxin-resistance BALB/C mouse. Toxicol. Rep. 2020, 7, 795–804. [Google Scholar] [CrossRef]
- Cheng, X.; Klaassen, C.D. Tissue distribution, ontogeny, and hormonal regulation of xenobiotic transporters in mouse kidneys. Drug Metab. Dispos. 2009, 37, 2178–2185. [Google Scholar] [CrossRef] [PubMed]
- Nigam, S.K.; Bush, K.T.; Martovetsky, G.; Ahn, S.Y.; Liu, H.C.; Richard, E.; Bhatnagar, V.; Wu, W. The organic anion transporter (OAT) family: A systems biology perspective. Physiol. Rev. 2015, 95, 83–123. [Google Scholar] [CrossRef] [PubMed]
- Thierry-Mieg, D.; Thierry-Mieg, J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006, 7 (Suppl. S1), S12. [Google Scholar] [CrossRef]
- Thomas, M. Manual annotation of the mouse genome: HAVANA and EUCOMM. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Kummu, M.; Sieppi, E.; Koponen, J.; Laatio, L.; Vahakangas, K.; Kiviranta, H.; Rautio, A.; Myllynen, P. Organic anion transporter 4 (OAT 4) modifies placental transfer of perfluorinated alkyl acids PFOS and PFOA in human placental ex vivo perfusion system. Placenta 2015, 36, 1185–1191. [Google Scholar] [CrossRef]
- Louisse, J.; Dellafiora, L.; van den Heuvel, J.; Rijkers, D.; Leenders, L.; Dorne, J.C.M.; Punt, A.; Russel, F.G.M.; Koenderink, J.B. Perfluoroalkyl substances (PFASs) are substrates of the renal human organic anion transporter 4 (OAT4). Arch. Toxicol. 2023, 97, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Serin Harmanci, A.; Harmanci, A.O.; Zhou, X. CaSpER identifies and visualizes CNV events by integrative analysis of single-cell or bulk RNA-sequencing data. Nat. Commun. 2020, 11, 89. [Google Scholar] [CrossRef]
- Rathi, A.V.; Cantalupo, P.G.; Sarkar, S.N.; Pipas, J.M. Induction of interferon-stimulated genes by Simian virus 40 T antigens. Virology 2010, 406, 202–211. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Johnson, W.W.; Shimada, T.; Ueng, Y.F.; Yamazaki, H.; Langouet, S. Activation and detoxication of aflatoxin B1. Mutat. Res. 1998, 402, 121–128. [Google Scholar] [CrossRef]
- McLean, M.; Dutton, M.F. Cellular interactions and metabolism of aflatoxin: An update. Pharmacol. Ther. 1995, 65, 163–192. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Reddel, R.R. SV40-induced immortalization of human cells. Crit. Rev. Oncog. 1994, 5, 331–357. [Google Scholar] [CrossRef]
- Langie, S.A.; Koppen, G.; Desaulniers, D.; Al-Mulla, F.; Al-Temaimi, R.; Amedei, A.; Azqueta, A.; Bisson, W.H.; Brown, D.G.; Brunborg, G.; et al. Causes of genome instability: The effect of low dose chemical exposures in modern society. Carcinogenesis 2015, 36 (Suppl. S1), S61–S88. [Google Scholar] [CrossRef]
- Hasty, P. Trex2 responds to damaged replication forks in diverse ways. Mol. Cell Oncol. 2021, 8, 1881394. [Google Scholar] [CrossRef]
- Paul, R.; Peraldi, R.; Kmita, M. The pioneering function of the hox transcription factors. Semin. Cell Dev. Biol. 2024, 152–153, 85–92. [Google Scholar] [CrossRef]
- Mori, J.O.; Keegan, J.; Flynn, R.L.; Heaphy, C.M. Alternative lengthening of telomeres: Mechanism and the pathogenesis of cancer. J. Clin. Pathol. 2024, 77, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Lezaja, A.; Altmeyer, M. Inherited DNA lesions determine G1 duration in the next cell cycle. Cell Cycle 2018, 17, 24–32. [Google Scholar] [CrossRef]
- Shen, D.W.; Pouliot, L.M.; Hall, M.D.; Gottesman, M.M. Cisplatin resistance: A cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012, 64, 706–721. [Google Scholar] [CrossRef]
- Romani, A.M.P. Cisplatin in cancer treatment. Biochem. Pharmacol. 2022, 206, 115323. [Google Scholar] [CrossRef]
- Selim, M.S.; Kassem, A.B.; El-Bassiouny, N.A.; Salahuddin, A.; Abu El-Ela, R.Y.; Hamza, M.S. Polymorphic renal transporters and cisplatin’s toxicity in urinary bladder cancer patients: Current perspectives and future directions. Med. Oncol. 2023, 40, 80. [Google Scholar] [CrossRef]
- Veiga-Matos, J.; Remiao, F.; Motales, A. Pharmacokinetics and Toxicokinetics Roles of Membrane Transporters at Kidney Level. J. Pharm. Pharm. Sci. 2020, 23, 333–356. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Zhao, B.; Chen, M.; Fu, X.; Zhang, Q.; Cui, Y.; Hu, X.; Zhou, W. Moving beyond cisplatin resistance: Mechanisms, challenges, and prospects for overcoming recurrence in clinical cancer therapy. Med. Oncol. 2023, 41, 9. [Google Scholar] [CrossRef] [PubMed]
- Calura, E.; Martini, P. Summarizing RNA-Seq Data or Differentially Expressed Genes Using Gene Set, Network, or Pathway Analysis. Methods Mol. Biol. 2021, 2284, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Rhee, S.Y. Interpreting omics data with pathway enrichment analysis. Trends Genet. 2023, 39, 308–319. [Google Scholar] [CrossRef]
- Sun, H.; Chen, G.; Guo, B.; Lv, S.; Yuan, G. Potential clinical treatment prospects behind the molecular mechanism of alternative lengthening of telomeres (ALT). J. Cancer 2023, 14, 417–433. [Google Scholar] [CrossRef]
- Hsieh, H.J.; Peng, G. Cellular responses to replication stress: Implications in cancer biology and therapy. DNA Repair 2017, 49, 9–20. [Google Scholar] [CrossRef]
- Mihevc, M.; Petreski, T.; Maver, U.; Bevc, S. Renal proximal tubular epithelial cells: Review of isolation, characterization, and culturing techniques. Mol. Biol. Rep. 2020, 47, 9865–9882. [Google Scholar] [CrossRef]
- Nieskens, T.T.; Peters, J.G.; Schreurs, M.J.; Smits, N.; Woestenenk, R.; Jansen, K.; van der Made, T.K.; Roring, M.; Hilgendorf, C.; Wilmer, M.J.; et al. A Human Renal Proximal Tubule Cell Line with Stable Organic Anion Transporter 1 and 3 Expression Predictive for Antiviral-Induced Toxicity. AAPS J. 2016, 18, 465–475. [Google Scholar] [CrossRef]
- Ishiguro, N.; Takahashi, E.; Arakawa, H.; Saito, A.; Kitagawa, F.; Kondo, M.; Morinaga, G.; Takatani, M.; Takahashi, R.; Kudo, T.; et al. Improvement of Protein Expression Profile in Three-Dimensional Renal Proximal Tubular Epithelial Cell Spheroids Selected Based on OAT1 Gene Expression: A Potential In Vitro Tool for Evaluating Human Renal Proximal Tubular Toxicity and Drug Disposition. Drug Metab. Dispos. 2023, 51, 1177–1187. [Google Scholar] [CrossRef]
- Klatt, O.C.; de Brouwer, L.; Hendriks, F.; Dehne, E.M.; Atac Wagegg, B.; Jennings, P.; Wilmes, A. Human and rat renal proximal tubule in vitro models for ADME applications. Arch. Toxicol. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Mahadeo, A.; Yeung, C.K.; Himmelfarb, J.; Kelly, E.J. Kidney microphysiological models for nephrotoxicity assessment. Curr. Opin. Toxicol. 2022, 30, 100341. [Google Scholar] [CrossRef] [PubMed]
- Frost, K.L.; Jilek, J.L.; Toth, E.L.; Goedken, M.J.; Wright, S.H.; Cherrington, N.J. Representative Rodent Models for Renal Transporter Alterations in Human Nonalcoholic Steatohepatitis. Drug Metab. Dispos. 2023, 51, 970–981. [Google Scholar] [CrossRef]
- Bai, M.; Shen, Q.; Wu, Y.; Ma, Z.; Wang, Y.; Chen, M.; Liu, D.; Zhou, L. Evaluation of transport mechanisms of methotrexate in human choriocarcinoma cell lines by LC-MS/MS. J. Pharm. Biomed. Anal. 2024, 247, 116268. [Google Scholar] [CrossRef] [PubMed]
- Poller, B.; Pearson, D.; Leuthold, L.A.; Fink, M.; Jullion, A.; Schweigler, P.; Tor Carreras, E.; Marvalin, C.; Loesche, C.; Weiss, H.M. Human Pharmacokinetics of LYS006, an Oral Leukotriene A4 Hydrolase Inhibitor Displaying Target-Mediated Drug Disposition. Drug Metab. Dispos. 2022, 50, 1472–1482. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xin, M.; Liang, S.; Xu, X.; Cai, T.; Dong, L.; Wang, C.; Wang, M.; Cui, Y.; Song, X.; et al. New insight into the management of renal excretion and hyperuricemia: Potential therapeutic strategies with natural bioactive compounds. Front. Pharmacol. 2022, 13, 1026246. [Google Scholar] [CrossRef]
- Ryu, S.; Yamaguchi, E.; Sadegh Modaresi, S.M.; Agudelo, J.; Costales, C.; West, M.A.; Fischer, F.; Slitt, A.L. Evaluation of 14 PFAS for permeability and organic anion transporter interactions: Implications for renal clearance in humans. Chemosphere 2024, 361, 142390. [Google Scholar] [CrossRef]
- Olde Hanhof, C.J.A.; Dilmen, E.; Yousef Yengej, F.A.; Latta, F.; Ammerlaan, C.M.E.; Schreurs, J.; Hooijmaijers, L.; Jansen, J.; Rookmaaker, M.B.; Orhon, I.; et al. Differentiated mouse kidney tubuloids as a novel in vitro model to study collecting duct physiology. Front. Cell Dev. Biol. 2023, 11, 1086823. [Google Scholar] [CrossRef]
- Voloshin, N.; Tyurin-Kuzmin, P.; Karagyaur, M.; Akopyan, Z.; Kulebyakin, K. Practical Use of Immortalized Cells in Medicine: Current Advances and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 12716. [Google Scholar] [CrossRef]
- de Bardet, J.C.; Cardentey, C.R.; Gonzalez, B.L.; Patrone, D.; Mulet, I.L.; Siniscalco, D.; Robinson-Agramonte, M.L.A. Cell Immortalization: In Vivo Molecular Bases and In Vitro Techniques for Obtention. BioTech 2023, 12, 14. [Google Scholar] [CrossRef]
- Dickson, M.A.; Hahn, W.C.; Ino, Y.; Ronfard, V.; Wu, J.Y.; Weinberg, R.A.; Louis, D.N.; Li, F.P.; Rheinwald, J.G. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 2000, 20, 1436–1447. [Google Scholar] [CrossRef]
- McConnell, B.B.; Gregory, F.J.; Stott, F.J.; Hara, E.; Peters, G. Induced expression of p16(INK4a) inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol. Cell Biol. 1999, 19, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Bozdogan, O.; Atasoy, P.; Batislam, E.; Basar, M.M.; Basar, H. Significance of p57(Kip2) down-regulation in oncogenesis of bladder carcinoma: An immunohistochemical study. Tumori 2008, 94, 556–562. [Google Scholar] [CrossRef]
- Molkentine, D.P.; Molkentine, J.M.; Bridges, K.A.; Valdecanas, D.R.; Dhawan, A.; Bahri, R.; Hefner, A.J.; Kumar, M.; Yang, L.; Abdelhakiem, M.; et al. p16 Represses DNA Damage Repair via a Novel Ubiquitin-Dependent Signaling Cascade. Cancer Res. 2022, 82, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Sen, M.; Akeno, N.; Reece, A.; Miller, A.L.; Simpson, D.S.; Wikenheiser-Brokamp, K.A. p16 controls epithelial cell growth and suppresses carcinogenesis through mechanisms that do not require RB1 function. Oncogenesis 2017, 6, e320. [Google Scholar] [CrossRef]
- Taub, M. Primary kidney cells. Methods Mol. Biol. 1997, 75, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.A.; Phadke, D.P.; Bostrom, M.A.; Shah, R.R.; Wright, G.M.; Wang, X.; Gordon, O.; Pelch, K.E.; Auerbach, S.S.; Paules, R.S.; et al. Arsenite malignantly transforms human prostate epithelial cells in vitro by gene amplification of mutated KRAS. PLoS ONE 2019, 14, e0215504. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Gel, B.; Serra, E. karyoploteR: An R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics 2017, 33, 3088–3090. [Google Scholar] [CrossRef]
- Dunnick, J.K.; Merrick, B.A.; Brix, A.; Morgan, D.L.; Gerrish, K.; Wang, Y.; Flake, G.; Foley, J.; Shockley, K.R. Molecular Changes in the Nasal Cavity After N, N-dimethyl-p-toluidine Exposure. Toxicol. Pathol. 2016, 44, 835–847. [Google Scholar] [CrossRef]





| Cell Type | Total Transcripts 1 | Transcripts 2 <5 BMCs | Transcripts 2 ≥5 BMCs | Total DEGs 3 | Up DEGs 3 | Down DEGs 3 | No Change 4 |
|---|---|---|---|---|---|---|---|
| hTERT | 55,401 | 32,027 | 23,374 | 6426 | 1460 | 4966 | 9998 |
| LgT | 55,401 | 31,813 | 23,588 | 5740 | 2138 | 3602 | 12,333 |
| Transporter Cluster | Total No. Transcripts in Cluster | Order from High to Low Expression per Cluster | No. Transcripts < 5 BMCs | ||
|---|---|---|---|---|---|
| Parent | LgT | hTERT | |||
| a. | 52 | hTERT > Parent > LgT | 0 | 0 | 0 |
| b. | 43 | hTERT > Parent & LgT | 3 | 2 | 0 |
| c. | 45 | LgT > hTERT > Parent | 1 | 0 | 1 |
| d. | 61 | Parent & LgT > hTERT | 1 | 1 | 11 |
| e. | 94 | Parent > LgT & hTERT | 0 | 6 | 24 |
| f. | 53 | Parent > hTERT > LgT | 0 | 7 | 3 |
| g. | 16 | Parent = LgT = hTERT | 0 | 1 | 2 |
| Total | 364 | 5 | 17 | 41 | |
| Gene Symbol a | Assay ID | Probe Fluorophore | Species |
|---|---|---|---|
| Exons 2–3 | dCNS754377321 | Hex | Mouse |
| Exons 3–4 | dCNS215834387 | Cy 5 | Mouse |
| Tbp | dMmuGEXS140712005 | Rox | Mouse |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merrick, B.A.; Brooks, A.M.; Foley, J.F.; Martin, N.P.; Fannin, R.D.; Gladwell, W.; Gerrish, K.E. hTERT and SV40LgT Renal Cell Lines Adjust Their Transcriptional Responses After Copy Number Changes from the Parent Proximal Tubule Cells. Int. J. Mol. Sci. 2025, 26, 3607. https://doi.org/10.3390/ijms26083607
Merrick BA, Brooks AM, Foley JF, Martin NP, Fannin RD, Gladwell W, Gerrish KE. hTERT and SV40LgT Renal Cell Lines Adjust Their Transcriptional Responses After Copy Number Changes from the Parent Proximal Tubule Cells. International Journal of Molecular Sciences. 2025; 26(8):3607. https://doi.org/10.3390/ijms26083607
Chicago/Turabian StyleMerrick, Bruce Alex, Ashley M. Brooks, Julie F. Foley, Negin P. Martin, Rick D. Fannin, Wesley Gladwell, and Kevin E. Gerrish. 2025. "hTERT and SV40LgT Renal Cell Lines Adjust Their Transcriptional Responses After Copy Number Changes from the Parent Proximal Tubule Cells" International Journal of Molecular Sciences 26, no. 8: 3607. https://doi.org/10.3390/ijms26083607
APA StyleMerrick, B. A., Brooks, A. M., Foley, J. F., Martin, N. P., Fannin, R. D., Gladwell, W., & Gerrish, K. E. (2025). hTERT and SV40LgT Renal Cell Lines Adjust Their Transcriptional Responses After Copy Number Changes from the Parent Proximal Tubule Cells. International Journal of Molecular Sciences, 26(8), 3607. https://doi.org/10.3390/ijms26083607






